Abstract
Background
The contribution of epigenetic factors, such as histone acetylation and DNA methylation, to the regulation of alcohol drinking behavior has been increasingly recognized over the last several years. GADD45b is a protein demonstrated to be involved in DNA demethylation at neurotrophic factor gene promoters, including at Brain-derived neurotrophic factor (Bdnf) which has been highly implicated in alcohol drinking behavior.
Methods
DNA methyltransferase-1 (Dnmt1), 3a, and 3b, and Gadd45a, b, and g mRNA were measured in the nucleus accumbens (NAc) and ventral tegmental areas (VTA) of high ethanol consuming C57BL/6J (C57) and low alcohol consuming DBA/2J (DBA) mice using qRT-PCR. In the NAc GADD45b protein was measured via immunohistochemistry and Bdnf9a mRNA using in situ PCR. Bdnf9a promoter histone H3 acetylated at lysines 9 and 14 (H3K9,K14ac) was measured using chromatin immunoprecipitation, and 5-methylcytosine (5MC) and 5-hydroxymethylcytosine (5HMC) using methylated DNA immunoprecipitation. Alcohol drinking behavior was evaluated in Gadd45b haplodeficient (+/−) and null mice (−/−) utilizing drinking in the dark (DID) and 2-bottle free-choice paradigms.
Results
C57 mice had lower levels of Gadd45b and g mRNA and GADD45b protein in the NAc relative to the DBA strain. C57 mice had lower NAc shell Bdnf9a mRNA levels, Bdnf9a promoter H3K9,K14ac, and higher Bdnf9a promoter 5HMC and 5MC. Acute ethanol increased GADD45b protein, Bdnf9a mRNA, and histone acetylation and decreased 5HMC in C57 mice. Gadd45b +/− mice displayed higher drinking behavior relative to wild-type littermates in both DID and 2-bottle free-choice paradigms.
Conclusions
These data indicate the importance of the DNA demethylation pathway and its interactions with histone post-translational modifications in alcohol drinking behavior. Further, we suggest that lower DNA demethylation protein GADD45b levels may affect Bdnf expression possibly leading to altered alcohol drinking behavior.
Keywords: GADD45b, histone, DNA methylation, Bdnf, alcoholism, nucleus accumbens
Introduction
Epigenetic pathways as possible explanatory mechanisms for addiction have gained increasing support over the last several years as they are able to account for both the heritable (genetic or epigenetic) and environmental factors widely believed to underlie these illnesses (Renthal and Nestler, 2009; Krishnan et al., 2014). However, among the drugs of abuse epigenetic mechanisms are uniquely pertinent to alcohol in that ethanol affects gene expression not only by altering neurotransmission on the cell surface, but can freely traverse all biological membranes including permeating the nucleus where it can directly alter epigenetic machinery (Pandey et al., 2008a; Warnault et al., 2013; Sakharkar et al., 2012; Zhong et al., 2010). Moreover, systemic inhibition of DNA methyltransferase (DNMT) activity decreases alcohol drinking and seeking behaviors in rodents (Warnault et al., 2013).
Histone post-translational modifications and DNA methylation are two important aspects of epigenetic gene regulation. Histone H3 acetylation generally induces an open chromatin state allowing for the binding of transcription factors, while DNMT-catalyzed methylation at the 5′ carbon of cytosine bases (5-methylcytosine (5MC)) reduces DNA access (Gavin and Floreani, 2014; Szyf, 2010). 5MC can be further processed into another stable cytosine modification, 5-hydroxymethylcytosine (5HMC). The effects of 5HMC on gene expression are complex and still under investigation with roles likely largely dependent on whether it precedes a guanine or adenine and its location relative to the gene transcription start site (TSS) (i.e., proximal to the promoter or in the gene body) (Kinde et al., 2015). 5HMC or 5MC are thought to be deaminated by a cytidine deaminase forming 5-hydroxymethyluracil (5HMU) or uracil. 5HMU or uracil is then believed to be removed by a DNA glycosylase. Although not enzymes themselves, Growth arrest and DNA Damage-inducible (GADD45) proteins (GADD45a, GADD45b, and GADD45g) are cornerstones of the DNA demethylation process in that they are capable of recruiting cytidine deaminases and DNA glycosylases to genomic loci (Cortellino et al., 2011; Guo et al., 2011; He et al., 2011).
Human and animal studies indicate dysregulation in both histone acetylation and DNA methylation in alcoholism. In humans with alcoholism, upregulation of histone methyltransferases SET domain containing 1A (SETD1A) and mixed-lineage leukemia 4 (MLL4) in the frontal cortex and basolateral nucleus of the amygdala, and decreased DNMT1 in the frontal cortex and basolateral and central nuclei of the amygdala have been reported (Ponomarev et al., 2012), while in psychotic subjects alcohol abuse appears to increase the cytidine deaminase Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC3C) mRNA in the parietal cortex (Guidotti et al., 2012). In rodents it was found that DNMT1 is increased in the nucleus accumbens (NAc) of mice undergoing cycles of excessive alcohol intake and withdrawal (Warnault et al., 2013). One hour after an acute ethanol injection there is a decrease in histone deacetylase (HDAC) activity and increase in histone H3 and H4 acetylation in rat amygdaloid brain regions. On the other hand, after an intermittent access paradigm or alcohol withdrawal there are decreases in histone acetylation (Pandey et al., 2008a; Sakharkar et al., 2012; Warnault et al., 2013).
