Abstract
Discriminating between bipolar disorder (BD) and major depressive disorder (MDD) is a major clinical challenge due to the absence of known biomarkers; hence a better understanding of their pathophysiology and brain alterations is urgently needed. Given the complexity, feature selection is especially important in neuroimaging applications, however, feature dimension and model understanding present serious challenges. In this study, a novel feature selection approach based on linear support vector machine with a forward-backward search strategy (SVM-FoBa) was developed and applied to structural and resting-state functional magnetic resonance imaging data collected from 21 BD, 25 MDD and 23 healthy controls. Discriminative features were drawn from both data modalities, with which the classification of BD and MDD achieved an accuracy of 92.1% (1,000 bootstrap resamples). Weight analysis of the selected features further revealed that the inferior frontal gyrus may characterize a central role in BD-MDD differentiation, in addition to the default mode network and the cerebellum. A modality-wise comparison also suggested that functional information outweighs anatomical by a large margin when classifying the two clinical disorders. This work validated the advantages of multimodal joint analysis and the effectiveness of SVM-FoBa, which has potential for use in identifying possible biomarkers for several mental disorders.
Index Terms: classification, feature selection, multimodal fusion, bipolar disorder, major depression
I. Introduction
Mood disorders have become the most costly brain diseases in the world [1], among which both bipolar disorder (BD) and unipolar or major depressive disorder (MDD) are characterized by depressive episodes. Currently, neither the clinical features of depression nor any known neuropsychological indicators readily differentiate trajectories of either MDD or BD, especially in the early course of illness [2]. Indeed, during the course of BD, depressive episodes usually present more often than manic or hypomanic symptoms, while sub-threshold manic symptoms can also be concealed in a mixed episode [3, 4]. Thus, around 80% of BD patients who are going through depressive episodes receive an incorrect diagnosis (mostly misdiagnosed as MDD) within the first years of seeking treatment [5, 6], leading to inappropriate and longer medication periods, poorer prognosis and greater health care expenses [7, 8]. Currently, there are no objective and clinically useful diagnostic markers for either disorder; hence, a better understanding of the pathophysiology of both mood disorders is urgently needed for developing more effective treatments and establishing the differential diagnosis.
On the other hand, feature selection can be preferable and may be imperative when it comes to neuroimaging data. The inherent characteristics of high feature dimension and small sample size of participants present severe challenges to researchers who wish to apply pattern classification. Recently, several neuroimaging studies have directly compared patients with BD and MDD using magnetic resonance imaging (MRI), which provides non-invasive observation of the structural characteristics and functional states of the brain. These studies exploited different measurements, including structural features such as gray matter volume [9], fractional anisotropy value of white matter [10], hyper intensities, and functional activity patterns during task or rest [11–14], suggesting that with appropriate analytical strategies, both structural and functional MRI data are capable of detecting brain abnormalities for distinguishing between BD and MDD. However, the above studies mostly focused on only one modality. As each imaging modality provides a different view of the brain, joint analysis of multimodal data may extend the limited information captured by isolated regional measures of structure or function and provide a better system-wide understanding of brain alterations related to mental illnesses [15, 16].
Most previous studies of BD and MDD tended to employ univariate approaches, despite the fact that the brain actually functions in a multivariate way [17]. Recent studies have suggested that, when analyzing fMRI data with sophisticated machine learning algorithms, informative fMRI patterns could be targeted to distinguish MDD patients [18] or even directly differentiate BD from MDD [19]. In this study, we improve the differential diagnosis between clinically diagnosed BD (type I) and MDD patients by implementing a novel feature selection method for use on multimodal MRI data.
The proposed method, called “SVM-FoBa”, is able to adaptively choose highly discriminative feature subsets by means of a forward-backward search strategy. Though a similar technique has been proposed with least square regression [20], the present method is fully integrated within a linear support vector machine (SVM) [21], whose multivariate objective function will be used to evaluate the effectiveness of a feature. The corresponding technical details are discussed in Section II.D.
The multimodal brain imaging features utilized here include the fractional amplitude of low-frequency fluctuation (fALFF) of resting-state functional MRI (fMRI) data, which has been suggested to reflect the intensity of regional spontaneous brain activity with high performance [22], and also voxel-wise gray matter (GM) volume obtained by voxel-based morphometry (VBM) from structural MRI [23] [24]. Inspecting two modalities at the same time should provide more comprehensive insight into brain disorder.
In short, the main goal of the present study was to identify the informative and biologically relevant features that can efficiently distinguish between patients with BD and MDD. With multimodal fusion effectively harnessed, the linear classifier can facilitate modal understanding by providing in-depth feature information (patterns from feature weights) in order to interpret results based on localized brain regions.
II. Materials and methods
A. Subject Inclusion and Exclusion
The research project was approved by the University of Western Ontario Research Ethics Board, in keeping with the Declaration of Helsinki. All research participants were provided with a written description of the study and had the opportunity to ask questions about the procedure. Written, informed consent was then obtained from willing participants.
A total of 21 participants with BDs and 25 age matched MDDs were recruited from the First Episode Mood and Anxiety Program in London, Ontario, Canada. Each participant met diagnostic criteria for bipolar disorder, type I, or major depressive disorder (MDD) using the structured clinical interview for DSM disorders-IV, research version (SCID-IV) or the diagnostic interview for genetic studies (DIGS). In addition, agreement between the clinical chart diagnosis and the SCID/DIGS diagnosis of MDD or BD-type I was required for the patient groups. Medications were unchanged for three weeks prior to scanning.
Patients were excluded if they had a history of head injury leading to unconsciousness for longer than a few seconds, or significant non-psychiatric medical illness. Individuals with an active substance use disorder (except possibly caffeine and/or nicotine abuse/dependence), posttraumatic stress disorder or obsessive-compulsive disorder were also excluded. Youth were excluded if they were in imminent danger to themselves or others, if they were actively psychotic, or if they had exclusions for MRI scanning. Any family history of BD in the MDD group was exclusionary.
For comparison purposes, 23 healthy participants were also included as the control group. The HC group had no history of significant head injury, major medical illness, psychiatric medication use or personal (as determined by the SCID or DIGS) or family history of psychiatric illness, with the exception of caffeine and/or nicotine abuse/dependence. There were no significant age differences between the three subject groups in these measures, but the bipolarity index (BPI) was significantly different between BD and MDD. A summary of demographic information is presented in TABLE I.
TABLE I.
Demographic and Clinical Data of Participants
| Group | BD | MDD | HC |
|---|---|---|---|
| Number | 21 | 25 | 23 |
| Age (years, mean±SD) | 21.6±2.9 | 20.1±2.8 | 20.5±1.9 |
| Gender (M:F) | 12:9 | 9:16 | 7:16 |
| YMRS (mean±SD) | 1.7±2.2 | 1.6±1.8 | 0.2±0.5 |
| BPI (mean±SD) | 77.2±15.2 | 27.1±6.4 | N/A |
SD, Standard Deviation; YMRS, Young Mania Rating Scale; BPI, Bipolarity Index, a scale measuring traits of bipolar disorder on a continuum.
B. Image Acquisition
All MRI imaging was collected using a 3.0 T MRI scanner (MAGNETOM Verio, Siemens, Erlangen, Germany) at the Lawson Health Research Institute, and a 32-channel phased-array head coil (Siemens). A T1-weighted, 3D magnetization-prepared rapid gradient echo sequence was used to collect anatomical images. Acquisition parameters were as follows: TR = 3000 ms, TE = 2.98 ms, flip angle = 9°, FOV = 256 mm × 256 mm, matrix size = 256 × 256, 176 sagittal slices, voxel size = 1 mm × 1 mm × 1 mm. Functional scans consisted of gradient-echo, echo-planar scans (TR = 2000 ms, TE = 30 ms, flip angle = 90°, FOV = 240 mm × 240 mm, matrix size = 80 × 80, 40 axial slices, thickness = 3 mm) with no parallel acceleration, covering whole brain with an isotropic spatial resolution of 3 mm for a total time of approximately 8 min (164 brain volumes). Participants were given the following instructions before scanning: “Lie comfortably, as still as possible with your eyes closed and let your mind wander without falling asleep.” No participant reported falling asleep during the scan when asked immediately after scanning.