The Brain-derived neurotrophic factor (Bdnf) gene is a target of epigenetic machinery, and its dysregulation has been highly implicated in alcoholism (Ma et al., 2009; Chen et al., 2003; Martinowich et al., 2003; McGough et al., 2004; Jeanblanc et al., 2009). Lower BDNF levels in rodent models is associated with increased ethanol consumption (Hensler et al., 2003; McGough et al., 2004; Yan et al., 2005; Jeanblanc et al., 2006; Moonat et al., 2011). The role of BDNF in alcohol drinking behavior is further supported by the fact that BDNF is an ethanol-responsive gene in humans and rodents (Pandey et al., 2008b; Joe et al., 2007; Huang et al., 2011). Acute ethanol, whether administered intraperitoneal (i.p.) or voluntarily consumed, increases Bdnf mRNA levels in the mouse dorsal striatum (McGough et al., 2004). While these studies indicate that ethanol increases BDNF expression leading to reduced ethanol consumption the mechanisms behind this response remain to be elucidated.
In the current study we examined baseline and ethanol-induced changes in epigenetic Bdnf regulation by comparing the high-drinking C57BL/6J (C57) and low-drinking DBA/2J (DBA) strains as well as mice with a deficiency in a DNA demethylating protein known to demethylate Bdnf, GADD45b (Misra and Pandey, 2003; Shelton and Grant, 2002; Kerns et al., 2005; Belknap et al., 1977; Belknap et al., 1993; Gupta et al., 2005; Ma et al., 2009; Gavin et al., 2012). We measured baseline and acute ethanol-induced changes in Gadd45 gene expression and the effects on Bdnf promoter 5MC, 5HMC, and histone H3 acetylation (H3K9,14ac) in these two strains. Subsequently, we used Gadd45b haploinsufficient and total knockout mice to further examine the direct role of GADD45b in ethanol drinking behaviors.
Methods & Materials
C57BL/6J vs. DBA/2J procedures
All experiments were conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. Male 12–16 week-old C57BL/6J (C57) and DBA/2J (DBA) mice were purchased from Jackson Laboratories (Bar Harbor, ME). Animals were group housed in a temperature-controlled room with a 12/12 hr light/dark cycle, with food and water provided ad lib. Mice received a single injection of 2 g/kg ethanol or normal saline i.p. and after 1 hr were injected with pentobarbital 50 mg/kg. Under sedation mice were either immediately decapitated and brain tissues were dissected and quickly frozen, or perfused with normal saline followed by 4% paraformaldehyde (PFA) solution prepared in 0.1 M phosphate buffer (pH 7.4). All brains were frozen and kept at −80 °C until further use. Blood was collected to measure ethanol levels using an Analox alcohol analyzer (Analox Instruments; Lunenburg, MA).
Breeding and genotyping of Gadd45b haploinsufficent and knockout mice
Gadd45b knock-out mice were obtained from the Liebermann laboratory at Temple University (Gupta et al., 2005) and backcrossed at least 5 generations to mice of a C57BL/6J genetic background. Wild-type, knockout and heterozygous mice were generated from heterozygous breeding pairs. Male mice (12–16 weeks old) were used in all experiments. Genotyping was performed by PCR. The DNA samples isolated from tail tips were used for PCR amplifications [95°C for 10 min; then 40 cycles of 95°C for 30 sec, 60°C for 1 min, and 72°C for 30 sec] to identify the wild-type (253 bp band), heterozygous (165 + 253 bp bands), and knockout (165 bp band) mice. The primers used were as follows: Gadd45b 1: 5′-TCAGGCAAGAGGAGACTGAGA-3′; Gadd45b 2: 5′-CAAGCGATCTGTCTTGCTCA-3′; PGK promoter: 5′-TAAAGCGCATGCTCCAGACT-3′.
Two bottle free-choice procedure
Twenty-four hour continuous access ethanol preference measurements were conducted using the two bottle free-choice paradigm (Thiele et al., 2000; Misra and Pandey, 2003). Singly housed mice were provided with two water bottles for two weeks with ad libitum access to food and water; a duration sufficient to habituate mice to drink equally from both bottles. Bottle positions were changed daily to avoid a positional habit. After two weeks one water bottle was replaced with a bottle containing ethanol with the position of the ethanol and water bottles being switched daily. Concentrations of ethanol were increased every 3 days from 3% to 6% to 9%, and finally 12% for the final 3 days. Consumption of ethanol and water was measured daily at 10AM and fresh water and ethanol (3, 6, 9, or 12%) solution in water were provided every day. The mean ethanol intake in g/kg, and water intake were calculated.
Drinking-in-the-Dark procedure
Drinking-in-the-Dark (DID) was performed as documented in previous publications (Rhodes et al., 2005). Mice were singly housed and allowed to acclimate to the reverse light-dark cycle for 2 weeks with ad libitum access to food and water. The light cycle was from 8:00PM to 8:00AM and the dark cycle from 8:00AM to 8:00 PM. Three hours after the beginning of the dark cycle (11:00AM), water bottles in home cages were replaced with bottles containing 20% ethanol in tap water. These bottles remained in place for 2 hrs (removed at 1:00PM) for the first 3 days. The experimental procedures on Days 4 and 5 were the same as those for Days 1, 2, and 3, except the ethanol containing bottles remained in place for 4 hrs (removed at 3:00PM). Ethanol consumption was assessed daily; average daily ethanol consumption across all genotypes was 5.58 g of ethanol per kg body weight (SEM=0.24).
Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR)
Total RNA was isolated using TRIZOL reagent (Life Technologies; Carlsbad, CA), and DNA removed using DNA-free™ DNA Removal Kit (Life Technologies; Carlsbad, CA). Target gene expression was normalized to Hypoxanthine Phosphoribosyltransferase 1 (Hprt1) (see Table 1 for primer sequences). For qRT–PCR, cDNA samples were analyzed using PikoReal Real-Time PCR (Thermo Fisher; Waltham, MA) and Maxima® SYBR Green/ROX qPCR Master Mix (Thermo Fisher; Waltham, MA). Primers were designed to span at least one intron–exon boundary, with the exception of Bdnf9a which lacks an intervening intron. The following cycling conditions were used: 10 min at 95 °C then 40 cycles at 95°C for 30 s, 60 °C for 1 min, and 72°C for 30 s.