C. Image Preprocessing and Preperation
The structural MRI data were preprocessed in SPM8 toolbox (http://www.fil.ion.ucl.ac.uk/spm/software/spm8) with the VBM8 (http://dbm.neuro.uni-jena.de/vbm/download/) and DARTEL package. The images were first segmented into gray matter, white matter (WM), cerebrospinal fluid (CSF), bone, and soft tissue. The DARTEL algorithm registered these tissue segmentations back and forth with the default templates based on the MNI152 brain, and finally generated population-specific templates. GM and WM density images of all subjects were subsequently normalized to the corresponding template. All these tissue images were corrected for individual head size in VBM8. After that, the modified gray matter volume of each region from the 116-area automated anatomical labeling (AAL) template [25] was extracted and locally averaged as an independent feature. Thus, a sample vector with a feature dimension of 116 was obtained for each subject. Gender was later regressed out to mitigate possible bias between groups. This feature from structural MRI will be henceforth called VBM-GM.
The preprocessing of the resting state fMRI data was carried out using DPARSF toolbox [26], including discarding the first 10 volumes of each functional time series to allow for magnetization equilibrium, correcting the slice timing for the remaining 154 images and realigning them to the first volume to provide for head motion correction, time series despiking, spatial smoothing with a Gaussian kernel of 6 mm full-width at half maximum (FWHM), normalizing the mean-based intensity, temporal band-pass filtering (0.01–0.10 Hz), removing linear and quadratic trends, performing linear and nonlinear spatial normalizing of the structural images to the MNI152 brain template, co-registering the anatomical volume with the mean functional volume, regressing out nuisance signals such as those from white matter, CSF, as well as six motion parameters, and resampling of the functional data into MNI space with the concatenated transformations. An fALFF index map was then generated to provide information regarding the amplitude of brain activity. Similar to the steps involved in the processing of GM, by separately averaging the fALFF values within each region of the 116 areas determined by the AAL mask, a functional sample set with 116 feature dimensions was obtained for later analysis. Gender factor was also regressed out for fALFF.
D. Feature Selection
Feature selection is often considered necessary for classifying neuroimaging data, especially when hundreds of thousands of features are presented as opposed to only dozens of samples, resulting in the curse of dimensionality [14]. In this study, though we have already reduced the number of features from ~70k to hundreds by using the AAL template, employing a feature selection process can further help precisely locate the regions of interest and facilitate data visualization. Also, a sparse feature set can provide substantial improvement to the performance of a more generalized classifier.
For neuroimaging data processing, conventional univariate feature selection approaches ignore the mutual information among features with certain independence or orthogonality assumptions, while in fact the intermingled activity pattern can serve as the optimal feature subset. Specifically, it is highly possible that some features that have little contribution to classification on their own (which will be filtered out using univariate methods) may yet provide a performance boost if combined with other features [27], i.e., unsynchronized fluctuations might emerge as an informative pattern [17].
Unlike univariate feature selection, multivariate feature selection takes relevancy of a group of features into account. Nonetheless, searching for the globally optimal subset with a given size is known to be NP-hard [28]. Exhaustive search can quickly become computationally infeasible when the dimensionality of the feature space becomes high. Although approximation of global optima can be achieved by some sophisticated methods, such as the genetic algorithm [29, 30], model understanding may present serious challenge. Consequently, researchers typically resort to algorithms aiming at obtaining the sub-optimal or locally optimal feature sets.
E. Development of SVM-FoBa
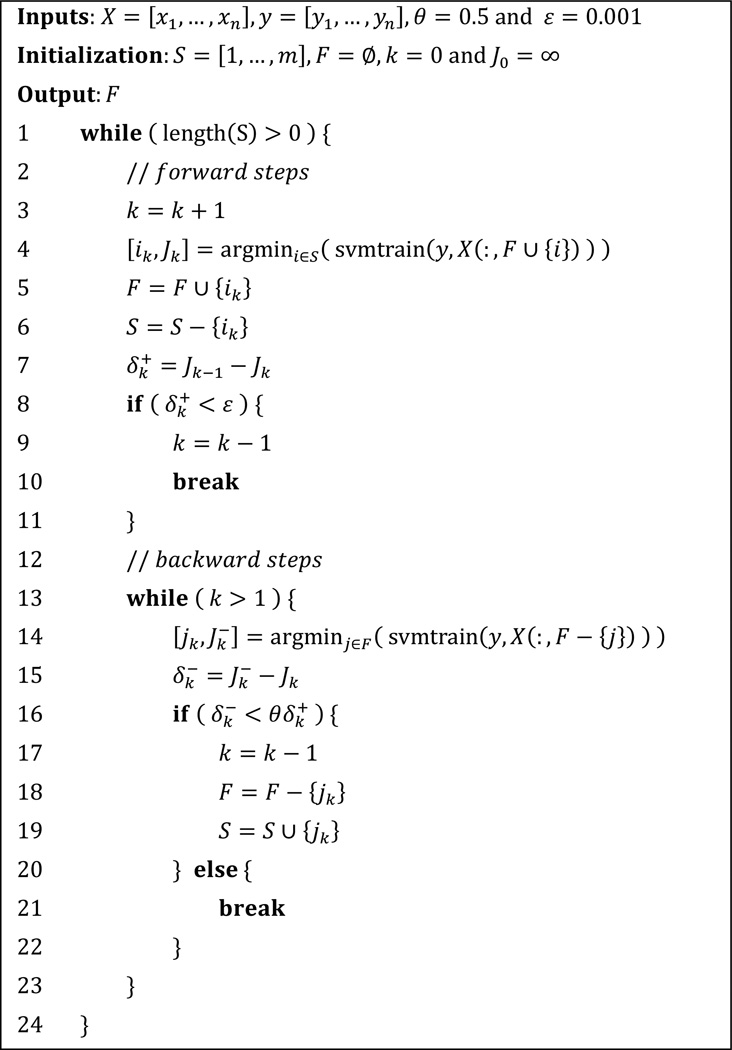
Known to be computationally advantageous, greedy search strategies normally serve in two directions: forward selection and backward elimination. Forward selection involves a bottom-up search model that starts with an empty set. During each iteration step, one feature is added to the current set F in order to aggressively reduce the loss function J. However, forward selection will yield nested set of features, in which features selected at step k are always included in the subset of step (k + 1). That is to say, the forward selection is not capable of inspecting other possible combinations of features than such additive ones.
Backward elimination, on the other hand, is the top-down analogy to forward selection. Starting with the complete set, features are removed one at a time so that the negative impact on performance shall be kept minimal. This iteration continues until the change of loss function J exceeds a certain threshold. Clearly, backward elimination generates nested subset as well. Furthermore, when dealing with high dimensional data, the backward approach can be computationally demanding since the combination of features will be checked from the universal set. Another major flaw of backward elimination is the propensity for overfitting. Should a perfect prediction be made the first time based on the complete feature set, removing one feature with the lowest increment to J would still result in overfitting.
In brief, both forward and backward strategies have their limitations. However, if the forward selection were to be combined with backward elimination, the error induced by earlier steps could have been adaptively corrected. Hence, we are motivated to develop “SVM-FoBa”, an SVM-based adaptive forward-backward greedy algorithm. During each iteration of SVM-FoBa, when a new feature is added by a forward step, backward step will step in to recheck the new feature subset as a whole, or even remove features with minor performance loss. This combination can be integrated with various prediction models for feature selection, such as least square regression [20]. In the current study, we invoked the value of the optimization objective function of the linear SVM as the loss function J, which can be written as:
where xi is the sample with yi corresponding to its label, w is the weight vector, b is the offset, C is the hyperparameter (1 by default), and [·]+ denotes the hinge function that [t]+ = max{0, 1 − t}.
Thus, by implementing SVM-FoBa on the brain imaging data of BD and MDD, the corresponding change of the objective function could directly characterize the impact of a feature on differentiating two types of patients.
As mentioned above, a forward step of SVM-FoBa always singles out the features with the largest decrement of the objective function, while the backward feature elimination will then be applied if needed. This procedure is repeated until the decrement of the objective function between consecutive steps has reduced below a given threshold ε. Obviously, balancing the forward and backward steps is the key to the algorithm. We certainly want to make reasonably aggressive backward steps to eliminate errors caused by earlier forward steps, while progress contributed by the forward steps should also be preserved as much as possible. We thus introduce the following definition, along with the notations, for clarity.
Definition
Assuming that at stage k, a feature ik is added to the feature subset Fk−1 by forward step, thereby generating a subset of Fk, with Jk being the corresponding value of the optimization objective function. The decrement of the objective function is denoted as , so that . Backward step then locates a feature jk ∈ Fk, whose elimination induces the smallest increment in the objective function, denoted as . With a given constant 0 < θ < 1, if , we define that at least one error has occurred in earlier forward steps.
Therefore, once errors induced by earlier forward steps are detected, backward elimination processes will automatically step in and repeat until all the errors are corrected. It is worth noting that the hyperparameter θ determines the bias between the forward and backward steps. As long as n forward steps have been carried out, the objective function J will be reduced by at least n(1 − θ)ε regardless of how many backward steps have been involved in the course, suggesting that the algorithm will stop after a finite number of forward steps. Thus, if the value of θ is small, the whole protocol will be toward the forward steps with less error correction, resulting in aggressive optimization; on the contrary, a large θ emphasize more on the backward procedures, suggesting slower convergence rate of the algorithm. In this work, we used θ = 0.5 as the trade-off value. Moreover, unlike forward selection or backward elimination alone, SVM-FoBa does not yield a series of nested subsets of features. As much more comprehensive combinations of features can be tested in a reasonable amount of time, we expected to see the features with the greatest discriminative power to be factored into the final classification model.
The pseudo-code for SVM-FoBa is demonstrated in Fig. 1. In this study, we adapted LibSVM [31] with SVM-FoBa algorithm to accomplish the feature selection. It is still worth noting that, with improvements over the conventional greedy methods in principle, SVM-FoBa does not necessarily excel in all aspects according to “no free lunch” theorem [32]; performance may also vary when it comes to different data sets, based upon which the classifiers will produce distinct biases.
Fig. 1.
Pseudo-code for SVM-FoBa algorithm.
III. Results
A. Comparison With Typical Feature Selection Methods
To validate the effectiveness of SVM-FoBa, we conducted a comparison study by testing different feature selection methods on public data sets. Other than SVM-FoBa, four conventional feature selection methods were included: the univariate T-test and Fisher score [14] ranking, the forward selection and the SVM-RFE algorithm [20]. The T-test and Fisher score ranking are commonly used in brain imaging studies. For comparison purpose, the forward selection method also utilized the multivariate objective function as its loss function. Standard backward elimination was not included due to insurmountable computational cost on high dimensional data. For each method, the selected feature set was entered into a linear SVM classifier for performance evaluation.
In simulation, we wanted to confirm the advantages of SVM-FoBa when starting with hundreds or thousands of features with limited samples that are similar to the real scenarios of neuroimaging study. Thus, public biomedical data sets with comparable number of instances were chosen for binary classification test, including the LSVT Voice Rehabilitation [33], the Colon Cancer [34] and the Leukemia Cancer [35] data sets (see TABLE II for detailed information). To avoid possible bias introduced by certain partitioning of training and test samples, we performed a leave-one-out cross-validation (LOOCV) procedure, which is known to be an unbiased estimator of the generalization performance of a classifier [36]. Specifically, on each fold of the LOOCV, all but one of the samples was used to train the classifier, while the remaining sample was saved for validating the results. This procedure proceeded until each sample had performed once as the test set. The classification accuracy, along with the sensitivity and specificity rates were then calculated based on how many correct predictions were made with all the folds added up against the sample size. The results are demonstrated in TABLE II.
TABLE II.
Performance of Different Feature Selection Methods on Three Public Data Sets
| LSVT Voice |
Colon Cancer |
Leukemia Cancer |
||
|---|---|---|---|---|
| Instances | 126 | 62 | 72 | |
| Features | 309 | 2,000 | 7,129 | |
| SVM-FoBa | No. features | 47 | 29–30 | 30–38 |
| Accuracy | 90.48% | 87.10% | 98.61% | |
| Sensitivity | 85.71% | 92.50% | 96.00% | |
| Specificity | 92.86% | 86.36% | 100% | |
| Forward selection | No. features | 83 | 37–39 | 39–43 |
| Accuracy | 87.30% | 85.48% | 97.22% | |
| Sensitivity | 85.71% | 90.00% | 96.00% | |
| Specificity | 90.48% | 77.27% | 100% | |
| SVM-RFE | No. features | 67–68 | 31 | 82–84 |
| Accuracy | 84.92% | 87.10% | 98.61% | |
| Sensitivity | 85.71% | 90.00% | 96.00% | |
| Specificity | 86.90% | 86.36% | 100% | |
| T-test ranking* | No. features | 47 | 29–30 | 30–38 |
| Accuracy | 88.89% | 87.10% | 95.83% | |
| Sensitivity | 83.33% | 87.50% | 96.00% | |
| Specificity | 91.67% | 86.36% | 95.74% | |
| Fisher score ranking* | No. features | 47 | 29–30 | 30–38 |
| Accuracy | 88.89% | 87.10% | 95.83% | |
| Sensitivity | 80.95% | 87.50% | 96.00% | |
| Specificity | 92.86% | 86.36% | 95.74% | |
T-Test and Fisher score method used the same number of features with that of SVM-FoBa for equivalent comparisons.
“LSVT Voice Rehabilitation” data set was obtained from the UCI Machine Learning Repository (http://archive.ics.uci.edu/ml/datasets.html).
Descriptions of “Colon Cancer” and “Leukemia Cancer” data can be found at http://www.inf.ed.ac.uk/teaching/courses/dme/html/datasets0405.html.
It is worth noting that the univariate feature indices (T-test and Fisher score) only provide a ranked list of features. Hence, for comparison purposes, we extracted the feature subset with the highest rank, where the set size was identical to the optimal number of features indicated by the SVM-FoBa algorithm. For example, as shown in TABLE II, with SVM-FoBa achieving its best prediction power for “Leukemia Cancer” data set when 30–38 features were selected (equal performance from 30 to 38 features), the same interval of top rated features were also chosen for T-test and Fisher score methods.
Generally, SVM-FoBa yielded better performance than the other four feature selection methods, particularly in sensitivity and specificity rates. According to Bayes' theorem, minor specificity decrement can cause considerable detection power loss. Thus, even if the improvement of SVM-FoBa is small in some cases, clinical diagnosis may ultimately benefit from this. Also, when compared to other multivariate feature selection approaches (the forward selection and SVM-RFE), SVM-FoBa extracted more compact feature subsets. This can be of great importance when trying to interpret the most prominent features, which, in the context of medical image data, might be the essential biomarkers; redundant features can harm the clinical judgment with irrelevant factors involved.
B. Feature Selection of fALFF and VBM-GM data
We applied SVM-FoBa with LOOCV on fALFF and VBM-GM data (each contained 116 features) for all three types of between-group classification conditions (BD vs. MDD, BD vs. HC, and MDD vs. HC). Similar to the strategy involved in Section II.A regarding public data sets, the corresponding accuracy rates and optimal number of features are calculated and listed in TABLE III.
TABLE III.
Determining the Optimal Discriminative Features Numbers Using SVM-FoBa in Single Modality
| Modality | Accuracy | Sensitivity | Specificity | No. of features |
|
|---|---|---|---|---|---|
| fALFF | BD vs. MDD | 73.91% | 71.43% | 80.00% | 16 |
| BD vs. HC | 63.64% | 57.14% | 69.57% | 5 | |
| MDD vs. HC | 64.58% | 60.00% | 73.91% | 28 | |
| VBM-GM | BD vs. MDD | 76.09% | 71.43% | 84.00% | 6 |
| BD vs. HC | 61.36% | 57.14% | 73.91% | 16 | |
| MDD vs. HC | 62.50% | 60.00% | 69.57% | 6 | |
The accuracy rates were calculated by dividing the number of the correct predictions across all folds of LOOCV by the number of samples. The sensitivity and specificity were obtained likewise.
Thus, with the optimal number of features provided, we were interested in which features contributed most to group discrimination. This was done by exploiting the quantitative advantage of the linear SVM classification model, whereby features with the greatest absolute weight value (averaged across all folds of LOOCV) could be factored out. We denote such important features as the “significant features.”