Table 1.
Primer sequences used in this study.
| mRNA Expression Primers | ||
|---|---|---|
| Genes | 5′ primer | 3′ primer |
| Bdnf9a | GCAGCTGGAGTGGATCAGTAA | TGGTCATCACTCTTCTCACCTG |
| Dnmt1 | AACCATCACCGTGCGAGACA | AGGGTGCTGAATTGCCTGGA |
| Dnmt3a | AGGAAGACCCCTGGAACTGCT | ACGTCCCCGACGTACATGATCT |
| Dnmt3b | GCGTCAGTACCCCATCAGTT | CACGAGGTCACCTATTCCAAA |
| Gadd45a | TGCGAGAACGACATCAACAT | TCCCGGCAAAAACAAATAAG |
| Gadd45b | GTTCTGCTGCGACAATGACA | TTGGCTTTTCCAGGAATCTG |
| Gadd45g | ATGACTCTGGAAGAAGTCCGT | CAGGGTCCACATTCAGGACT |
| Hprt1 | GCCGAGGATTTGGAAAAAGT | ACAGAGGGCCACAATGTGAT |
| Methylated DNA Immunoprecipitation Primers | ||
| Genes | 5′ primer | 3′ primer |
| Bdnf9a −171 to +32 | CATGAGACCGGGCAAGTC | CCTTGGGAGGAATGTGTGAT |
GADD45b immunolabeling
Protein levels of GADD45b were determined by the gold-immunolabeling histochemical procedure as described previously (Pandey et al., 2004). Mice were anesthetized, perfused intracardially with normal saline (40 ml), and then 100 ml of 4% ice-cold paraformaldehyde fixative. Brains were removed and placed in fixative for 20 hr at 4°C. After fixation, brains were incubated in 10%, 20% then 30% sucrose (prepared in 0.1 M phosphate buffer, pH 7.4). Twenty micron sections were used for gold immunolabelling. Anti-GADD45b antibody (Aviva Systems Biology; San Diego, CA) in a 1:300 dilution in 1% BSA in 0.01 M PBS was incubated overnight at room temperature followed by incubation with conjugated anti-rabbit secondary antibody (1:200 dilution in 1% BSA in 0.01 M PBS) for 1 hr at room temperature. The quantification of gold-immunolabeled protein was performed using an Image Analysis System connected to a light microscope that calculated the staining intensity based on the number of immunogold particles per 100 μm2 area of a defined brain structure at high magnification (100×). Immunogold particles in the defined brain structures of three adjacent brain sections of each mouse were counted then averaged for each mouse.
In situ RT-PCR for Bdnf9a mRNA measurements
Mouse brain sections were used to determine mRNA levels using in situ RT-PCR, as reported previously (Pandey et al., 2004). Floating brain sections (40 μm thickness) were treated with proteinase K (1 μg/ml in 1× PBS containing 0.05% Triton X-100) for 15 min at 37°C and then subjected to DNase digestion. After the sections were washed with 1× PBS, they were transferred to PCR tubes containing 100 μl of PCR mixture (Applied Biosystems, Foster city, CA) and reverse transcribed for 1 hr at 42°C with reverse transcriptase in the presence of oligo-dT. In the negative sections, reverse transcriptase enzyme was not added. The PCR was performed with Taq DNA polymerase enzyme and 36–38 pmol of each Bdnf9a primers (primers 5′-GCAGCTGGAGTGGATCAGTAA-3′ and 5′-TGGTCATCACTCTTCTCACCTG-3′) and 1 mM each of NTP and dTTP, except that the dTTP was replaced by digoxigenin (DIG)-11-dUTP PCR conditions for Bdnf9a: 95°C for 5 min; 95°C for 30 sec; 60°C for 30 sec; 72°C for 30 sec (total of 28 cycles), and then 72°C for 7 min]. After PCR, sections were mounted on slides, and Bdnf9a-positive cell bodies were detected using alkaline phosphatase-conjugated anti-DIG Ab and subsequent staining of the complex with the specific substrate nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indoyl-phosphate (Roche Molecular Biochemical; Mannheim, Germany). The optical density (OD) of Bdnf9a positive cell bodies was calculated with an Image analyzer (Loats Associates; Westminster, MD). The OD of the negative brain sections was subtracted from the positive brain sections. The OD of positive cell bodies in the defined brain structures of three adjacent brain sections of each mouse was calculated, and values were averaged for each mouse. The results are represented as the mean OD per 100 pixel area.
Methylated DNA Immunoprecipitation (MeDIP) or hydroxymethylated MeDIP (hMeDIP)
MeDIP and hMeDIP were performed as previously published by our group (Gavin et al., 2015; Gavin et al., 2012). Tissue samples were sonicated to 200–500 bp segments in SDS lysis buffer. DNA was extracted using proteinase K 37°C overnight followed by phenol-chloroform. Two micrograms of DNA were resuspended in 250μL of ChIP dilution buffer, and incubated with 5 μL of 5-methylcytosine (5MC) monoclonal mouse (Diagenode; Denville, NJ) or 2 μL of rabbit polyclonal 5-hydroxymethylcytosine (Active Motif; Carlsbad, CA) antibodies overnight on a rotator at 4°C. Antibody-DNA complexes were then precipitated using Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology; Dallas, TX) for 3 hrs on a rotator at 4°C. Following low salt, high salt, LiCl, and TE washes DNA was eluted from beads using elution buffer at 67°C for 2 hrs. DNA was then extracted using proteinase K 37°C overnight followed by phenol-chloroform. Samples were resuspended in nuclease-free water and measured via qRT-PCR relative to input DNA. IP samples were normalized to input. Values presented are calculated as percent input.