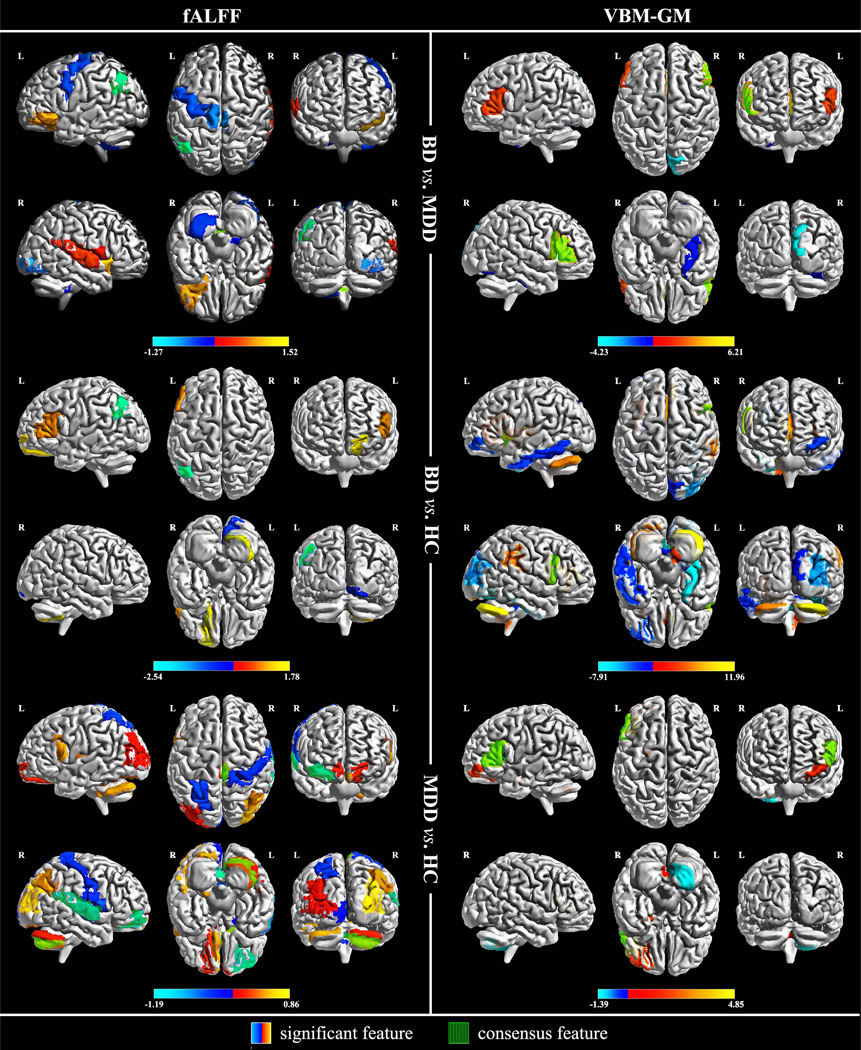
It should be noted that, in addition to the absolute weight value that represents the contribution made to the classifier, the sign of the weight can also provide critical information due to the linearity of the binary decision function. Namely, the sign of a weight indicates into which label a sample will be classified. For example, as in the context of “BD vs. MDD”, BD is deemed as the negative sample with the label “−1”, whereas MDD is assigned with “+1”. In this case, a feature with positive weight should reflect its negative connection to BD, since a test sample with larger value along that dimension will imbue the decision function with greater impetus to be positive, i.e., to classify this sample as MDD. Conversely, a BD sample should present with a smaller value along that specific dimension. Thus, for each feature selection condition in TABLE III, we drew a set of the top n significant features, with n being equal to the optimal number of features that SVM-FoBa suggested to select. These most discriminative features are listed in TABLE IV. Using the BrainNet Viewer toolbox [37], a visualization of these features is also generated by mapping their weight values to the corresponding brain regions, as is shown in Fig. 2.
TABLE IV.
Significant Features Utilizing Two Different Modalities
| fALFF data | |||||||||||
| BD vs. MDD (16 significant features) | BD vs. HC (5 significant features) | MDD vs. HC (28 significant features) | |||||||||
| AAL No. | Brain area | Weight | AAL No. | Brain area | Weight | AAL No. | Brain area | Weight | |||
| ● | 116 | Vermis_10 | 1.5199 | ● | 65 | Angular_L | −2.5417 | ● | 114 | Vermis_8 | −1.186 |
| ● | 75 | Pallidum_L | −1.2746 | 102 | Cerebelum_7b_R | 1.7775 | ● | 10 | Frontal_Mid_Orb_R | −0.889 | |
| 30 | Insula_R | 1.2559 | 5 | Frontal_Sup_Orb_L | 1.6366 | ● | 70 | Paracentral_Lobule_R | 0.8612 | ||
| ● | 109 | Vermis_1_2 | −1.2051 | 13 | Frontal_Inf_Tri_L | 1.1592 | ● | 82 | Temporal_Sup_R | −0.8009 | |
| ● | 65 | Angular_L | −1.1779 | 48 | Lingual_R | −0.9787 | 52 | Occipital_Mid_R | 0.7818 | ||
| 15 | Frontal_Inf_Orb_L | 1.0519 | 96 | Cerebelum_3_R | 0.761 | ||||||
| 70 | Paracentral_Lobule_R | −0.9367 | ● | 94 | Cerebelum_Crus2_R | 0.673 | |||||
| 54 | Occipital_Inf_R | −0.8713 | ● | 22 | Olfactory_R | 0.6635 | |||||
| 69 | Paracentral_Lobule_L | −0.7827 | 27 | Rectus_L | 0.6544 | ||||||
| 76 | Pallidum_R | −0.7475 | 91 | Cerebelum_Crus1_L | 0.6384 | ||||||
| 82 | Temporal_Sup_R | 0.5871 | 107 | Cerebelum_10_L | 0.6229 | ||||||
| 1 | Precentral_L | −0.568 | 11 | Frontal_Inf_Oper_L | 0.6149 | ||||||
| 103 | Cerebelum_8_L | −0.4937 | 66 | Angular_R | 0.611 | ||||||
| 80 | Heschl_R | 0.4182 | 18 | Rolandic_Oper_R | −0.6016 | ||||||
| 18 | Rolandic_Oper_R | 0.3262 | 116 | Vermis_10 | −0.5491 | ||||||
| 108 | Cerebelum_10_R | −0.2163 | 79 | Heschl_L | 0.5066 | ||||||
| 102 | Cerebelum_7b_R | 0.4695 | |||||||||
| 59 | Parietal_Sup_L | −0.4365 | |||||||||
| 58 | Postcentral_R | −0.3897 | |||||||||
| 71 | Caudate_L | 0.3272 | |||||||||
| 5 | Frontal_Sup_Orb_L | 0.2623 | |||||||||
| 26 | Frontal_Mid_Orb_R | 0.2181 | |||||||||
| 109 | Vermis_1_2 | 0.2091 | |||||||||
| 92 | Cerebelum_Crus1_R | 0.1977 | |||||||||
| 43 | Calcarine_L | −0.1931 | |||||||||
| 42 | Amygdala_R | −0.1826 | |||||||||
| 33 | Cingulum_Mid_L | 0.1711 | |||||||||
| 51 | Occipital_Mid_L | 0.17 | |||||||||
| VBM-GM data | |||||||||||
| BD vs. MDD (6 significant features) | BD vs. HC (16 significant features) | MDD vs. HC (6 significant features) | |||||||||
| AAL No. | Area name | Weight | AAL No. | Area name | Weight | AAL No. | Area name | Weight | |||
| ● | 14 | Frontal_Inf_Tri_R | 6.2067 | 92 | Cerebelum_Crus1_R | 11.7555 | ● | 13 | Frontal_Inf_Tri_L | 4.8525 | |
| 46 | Cuneus_R | −4.2253 | ● | 12 | Frontal_Inf_Oper_R | 9.7199 | 9 | Frontal_Mid_Orb_L | 1.9852 | ||
| ● | 12 | Frontal_Inf_Oper_R | 3.617 | 32 | Cingulum_Ant_R | 9.236 | 41 | Amygdala_L | 1.8239 | ||
| 13 | Frontal_Inf_Tri_L | 2.6796 | 56 | Fusiform_R | −7.9112 | 104 | Cerebelum_8_R | −1.3948 | |||
| ● | 31 | Cingulum_Ant_L | 2.6275 | 91 | Cerebelum_Crus1_L | 7.6179 | 73 | Putamen_L | 1.3436 | ||
| 56 | Fusiform_R | −0.6357 | 64 | SupraMarginal_R | 7.2463 | 13 | Frontal_Inf_Tri_L | 0.9092 | |||
| 52 | Occipital_Mid_R | −5.7348 | |||||||||
| 106 | Cerebelum_9_R | 5.3595 | |||||||||
| ● | 29 | Insula_L | 3.7479 | ||||||||
| 9 | Frontal_Mid_Orb_L | −3.2096 | |||||||||
| 31 | Cingulum_Ant_L | 2.921 | |||||||||
| ● | 114 | Vermis_8 | −2.9008 | ||||||||
| 89 | Temporal_Inf_L | −2.6185 | |||||||||
| 98 | Cerebelum_4_5_R | −2.2865 | |||||||||
| 115 | Vermis_9 | −2.0829 | |||||||||
| 46 | Cuneus_R | −1.7973 | |||||||||
“AAL No.” stands for the area order of the AAL templates a feature corresponds to, while “Area name” is the abbreviated name of the brain region related to that specific number.
The weights of the significant features dedicated to 6 classification conditions are listed in the “Weight” columns. Denoted with black dots, all the consensus features are included by the significant features.
Fig. 2.