Chromatin Immunoprecipitation (ChIP)
ChIP experiments were performed as previously published by our group (Gavin et al., 2009). Cross-linking was performed by adding a final concentration of 1.5% formaldehyde to tissue homogenized in RPMI media (Life Technologies; Carlsbad, CA) for 15 minutes. Cross-linking was quenched using a final concentration of 130mM of glycine. Cells were washed with PBS in the presence of protease inhibitors then sonicated to 200–500 bp segments in SDS lysis buffer. Sonicated chromatin was resuspended at 1:10 in ChIP dilution buffer. IP samples were pre-cleared with 20 μL Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology; Dallas, TX) for 2 hrs on a rotator at 4°C. Subsequently, samples were incubated overnight with rabbit polyclonal anti-H3K9,K14ac antibody (EMD Millipore; Kankakee, IL) on a rotator at 4°C. Antibody-DNA complexes were precipitated using Protein A/G PLUS-Agarose beads (Santa Cruz Biotechnology; Dallas, TX) for 3 hrs on a rotator at 4°C. Following low salt, high salt, lithium chloride, and TE washes complexes were eluted from beads at 67°C for 2 hrs using elution buffer then IP and input (33% of IP by volume) samples were uncrosslinked using a final concentration of 0.2 M NaCl. DNA was then extracted using proteinase K at 37 °C overnight followed by phenol-chloroform extraction and resuspended in water. Samples were measured using qPCR. IP samples were normalized to input. Values presented are calculated as percent input.
Statistics
Data were analyzed with Student’s t-test or one-way analysis of variance (ANOVA) with post hoc Bonferroni tests as appropriate. Ethanol consumption data (both two-bottle choice and DID) were analyzed using two-way repeated measures (RM) ANOVA for genotype and duration of administration for DID or genotype and time for two-bottle free choice. Post hoc comparisons for two-way ANOVA were performed using Bonferroni. All statistical analyses were performed using GraphPad Prism (version 15.0 for Windows) (Graphpad; La Jolla, CA). All statistical tests were two sided.
Results
C57 mice have lower GADD45b mRNA and protein expression in the nucleus accumbens (NAc) compared to the DBA strain
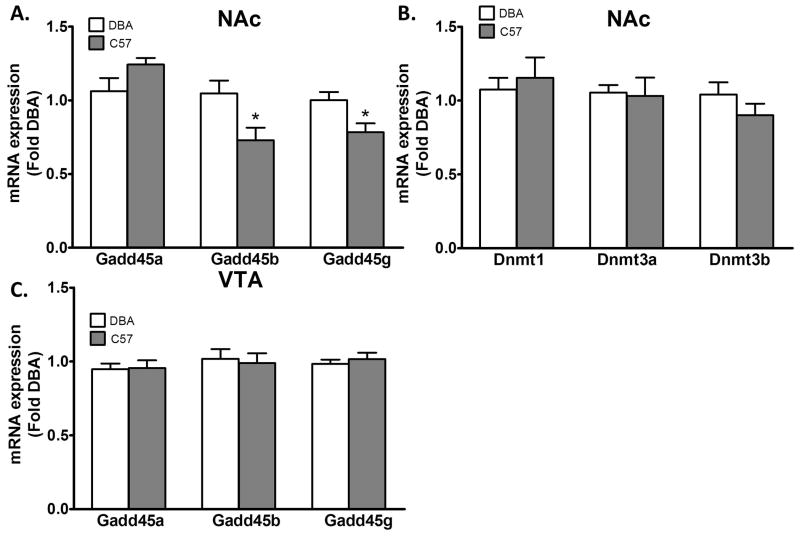
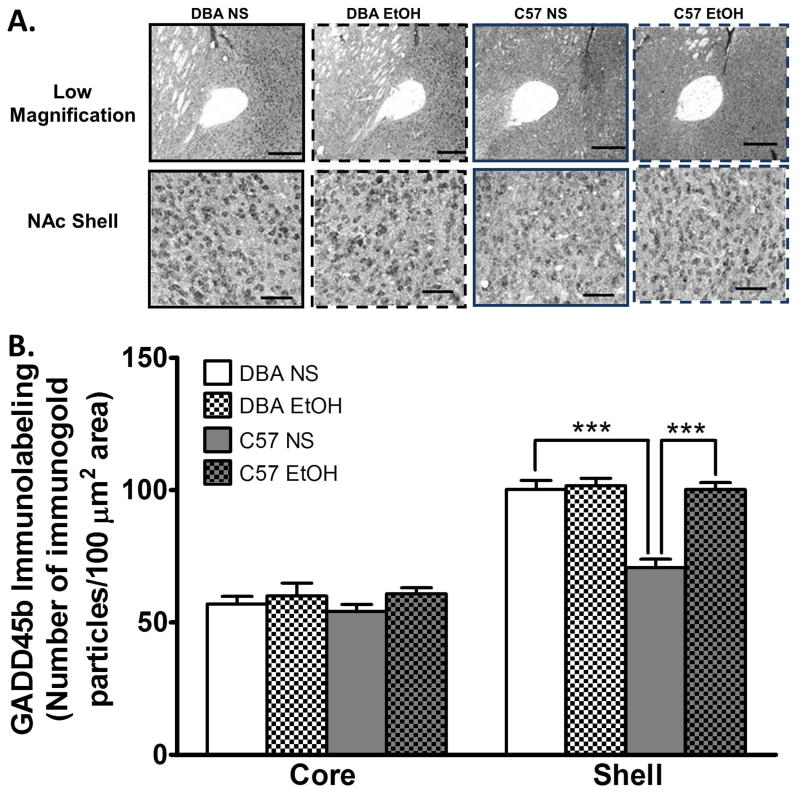
We compared mRNA expression encoding key components of DNA demethylation, the Gadd45 genes (Gadd45a, b, and g). We found that relative to DBA mice C57 mice had lower mRNA expression of Gadd45b (t16 = 2.618, p = 0.02) and Gadd45g (t16 = 2.672, p = 0.02) in the NAc (Figure 1A). By comparison, a similar difference was not found in the ventral tegmental area (VTA) (Figure 1C). We did not find a difference in the mRNA expression levels of the DNA methylating enzymes, Dnmt1, 3a, and 3b in the entire NAc (Figure 1B). The lower level of NAc Gadd45b mRNA expression we found translated to lower GADD45b protein levels in the NAc shell (ANOVA F (3,19)= 24.30, Bonferroni post hoc test, P < 0.0001), but not NAc core of normal saline treated mice. Moreover, an acute i.p. injection of ethanol 2 g/kg increased GADD45b protein expression in the NAc shell of C57 mice, but did not affect expression in DBA mice relative to saline treated controls (Figure 2). This was despite equivalent blood alcohol concentrations achieved in the two strains (C57 Mean±SEM 179.2±6.2 mg/dl vs. DBA Mean±SEM 193.8±9.1 mg/dl, n = 12 per group).