Visualization of the significant features. Weight values were assigned to their corresponding AAL brain areas as mapped in color bar. The fALFF modality data are in the left column, whilst the VBM-GM modality is on the right. From top to bottom, classification conditions are arranged in this order: BP vs. UP, BP vs. HC, UP vs. HC. Consensus features are displayed with vertical green stripes overlapped on original color mapping of the significant features (see terms with black dot shown in TABLE IV).
On each fold of the LOOCV, the SVM-FoBa algorithm selected a set of highly discriminative features. However, since the training process was based on a slightly different subset of samples from fold to fold, the chosen feature set also varied each time. For instance, according to TABLE III, while 16 was decided as the optimal feature number when classifying BD and MDD subjects on fALFF data, that did not necessarily mean that the same 16 features were drawn across all the 46 cross-validations (CV); rather, as each CV fold chose its own 16 features, the average classification accuracy ranked the highest among all possible feature sizes. Therefore, we would particularly like to know which features were constantly chosen during the whole CV procedure, which we call “consensus features” hereafter. By recording the frequency of features being selected in the training process, we extracted the consensus feature(s) prevailing in all cross-validation iterations for each of the 6 conditions (two modalities × three types of classification). Besides, since the consensus features were constantly selected by the algorithm, consistent and significant contributions should have been made by these features across the entire CV process. It is not surprising, therefore, that for each classification condition, the consensus features constitute a subset of the significant features (see terms with black dot in TABLE IV, as well as the areas shaded with vertical green stripes in Fig.2).
Combining the results of both fALFF and VBM-GM, it can be observed that sub-regions related to the bilateral inferior frontal gyri (IFG) were frequently chosen with large weights among all 6 conditions (see Fig. 2). Considering that BD was always the negative sample, and all the significant features concerning IFG were of positive weights, a negative correlation between BD and other subject groups regarding the status of IFG is implied. Roughly speaking, a smaller IFG index value, either by fALFF or VBM-GM, would suggest a larger likelihood of BD classification. Interestingly, a series of the significant features with respect to discriminating BD patients also falls within the default mode network (DMN), including the angular gyrus, the anterior cingulate cortex, the cuneus area, and the fusiform and precentral gyri. Additionally, the cerebellum plays an important role in discriminating different subject groups in 5 out of 6 conditions. In particular, the vermis was identified, consistent with previous findings [38–40].
Aside from the common discoveries observed above, we were also curious to identify which specific group-discriminating information could be found using each modality separately. By analyzing the weights of the significant features, we found that the left angular gyrus, exclusively selected with fALFF data, was not only a consensus feature, but also provided significant predictive power to differentiate BD from MDD or HC. A recent multimodal research study suggested that the angular gyrus could play a role in discriminating BD and schizophrenia [41]. In addition, the bilateral pallidum and paracentral lobules, as well as the left inferior occipital cortex were identified by fALFF with negative weights only in the BD vs. MDD condition. Likewise, the right rolandic operculum and the right superior temporal gyrus were positively associated with the classification of MDD in fALFF (positive weights in BD vs. MDD, negative weights in MDD vs. HC), while the right Heschl’s gyrus was uniquely chosen to distinguish MDD from BD with positive weight.
On the other hand, for VBM-GM, the right fusiform and the right cuneus areas showed negative weights when differentiating BD from MDD and HC. Moreover, the left anterior cingulate cortex appeared with positive weights in both BD vs. MDD and BD vs. HC conditions.
Together, we discovered a series of significant features (brain areas) that may help distinguish BD from MDD with either structural or functional information, which will be discussed in detail in Section IV.
C. Classification with the Selected Features
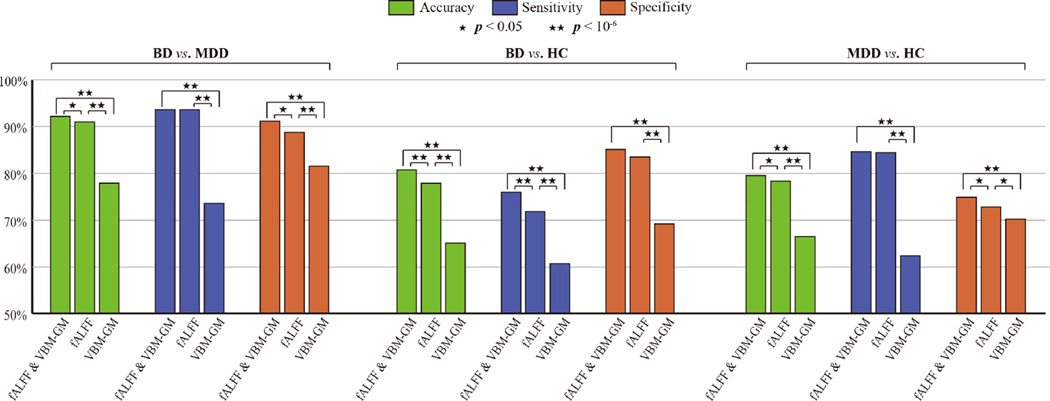
With the significant features in hand, it is natural to ask how “significant” these features really were. In other words, we would like to evaluate the classification performance using the significant features solely. Thus, we extracted 6 new sample sets with only significant features from the previous data according to TABLE III. To further investigate the multimodal performance, we also created three union sets of fALFF and VBM-GM for each of the classification conditions. Accordingly, a total of 9 sets of samples were generated, with three modality combinations (fALFF, VBM-GM, and fALFF and VBM-GM) × three conditions (BD vs. MDD, BD vs. HC, and MDD vs. HC). In order to yield more generalized results, these new data were entered into a linear SVM classifier using bootstrap aggregating [42]. That is, after a full randomization of sample order for each iteration, approximately 63.2% of the samples were used as the training set, while the rest of the samples served as the test set. In this study, we conducted 1,000 bootstrap resamples for each of the 9 data sets; classification accuracy, sensitivity and specificity rates were calculated with these 1,000 results summarized. An illustration comparing these results is provided in Fig. 3.
Fig. 3.
Performance of the linear SVM classifier using the significant features (1,000 bootstrap resamples). Paired T-tests were conducted to show the significance of differences between performance indices with two level of p-value provided. For all three classification conditions, the combined feature set outperform each separate modality alone, with the classification accuracy for BP vs. MDD being 92.07%, BP vs. HC 80.78%, and MDD vs. HC 79.51%. Sensitivity and specificity rates share the same propensity. Moreover, functional features yielded better results than structural ones.
Results showed that the performance utilizing multimodal features was always better than results using a single modality. Using two modalities, classification accuracies were 92.07% for BD vs. MDD, 80.78% for BD vs. HC, and 79.51% for MDD vs. HC. The sensitivity and specificity rates also showed similar tendencies. This makes sense as distinct modalities may offer complementary anatomical or functional information relevant to the classification of two clinical disorders.
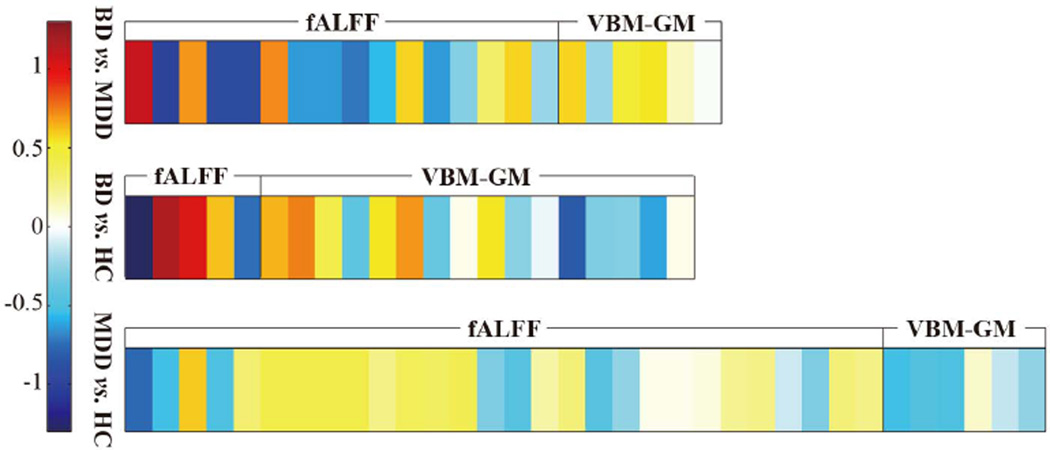
Additionally, it can be seen that the significant features of fALFF outperform those of VBM-GM by a large margin, to the extent that only very small performance gap could be observed between fALFF and the fusion of two modalities Specifically, for the BD vs. MDD condition, the classification accuracy of fALFF is 90.89%, against the one of VBM-GM at 77.83%. We thus conclude that, while still being important, structural features might supply limited information in the context of distinguishing between BD and MDD. To confirm this, we averaged the weight vectors learned by 1,000 linear SVM classifiers and created a weight map for each condition, as is shown in Fig. 4. Dimensions belonging to the fALFF yielded greater weight values than VBM-GM. In general, the most significant features in separate modality analyses also contributed the greatest to the final classification for combined modalities.