Figure 1. Baseline differences in Gadd45 gene expression in the entire nucleus accumbens (NAc) in C57BL/6J (C57) mice compared to DBA/2J (DBA).
A. Gadd45b and Gadd45g mRNA expression is lower in the NAc of C57 mice (n = 9) compared to the DBA strain (n = 9). B. We find no significant differences in Dnmt1, 3a, or 3b mRNA expression in the NAc in these strains. C. We also find no differences in Gadd45 gene expression in the VTA in C57 and DBA mice. Values are mean±SEMs. *p<0.05.
Figure 2. Lower GADD45b protein expression in the C57BL/6J (C57) nucleus accumbens (NAc) shell compared to DBA/2J (DBA) mice is reversed by acute ethanol.
A. Representative photomicrographs at low magnification(scale bar = 200 μm) and higher magnification (scale bar = 50 μm) of GADD45b gold-immunolabeling in the core and shell of the NAc of C57 and DBA mice treated with normal saline (NS) or ethanol 2 g/kg after 1 h (EtOH). B. GADD45b protein expression is lower in the NAc shell of C57 mice compared to DBA in NS treated mice. EtOH increased GADD45b in C57 mice (n = 5 per group). Values are mean±SEMs. ***p<0.001.
Lower Bdnf9a expression in the NAc shell of C57 mice relative to DBA
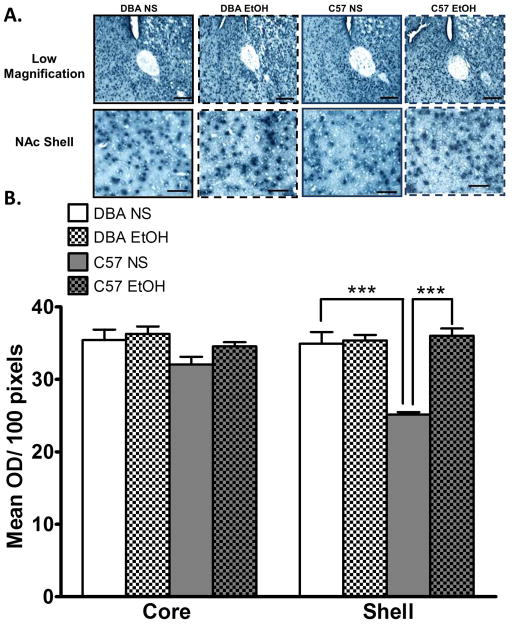
Considering the regionally lower levels in GADD45b protein expression we examined mRNA expression of a known target of GADD45b, Bdnf9a. We found significantly lower Bdnf9a mRNA expression in the NAc shell but not core of normal-saline treated C57 mice relative to DBA (ANOVA F(3,19)= 23.04, Bonferroni post hoc test, P < 0.0001). This difference between the strains was reduced by acute ethanol treatment (Figure 3).
Figure 3. Lower Bdnf9a mRNA expression in the C57BL/6J (C57) nucleus accumbens (NAc) shell compared to DBA/2J (DBA) mice is reversed by acute ethanol.
A. Representative photomicrographs at low magnification (scale bar = 200 μm) and higher magnification (scale bar = 50 μm) of Bdnf9a mRNA in the core and shell of the NAc of C57 and DBA mice treated with normal saline (NS) or ethanol 2 g/kg after 1 h (EtOH). B. Bdnf9a mRNA expression is lower in the NAc shell of C57 mice compared to DBA in NS treated mice. EtOH increased Bdnf9a in C57 mice (n = 5 per group). Values are mean±SEMs. ***p<0.001.
Epigenetic differences in the NAc of C57 vs. DBA mice at Bdnf9a
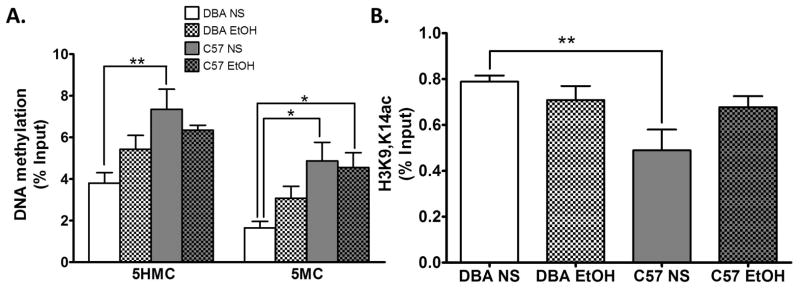
GADD45b has been shown to be involved in demethylating the Bdnf9a promoter (Ma et al., 2009; Gavin et al., 2015). We measured 5-hydroxymethylcytosine (5HMC) and 5-methylcytosine (5MC) at the Bdnf9a promoter in C57 and DBA mice. We found a significant increase in both 5HMC (ANOVA F(3,28) = 4.660, Bonferroni post hoc test, P = 0.01) and 5MC (ANOVA F(3,30) = 4.628, Bonferroni post hoc test, P = 0.01) in normal saline treated C57 mice relative to DBA. Moreover, acute ethanol treatment reduced the 5HMC difference between the strains. On the other hand, 5MC remained significantly elevated in the C57 strain relative to DBA following ethanol treatment (Figure 4A).
Figure 4. Increased 5-hydroxymethylcytosine (5HMC) and 5-methylcytosine (5MC) and lower histone H3 acetylation (H3K9,14ac) in the C57BL/6J (C57) entire nucleus accumbens (NAc) compared to DBA/2J (DBA) mice is reversed by acute ethanol.
A. We measured 5HMC and 5MC at the Bdnf9a promoter via hydroxymethylayted (hMeDIP) and methylated DNA immunoprecipitation (MeDIP). We find that C57 mice have increased levels of 5HMC and 5MC at the Bdnf9a promoter in the NAc compared to DBA mice. One hour after an i.p. administration of ethanol 2 g/kg (EtOH) the 5HMC difference between C57 and DBA mice is no longer significant (n = 6–8 per group). B. H3K9,14ac was measured at the Bdnf9a promoter via chromatin immunoprecipitation (ChIP). We find that C57 mice have lower levels of H3K9,14ac at the Bdnf9a promoter in the NAc compared to DBA mice. One hour after an i.p. administration of EtOH 2 g/kg H3K9,14ac levels no longer differ between C57 and DBA mice (n = 7–8 per group). Values are mean±SEMs. *p<0.05, **p<0.01.
In addition, to measuring DNA methylation we measured histone H3 acetylation (H3K9,14ac). This was in part based on a previous publication from our group in which it was noted that C57 mice have lower levels of CREB protein and its activated form, phosphorylated CREB (pCREB), which is involved in histone acetylation in the NAc, compared to DBA mice (Misra and Pandey, 2003). In accord with the lower CREB and pCREB levels we found lower H3K9,14ac at the Bdnf9a promoter in normal saline treated C57 mice relative to the DBA strain which was no longer significantly different from DBA following acute ethanol (ANOVA F(3,30) = 4.286, Bonferroni post hoc test, P = 0.01) (Figure 4B).
Baseline Gadd45 mRNA expression differences in Gadd45b deficient mice
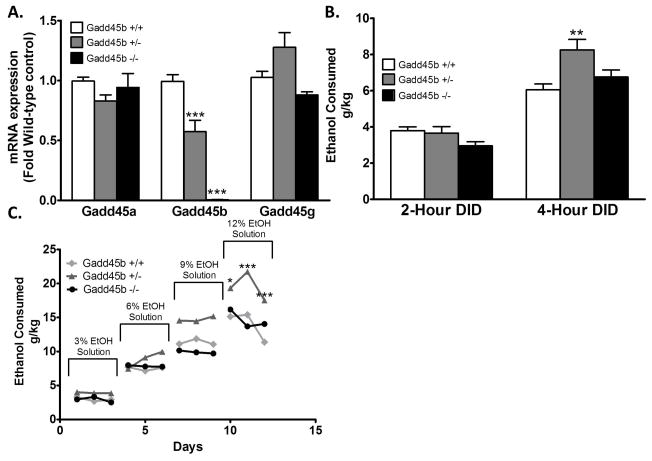
We measured Gadd45 mRNA transcripts in the entire NAc of Gadd45b +/+, Gadd45b +/−, and Gadd45b −/−. We found no change in Gadd45a or Gadd45g in Gadd45b +/− mice relative to Gadd45b+/+ at baseline (ANOVA F(2,15) = 7.793, Bonferroni post hoc test, P = 0.01). Predictably, Gadd45b was reduced in Gadd45b +/− and Gadd45 −/− mice relative wild-type control (ANOVA F(2,15) = 63.42, Bonferroni post hoc test, P < 0.0001) (Figure 5A).
Figure 5. Increased binge-like and 2-bottle free-choice ethanol consumption in Gadd45b haploinsufficient (+/−) mice.
A. In the entire NAc there is no significant difference in Gadd45a or Gadd45g expression between wild-type (Gadd45b +/+) and haploinsufficient (Gadd45b +/−) or complete knockout (Gadd45b −/−) mice. Predictably, Gadd45b mRNA expression is lower in Gadd45b +/− mice and absent in Gadd45b −/− mice (n = 5–6). B. Mean ethanol consumption in g/kg during first three days of 2-h drinking-in-the-dark (DID) sessions and last two days of 4-h DID sessions. Gadd45b +/− mice (n = 10) drink significantly more ethanol during the 4-day sessions than wild-type (+/+) (n = 11) or knockout mice (−/−) (n = 9). C. In a two-bottle free-choice paradigm Gadd45b +/− mice consume more ethanol than either Gadd45b +/+ or Gadd45b −/− at 12% ethanol (n = 8 per group). Values are mean±SEMs. *p<0.05, **p<0.01; ***p<0.001.
GADD45b deficiency alters ethanol drinking behaviors
In order to clarify the role of GADD45b in ethanol drinking behaviors we assessed ethanol intake using the drinking in the dark (DID) paradigm and 2-bottle free choice paradigms of Gadd45b haploinsufficient (Gadd45b +/−) and complete knockout animals (Gadd45b −/−), which had been backcrossed into a C57 genetic background.
In the DID paradigm, during the 4 hr drinking sessions on days 4 and 5, Gadd45b +/−, but not Gadd45b −/− mice, drank significantly more ethanol than Gadd45b +/+ controls (two-way RM ANOVA, effect of genotype: F2,52 = 3.367, P = 0.04; duration: F1,52 = 111.5, P < 0.0001; interaction: F2,27 = 4.191, P = 0.02) (Figure 5B).