Fig. 4.
Visualization of the weight vector learned by linear SVM, wherein the values were averaged across 1,000 bootstrap resamples (not the weighs shown in TABLE IV). Darker color represents a larger absolute value of feature weight. The fALFF modality elicits greater weights when compared to the VBM-GM.
IV. Discussion
To the best of our knowledge, this is the first study to combine both anatomical and functional information to distinguish BD-I, MDD and HC during late adolescents/young adulthood (mean age = 20–21), in the early course of affective illness, using an advanced multivariate technique.
A known challenge in brain imaging data classification is the reduction of feature dimensionality, without which overfitting can occur, so that the generalization performance of a classifier may be compromised. One possible solution is the incorporation of feature reconstruction, whereby existing features will be transformed into a new space, revealing the discriminative information that is not evident in the original space, such as clustering, basic linear transforms (e.g., principal component analysis and linear discriminant analysis), and nonlinear transforms [43]. However, as none of the original input features will be discarded, model understanding can be rendered impractical, especially for neuroimaging data. In this study, we proposed a novel feature selection method by incorporating a forward-backward search strategy integrated with the linear SVM, aiming to capture significant features and to identify the potential imaging biomarkers of two mood disorders that are, in clinical practice, often challenging to differentiate early in their course. It is worth mentioning that, before applying SVM-FoBa to neuroimaging data, we conducted a series of experiments to verify whether this algorithm was advantageous to other typical feature selection methods on public data sets. Results demonstrated that not only did SVM-FoBa yield superior classification accuracy in most cases, but more feature combinations may also be comprehensively investigated, achieving higher sensitivity and specificity rates.
As to feature selection, other algorithms that integrate a sparse prior in the optimization objective function of SVM may also be feasible. For example, Bayesian SVM [44, 45] seeks to estimate the optimal hyperparameters with probabilistic framework employed, which mostly incorporates Gaussian kernel; The genetic algorithm [29, 30] has been widely used as an approximation of global optima, but has an extra layer of pseudo-biology with a series of hyperparameters that may complicate the result interpretation, let alone the slow convergence on nontrivial problems [46]. In this study, the ultimate question that we were trying to answer was which features — biomarkers — differentiated BD and MDD diseases. Known as the methodology of decoding [47], the key to model understanding is inverse inference; with the linearity of SVM-FoBa exploited, the contribution of features can be quantified, compared and interpreted with their corresponding weights.
Regarding our methods, two caveats should be considered. The first is the feature selection criteria. Different classifiers generate different biases; a feature subset optimal for one classifier might be less useful for another. Instead of using misclassification rate as the loss function, we directly incorporated the value of the objective function of the trained SVM, by which the discriminative power of a feature set can be dynamically measured. In other words, we considered feature selection as part of the training process. Since the cost to collect medical imaging samples is usually high, this approach may promise a better use of the available data at hand. Furthermore, the intrinsic SVM regularization of the objective function can prevent the classifier from overfitting the training set.
The second caveat is search strategy. Although the conventional greedy method may aggressively generate sparse solutions, the selected nested subsets of features would cause the errors produced in earlier steps to be uncorrected. To overcome this problem, we therefore developed the SVM-FoBa algorithm to adaptively select a significant feature set for each of the classification conditions. It is important to note that the proposed algorithm involves a parameter ε that determines the threshold of the loss functional value between steps. This can be regarded as the issue of model selection in machine learning theory. Since a good model selection criteria can be of great importance [20], we used the LOOCV as the outer loop to measure the performance, while a 10-fold CV procedure was also implemented in the inner loop to decide the optimal value of ε.
On the clinical side, subareas of the inferior frontal cortex established themselves as the critical, common significant discriminative features across diagnoses and modalities. As mentioned before, a consistent result was that BD was negatively correlated with the features concerning IFG subareas, no matter which modality was considered. It has been reported that shrinkage of the gray matter volume in inferior frontal gyrus can be observed in BD patients [48, 49]. Clearly, our BD vs. HC result with respect to the VBM-GM modality offers confirmation of this view. Previous studies also suggest that MDD patients will show tissue loss in IFG [50]. Again, this was corroborated by the VBM-GM result pertaining to condition MDD vs. HC, where a positive weight was assigned to the consensus feature of pars triangularis of IFG. Considering that with the same data modality two consensus features of IFG were trained with positive weights in the BD vs. MDD condition, we speculate that, although both types of clinical disorders could cause gray matter loss in IFG, BD patients might suffer from a greater shrinkage in volume. For the fALFF data, a similar trend also appeared such that the BD group obtained positive feature weights in IFG in the BD vs. MDD and BD vs. HC conditions, suggesting more severe functional deficits in IFG for BD patients when compared to depressive or healthy individuals. In sum, BD appeared to elicit abnormalities in IFG for both structural and functional modalities that MDD patients did not. Further analysis should be conducted to verify if the IFG area is a robust predictor for distinguishing BD and MDD.
In addition, sub-regions of the cerebellum, especially the vermis, were identified as an important factor to classify BD, MDD and HC in most conditions. The cerebellum has shown to be both anatomically and functionally connected with the prefrontal cortex and subcortical limbic structures. One meta-analysis study proposed that the cerebellum might play a functional role in normal and abnormal mood regulation, such that the abnormality could be the underlying cause of psychosis, depression and mania [38]. In particular, vermal deficits have been reported for both individuals with BD [40, 51–54] and MDD [55–57] in many reports. Our results further confirmed these findings for both anatomy and function. While often overlooked by researchers, it is advised that dedicated multivariate studies concerning mental disorders should scrutinize the substructures and functions of the cerebellum in the future.
Furthermore, a group of brain regions belonging to the DMN were selected as the significant features to discriminate BD from other subject groups. The right inferior occipital area and the right fusiform area were selected with significant weights to classify samples into BD, based on either fALFF or VBM-GM data (see Section III.B). Previous imaging studies have suggested several unusual DMN features for BD patients: incorporation of atypical regions such as the occipital cortex, lateral parietal, striatum and pontine areas, and also reduced correlation between typical DMN nodes such as the fusiform and hippocampal regions [14, 58]. In contrast to more general abnormalities of the medial prefrontal cortex that are also seen in other mental disorders (e.g., depression and schizophrenia), these disturbances might be unique for BD. Hence, our findings provide evidence to support this view.
In this study, we conducted classification experiments using both single modalities and their combination. The two data modalities have yielded separate discoveries, with one enhancing or complementing the other; more importantly, the combination of fALFF and VBM-GM features improved the classification accuracy between two mood disorders that may be easily confused in their first episode. Also, during our dedicated classification test of the significant features, the fALFF modality outperformed VBM-GM with considerable margin (see Fig. 3 and Fig. 4 for detail), while also being very closed to the accuracy of two modalities merged together. Despite the fact that the VBM-GM data alone has provided a large amount of useful information regarding early stage of the illness of the patients studied, functional features appear to be more informative when separating BD and MDD. Although abnormalities in anatomical structures can ultimately be the cause of dysfunctions [59], it does not necessarily mean that all crucial structural deficits can be efficiently detected by means of current brain imaging technologies and techniques. Moreover, even while the mechanisms underpinning mental disorders are of great interest, a key difficulty lies in the clinical diagnosis of BD patients. In that sense, reliable biomarkers, or rather effective neuroimaging predictors are of primary importance. Our results suggest that more attention should be drawn to examine various functional indices (e.g., functional connectivity) to discriminate BD from MDD.
Finally, several general limitations have to be acknowledged. First, as SVM-FoBa effectively addresses some intrinsic issues related to typical greedy methods, it is essentially an algorithm seeking the local optima. Furthermore, our sample size was relatively small and similar analyses should be performed with a larger number of subjects. In addition, due to the lack of additional data, we conducted the final classification test of significant features on the original samples, which might introduce a bias. Future studies with independent subject data are preferable. Moreover, the fALFF and VBM-GM features were obtained by directly averaging each subarea of AAL template. This operation eliminated the fine voxel-wise details within each area, which might contain potentially useful information for group discrimination. Another concern is reflected by the medication status of the subjects. Although previous studies suggest that medication effect might be negligible for fMRI data [60], it is important to study medication naïve patients to ensure medication effects are not playing a role.