In the 2-bottle free choice paradigm mice were provided with two water bottles for two weeks. Subsequently, we provided mice with one bottle of 3% ethanol and a bottle of water for 3 days, then 6% ethanol for the next 3 days, 9% for 3 days, and finally 12% ethanol. The locations of the water and ethanol bottles were switched daily. Similar to the DID paradigm we found an increase in ethanol consumption in the Gadd45b +/− mice relative to the Gadd45b+/+ wild-type control on the 12% ethanol concentration days, which was not found in the total knockout Gadd45b −/− (two-way RM ANOVA, effect of genotype: F2,252 = 31.81, P < 0.0001; time: F11,252 = 136.4, P < 0.0001; interaction, F22,252 = 1.929, P = 0.009) (Figure 5C). No significant differences were found in water consumption during the 12% ethanol consuming days (Table 2).
Table 2.
| Gadd45b +/+ (n=8) | Gadd45b +/− (n=8) | Gadd45b −/− (n=8) | Sig. | |
|---|---|---|---|---|
| Weight (kg) | 0.029±0.001 | 0.028±0.001 | 0.028±0.001 | NS |
| Total Fluid Intake (mL/kg) | 179.0±8.67 | 215.9±11.6 | 179.4±10.9 | NS |
| Water Intake (mL/kg) | 58.0±10.0 | 50.5±9.5 | 54.9±10.4 | NS |
| Percent Water Intake | 31.8±4.5 | 23.1±4.6 | 29.4±5.6 | NS |
| Percent Ethanol Intake | 68.2±4.5 | 76.9±4.6 | 70.6±5.6 | NS |
Discussion
In the current study we used the high-alcohol consuming C57BL/6J (C57) and low consuming DBA/2J (DBA) mouse strains to study potential pathways leading to increased ethanol consumption based on numerous reports documenting the higher ethanol consumption of C57 mice compared to DBA in a variety of drinking paradigms (Yoneyama et al., 2008; Belknap et al., 1993; Rhodes et al., 2005). We found lower Gadd45b and Gadd45g mRNA expression in the entire NAc and GADD45b protein in the NAc shell of C57 mice compared to DBA, but no differences in Dnmt expression and no VTA Gadd45 gene expression differences. As perhaps a downstream consequence of lower NAc GADD45b and g we also found lower Bdnf9a mRNA expression in the NAc shell. GADD45b has been documented to play an important role in DNA demethylation at the Bdnf9a promoter, thereby having a positive modulatory function (Gavin et al., 2012; Ma et al., 2009; Gavin et al., 2015). In agreement with this reported role we found higher 5HMC and 5MC at the Bdnf9a promoter in the NAc of C57 mice as compared with DBA. In conjunction with higher 5HMC and 5MC at the Bdnf9a promoter there was lower H3K9,14ac in C57 vs. DBA mice.
The role of ethanol as an HDAC inhibitor has been well-documented. Acute ethanol has been reported to increase histone acetylation in the brain likely in part through its HDAC inhibitory activity, including in the NAc (Pandey et al., 2008a; Sakharkar et al., 2012; Botia et al., 2012). We also found that acute ethanol injection increased NAc histone acetylation and reduced 5HMC at the Bdnf gene in the NAc of C57 mice without producing any effects in DBA mice. We recently reported in primary neuron cultures 5HMC is highly responsive to exogenous stimuli (Gavin et al., 2015). Further, the ability of an HDAC inhibitor, in this case acute ethanol, to induce DNA demethylation has been previously documented (Detich et al., 2003; Dong et al., 2007).
Over the last two decades studies have implicated BDNF in the survival and function of adult dopamine neurons, learning, memory, synaptic plasticity and regulation of dendritic morphology (Russo et al., 2009). In humans, BDNF polymorphisms have been associated with vulnerability to alcohol dependence and relapse following treatment. Decreased plasma BDNF levels have been observed in alcoholics, particularly those with a family history of alcoholism (Joe et al., 2007; Zanardini et al., 2011). Additionally, gene microarrays in humans indicate BDNF as an ethanol-responsive gene (Davis, 2008). In rodent models, microarray data indicate differential effects of acute ethanol on Bdnf signaling in the NAc between C57 and DBA strains (Kerns et al., 2005). Further, i.p. injection or self-administration of ethanol increases BDNF expression (Jeanblanc et al., 2009; Pandey et al., 2008b). Moreover, Bdnf haploinsufficient mice (Bdnf+/−) exhibit a preference for voluntary ethanol consumption (Jeanblanc et al., 2009), and alcohol preferring rats have innately lower levels of BDNF in the bed nucleus of the stria terminalis and in the central extended and medial amygdala as compared with alcohol non-preferring rats (Prakash et al., 2008). On the other hand, BDNF infusion decreases ethanol intake (Jeanblanc et al., 2009; Pandey et al., 2006). Even among C57 mice, BDNF expression in the NAc is inversely correlated with alcohol-drinking behavior (Wolstenholme et al., 2011). Taken together, these data indicate the importance of BDNF in alcohol use disorders as a potential contributor through both the reward and anxiety circuity.