V. Conclusion
To the best of our knowledge, this is the first study to utilize a multivariate feature selection approach to classify BD and MDD with multiple modalities. The present work demonstrated that, with the help of a novel feature selection approach involving forward-backward strategy, highly discriminative features could be extracted from the data of both functional and structural modalities for bipolar and major depressive disorders, whereby high classification accuracy could be achieved. A large portion of the most informative features belonged to the IFG, DMN and the cerebellar vermis. Specifically, when compared to HC and MDD, BD patients elicited the lowest fALFF and VBM-GM value in IFG. Moreover, a trend was found that default mode network, with the adoption of atypical nodes, was helpful exclusively for identifying bipolar patients. These findings, which were predominantly driven by the functional information, suggest possible biomarkers for differential diagnosis of BD versus MDD. This should be confirmed in future studies.
Acknowledgment
This work was partially supported by “100 Talents Plan” of Chinese Academy of Sciences, Chinese National Science Foundation No. 81471367 and the State High-Tech Development Plan (863), grant No. SQ2015AA0200506 (to J. Sui); the Strategic Priority Research Program of the Chinese Academy of Sciences, grant No. XDB02030300 and National Key Basic Research and Development Program (973), grant No. 2011CB707800, (to T.-Z. Jiang); National Institutes of Health grants R01EB006841, R01EB005846, and P20GM103472 (to V.D. Calhoun); the Lawson Health Research Institute, grant No. LHR D1374 (to E.A. Osuch); the Pfizer Independent Investigator Award, grant No. WS2249136 (to E.A. Osuch); and PhD Research Startup Foundation of Jiangxi Normal University, grant No. 6247 (to M.-H. Zhu).
Contributor Information
Nan-Feng Jie, Email: nfjie@nlpr.ia.ac.cn, Brainnetome center and the National Laboratory of Pattern Recognition, Institute of Automation, Chinese Academy of Sciences, Beijing, China.
Mao-Hu Zhu, Email: maohzhu@hotmail.com, Elementary Educational College, Jiangxi Normal University, Nanchang, China.
Xiao-Ying Ma, Email: xiaoying.ma.ccm@gmail.com, School of Information Science and Engineering, Lanzhou University, Lanzhou, China.
Elizabeth A Osuch, Email: elizabeth.osuch@lhsc.on.ca, Dept. of Psychiatry, University of Western Ontario and the Lawson Health Research Institute Imaging Division, London, Ontario, Canada.
Michael Wammes, Dept. of Psychiatry, University of Western Ontario and the Lawson Health Research Institute Imaging Division, London, Ontario, Canada.
Jean Théberge, Email: jtheberge@lawsonimaging.ca, Dept. of Medical Biophysics, University of Western Ontario and the Lawson Health Research Institute Imaging Division, London, Ontario, Canada.
Huan-Dong Li, Brainnetome center and the National Laboratory of Pattern Recognition, Institute of Automation, Chinese Academy of Sciences, Beijing, China.
Yu Zhang, Brainnetome center and the National Laboratory of Pattern Recognition, Institute of Automation, Chinese Academy of Sciences, Beijing, China.
Tian-Zi Jiang, Brainnetome center and the National Laboratory of Pattern Recognition, Institute of Automation, Chinese Academy of Sciences, Beijing, China.
Jing Sui, Brainnetome center and the National Laboratory of Pattern Recognition, Institute of Automation, Chinese Academy of Sciences, Beijing, China; Mind Research Network/LBERI.
Vince D Calhoun, Email: vcalhoun@unm.edu, Mind Research Network/LBERI; Dept. of ECE, University of New Mexico, Albuquerque, NM, 87106.
References
- 1.Luckenbaugh DA, Ameli R, Brutsche NE, et al. Rating depression over brief time intervals with the Hamilton Depression Rating Scale: Standard vs. abbreviated scales. J Psychiatr Res. 2015 Feb;61:40–45. doi: 10.1016/j.jpsychires.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuellar AK, Johnson SL, Winters R. Distinctions between bipolar and unipolar depression. Clinical psychology review. 2005;25(3):307–339. doi: 10.1016/j.cpr.2004.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardoso de Almeida JR, Phillips ML. Distinguishing between unipolar depression and bipolar depression: current and future clinical and neuroimaging perspectives. Biol Psychiatry. 2013 Jan 15;73(2):111–118. doi: 10.1016/j.biopsych.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirschfeld RM, Vornik LA. Bipolar disorder--costs and comorbidity. Am J Manag Care. 2005 Jun;11(3 Suppl):S85–S90. [PubMed] [Google Scholar]
- 5.Hirschfeld RMA, Vornik LA. Bipolar disorder - Costs and comorbidity. American Journal of Managed Care. 2005 Jun;11(3):S85–S90. [PubMed] [Google Scholar]
- 6.Hirschfeld RM, Lewis L, Vornik LA. Perceptions and impact of bipolar disorder: how far have we really come? Results of the national depressive and manic-depressive association 2000 survey of individuals with bipolar disorder. The Journal of clinical psychiatry. 2003 Feb;64(2):161–174. [PubMed] [Google Scholar]
- 7.Kupfer DJ. The increasing medical burden in bipolar disorder. JAMA : the journal of the American Medical Association. 2005 May 25;293(20):2528–2530. doi: 10.1001/jama.293.20.2528. [DOI] [PubMed] [Google Scholar]
- 8.Dudek D, Siwek M, Zielinska D, et al. Diagnostic conversions from major depressive disorder into bipolar disorder in an outpatient setting: results of a retrospective chart review. Journal of affective disorders. 2013 Jan 10;144(1–2):112–115. doi: 10.1016/j.jad.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 9.MacMaster FP, Leslie R, Rosenberg DR, et al. Pituitary gland volume in adolescent and young adult bipolar and unipolar depression. Bipolar Disord. 2008 Feb;10(1):101–104. doi: 10.1111/j.1399-5618.2008.00476.x. [DOI] [PubMed] [Google Scholar]
- 10.Versace A, Almeida JR, Quevedo K, et al. Right orbitofrontal corticolimbic and left corticocortical white matter connectivity differentiate bipolar and unipolar depression. Biol Psychiatry. 2010 Sep 15;68(6):560–567. doi: 10.1016/j.biopsych.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Versace A, Thompson WK, Zhou D, et al. Abnormal left and right amygdala-orbitofrontal cortical functional connectivity to emotional faces: state versus trait vulnerability markers of depression in bipolar disorder. Biol Psychiatry. 2010 Mar 1;67(5):422–431. doi: 10.1016/j.biopsych.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Almeida JR, Versace A, Hassel S, et al. Elevated amygdala activity to sad facial expressions: a state marker of bipolar but not unipolar depression. Biol Psychiatry. 2010 Mar 1;67(5):414–421. doi: 10.1016/j.biopsych.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor Tavares JV, Clark L, Furey ML, et al. Neural basis of abnormal response to negative feedback in unmedicated mood disorders. Neuroimage. 2008 Sep 1;42(3):1118–1126. doi: 10.1016/j.neuroimage.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duda RO, Hart PE, Stork DG. Pattern classification. John Wiley & Sons; 2012. [Google Scholar]
- 15.Sui J, Adali T, Yu Q, et al. A review of multivariate methods for multimodal fusion of brain imaging data. Journal of neuroscience methods. 2012;204(1):68–81. doi: 10.1016/j.jneumeth.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calhoun VD, Adali T. Feature-based fusion of medical imaging data. IEEE Trans Inf Technol Biomed. 2009 Sep;13(5):711–720. doi: 10.1109/TITB.2008.923773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norman KA, Polyn SM, Detre GJ, et al. Beyond mind-reading: multi-voxel pattern analysis of fMRI data. Trends Cogn Sci. 2006 Sep;10(9):424–430. doi: 10.1016/j.tics.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Zeng LL, Shen H, Liu L, et al. Identifying major depression using whole-brain functional connectivity: a multivariate pattern analysis. Brain. 2012 May;135(Pt 5):1498–1507. doi: 10.1093/brain/aws059. [DOI] [PubMed] [Google Scholar]
- 19.Grotegerd D, Suslow T, Bauer J, et al. Discriminating unipolar and bipolar depression by means of fMRI and pattern classification: a pilot study. Eur Arch Psychiatry Clin Neurosci. 2013 Mar;263(2):119–131. doi: 10.1007/s00406-012-0329-4. [DOI] [PubMed] [Google Scholar]
- 20.Guyon I, Weston J, Barnhill S, et al. Gene selection for cancer classification using support vector machines. Machine Learning. 2002;46(1–3):389–422. [Google Scholar]
- 21.Cortes C, Vapnik V. Support-Vector Networks. Machine Learning. 1995 Sep;20(3):273–297. [Google Scholar]
- 22.Zou Q-H, Zhu C-Z, Yang Y, et al. An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: fractional ALFF. Journal of neuroscience methods. 2008;172(1):137–141. doi: 10.1016/j.jneumeth.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashburner J, Friston KJ. Voxel-based morphometry—the methods. Neuroimage. 2000;11(6):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 24.Kubicki M, Shenton M, Salisbury D, et al. Voxel-based morphometric analysis of gray matter in first episode schizophrenia. Neuroimage. 2002;17(4):1711–1719. doi: 10.1006/nimg.2002.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 26.Yan C, Zang Y. DPARSF: a MATLAB toolbox for “pipeline” data analysis of resting-state fMRI. Frontiers in systems neuroscience. 2010;4 doi: 10.3389/fnsys.2010.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calhoun VD, Adali T. Feature-based fusion of medical imaging data. Information Technology in Biomedicine, IEEE Transactions on. 2009;13(5):711–720. doi: 10.1109/TITB.2008.923773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohavi R, John GH. Wrappers for feature subset selection. Artificial Intelligence. 1997 Dec;97(1–2):273–324. [Google Scholar]
- 29.Davis L. Handbook of genetic algorithms. Van Nostrand Reinhold New York: 1991. [Google Scholar]
- 30.Frohlich H, Chapelle O, Scholkopf B. Feature selection for support vector machines by means of genetic algorithm. :142–148. [Google Scholar]
- 31.Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST) 2011;2(3):27. [Google Scholar]
- 32.Wolpert DH, Macready WG. No free lunch theorems for optimization. Evolutionary Computation, IEEE Transactions on. 1997;1(1):67–82. [Google Scholar]
- 33.Tsanas A, Little MA, Fox C, et al. Objective automatic assessment of rehabilitative speech treatment in Parkinson's disease. Neural Systems and Rehabilitation Engineering, IEEE Transactions on. 2014;22(1):181–190. doi: 10.1109/TNSRE.2013.2293575. [DOI] [PubMed] [Google Scholar]
- 34.Alon U, Barkai N, Notterman DA, et al. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proceedings of the National Academy of Sciences. 1999;96(12):6745–6750. doi: 10.1073/pnas.96.12.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 36.Rakotomamonjy A. Variable selection using svm based criteria. The Journal of Machine Learning Research. 2003;3:1357–1370. [Google Scholar]
- 37.Xia M, Wang J, He Y. BrainNet Viewer: a network visualization tool for human brain connectomics. PLoS One. 2013;8(7):e68910. doi: 10.1371/journal.pone.0068910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konarski JZ, McIntyre RS, Grupp LA, et al. Is the cerebellum relevant in the circuitry of neuropsychiatric disorders? Journal of Psychiatry and Neuroscience. 2005;30(3):178. [PMC free article] [PubMed] [Google Scholar]
- 39.Peng J, Liu J, Nie B, et al. Cerebral and cerebellar gray matter reduction in first-episode patients with major depressive disorder: a voxel-based morphometry study. European journal of radiology. 2011;80(2):395–399. doi: 10.1016/j.ejrad.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 40.DelBello MP, Strakowski SM, Zimmerman ME, et al. MRI analysis of the cerebellum in bipolar disorder: a pilot study. Neuropsychopharmacology. 1999;21(1):63–68. doi: 10.1016/S0893-133X(99)00026-3. [DOI] [PubMed] [Google Scholar]
- 41.Sui J, Pearlson G, Caprihan A, et al. Discriminating schizophrenia and bipolar disorder by fusing fMRI and DTI in a multimodal CCA+ joint ICA model. Neuroimage. 2011;57(3):839–855. doi: 10.1016/j.neuroimage.2011.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauer E, Kohavi R. An empirical comparison of voting classification algorithms: Bagging, boosting, and variants. Machine Learning. 1999;36(1–2):105–139. [Google Scholar]
- 43.Roweis ST, Saul LK. Nonlinear dimensionality reduction by locally linear embedding. Science. 2000;290(5500):2323–2326. doi: 10.1126/science.290.5500.2323. [DOI] [PubMed] [Google Scholar]
- 44.Sollich P. Bayesian methods for support vector machines: Evidence and predictive class probabilities. Machine learning. 2002;46(1–3):21–52. [Google Scholar]
- 45.Zhang Z, Jordan MI. Bayesian multicategory support vector machines. arXiv preprint arXiv:1206.6863. 2012 [Google Scholar]
- 46.Skiena SS. The algorithm design manual: Text. Springer Science & Business Media; 1998. [Google Scholar]
- 47.Haynes JD, Rees G. Decoding mental states from brain activity in humans. Nat Rev Neurosci. 2006 Jul;7(7):523–534. doi: 10.1038/nrn1931. [DOI] [PubMed] [Google Scholar]
- 48.Lyoo IK, Kim MJ, Stoll AL, et al. Frontal lobe gray matter density decreases in bipolar I disorder. Biol Psychiatry. 2004;55(6):648–651. doi: 10.1016/j.biopsych.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 49.Farrow TF, Whitford TJ, Williams LM, et al. Diagnosis-related regional gray matter loss over two years in first episode schizophrenia and bipolar disorder. Biol Psychiatry. 2005;58(9):713–723. doi: 10.1016/j.biopsych.2005.04.033. [DOI] [PubMed] [Google Scholar]
- 50.Li C-T, Lin C-P, Chou K-H, et al. Structural and cognitive deficits in remitting and non-remitting recurrent depression: a voxel-based morphometric study. Neuroimage. 2010;50(1):347–356. doi: 10.1016/j.neuroimage.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 51.Kim D, Byul Cho H, Dager SR, et al. Posterior cerebellar vermal deficits in bipolar disorder. J Affect Disord. 2013;150(2):499–506. doi: 10.1016/j.jad.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cecil KM, DelBello MP, Sellars MC, et al. Proton magnetic resonance spectroscopy of the frontal lobe and cerebellar vermis in children with a mood disorder and a familial risk for bipolar disorders. J Child Adolesc Psychopharmacol. 2003;13(4):545–555. doi: 10.1089/104454603322724931. [DOI] [PubMed] [Google Scholar]
- 53.Strakowski SM, Adler CM, DelBello MP. Volumetric MRI studies of mood disorders: do they distinguish unipolar and bipolar disorder? Bipolar Disord. 2002;4(2):80–88. doi: 10.1034/j.1399-5618.2002.01160.x. [DOI] [PubMed] [Google Scholar]
- 54.Mills NP, DelBello MP, Adler CM, et al. MRI analysis of cerebellar vermal abnormalities in bipolar disorder. American Journal of Psychiatry. 2005;162(8):1530–1533. doi: 10.1176/appi.ajp.162.8.1530. [DOI] [PubMed] [Google Scholar]
- 55.Shah S, Doraiswamy P, Husain M, et al. Posterior fossa abnormalities in major depression: a controlled magnetic resonance imaging study. Acta Psychiatr Scand. 1992;85(6):474–479. doi: 10.1111/j.1600-0447.1992.tb03214.x. [DOI] [PubMed] [Google Scholar]
- 56.Dolan R, Bench CJ, Brown R, et al. Regional cerebral blood flow abnormalities in depressed patients with cognitive impairment. Journal of Neurology, Neurosurgery & Psychiatry. 1992;55(9):768–773. doi: 10.1136/jnnp.55.9.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sweeney JA, Strojwas MH, Mann JJ, et al. Prefrontal and cerebellar abnormalities in major depression: evidence from oculomotor studies. Biol Psychiatry. 1998;43(8):584–594. doi: 10.1016/s0006-3223(97)00485-x. [DOI] [PubMed] [Google Scholar]
- 58.Öngür D, Lundy M, Greenhouse I, et al. Default mode network abnormalities in bipolar disorder and schizophrenia. Psychiatry Research: Neuroimaging. 2010;183(1):59–68. doi: 10.1016/j.pscychresns.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Passingham RE, Stephan KE, Kötter R. The anatomical basis of functional localization in the cortex. Nature Reviews Neuroscience. 2002;3(8):606–616. doi: 10.1038/nrn893. [DOI] [PubMed] [Google Scholar]
- 60.Phillips M, Travis M, Fagiolini A, et al. Medication effects in neuroimaging studies of bipolar disorder. American Journal of Psychiatry. 2008;165(3):313–320. doi: 10.1176/appi.ajp.2007.07071066. [DOI] [PMC free article] [PubMed] [Google Scholar]