Considering the lower GADD45b protein levels in the higher ethanol consuming C57 strain, we examined drinking behavior in Gadd45b +/− and Gadd45b −/− mice compared to wild-type control to directly implicate GADD45b in alcohol drinking behaviors. Interestingly, Gadd45b +/− mice but not Gadd45b −/− consumed more ethanol in both DID and 2-bottle free choice paradigms compared to wild-type controls. Although it is true that some genetic differences are additive (i.e., the null mutant is more different from wild-type control than the heterozygote) other genetic differences produce phenotypes in which there is dominance or overdominance. Overdominance (the haploinsufficient genotype showing a more robust phenotype than either the null mutant or wild-type) is commonly found in the alcohol literature. On a multi-genic level, F1 hybrid mouse strains produced from breeding inbred strains often display increased drinking behavior compared to either parent strain (Blednov et al., 2010). Gene specific examples exist as well. Transcription factor Lmo3 null mice have been reported to be no different than their wild-type littermates, while Lmo3 partial knockdowns drink more (Savarese et al., 2014; Lasek et al., 2011). Similarly, dopamine transporter, GABA transporter, and synaptic vesicle protein Rab3A haploinsufficient mice voluntarily consume more ethanol compared to their wild-type and homozygous null littermates (Savelieva et al., 2002; Cai et al., 2006; Kapfhamer et al., 2008). Further, all three Gadd45 proteins share partial sequence homology and Gadd45a, like Gadd45b, has also been shown to be involved in DNA demethylation (Gavin et al., 2012; Jung et al., 2007; Barreto et al., 2007). Therefore, it is possible that there is redundancy in aspects of their functions. Nonetheless, these data along with the findings of lower expression of Gadd45g and b in the NAc of C57 mice suggests the involvement of Gadd45b in the regulation of alcohol drinking behaviors.
Gadd45b −/− mice have been shown to have deficits in dendritic arborization and neurogenesis following electroconvulsive shock (Ma et al., 2009). Behaviorally, Gadd45b −/− mice have been reported to have both deficits and enhanced hippocampus dependent learning, and no differences in amygdala-dependent learning or increased measures of anxiety (Leach et al., 2012; Sultan et al., 2012). The current study is the first to our knowledge to examine Gadd45b in the reward pathway and to demonstrate its potential role in alcohol addiction.
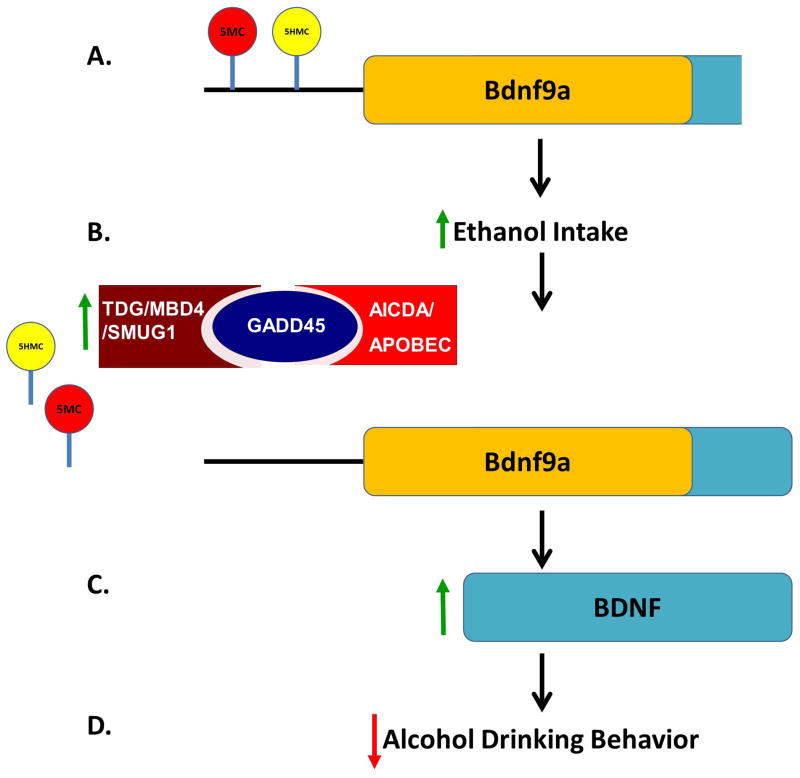
Several studies indicate that ethanol treatment alters DNA methylation homeostasis. In cultured astrocytes prolonged ethanol treatment reduces DNMT activity, while intermittent alcohol treatment of mice increases NAc DNMT1 protein levels (Warnault et al., 2013; Zhang et al., 2014). In human alcoholics DNMT1 expression is reduced in the basolateral and central nuclei of the amygdala and the cortex (Ponomarev et al., 2012). Our own data indicate increased NAc DNA methylation at the Bdnf9a gene promoter and lower levels of a DNA demethylating protein, GADD45b, predisposed to higher ethanol drinking behavior. Taken together it appears that an imbalance of DNA methylating and demethylating factors, favoring increased DNA methylation, may predispose to increased alcohol drinking behavior, while alcohol reverses this imbalance (Figure 6).
Figure 6. Hypothetical pathway of ethanol-induced Bdnf induction through DNA demethylation.
A. The Bdnf9a gene promoter is both methylated (5MC) and hydroxymethylated (5HMC) leading to less expression. A deficiency in BDNF may contribute to increased alcohol consumption. B. Acute alcohol induces the DNA demethylation machinery including GADD45b which leads to the removal of 5HMC and 5MC. C. The removal of methylated cytosines allows for increased Bdnf9a mRNA and consequently peptide expression. D. Increased BDNF possibly reduces further ethanol drinking.
Here, we demonstrate that a deficiency in GADD45b in the NAc altered the epigenetic milieu at the Bdnf9a promoter, thereby affecting Bdnf expression. Lower GADD45b levels corresponded to increased alcohol drinking behavior whether measured by DID or 2-bottle free choice paradigm. We reported earlier that epigenetic regulation of Bdnf expression in the amygdala regulates alcohol drinking behaviors (Moonat et al., 2013). Based on the results of present studies, we hypothesize that an epigenetically-mediated reduction in Bdnf predispose to higher ethanol consumption. This study implicates DNA methylation dynamics in the NAc as important variables leading to increased alcohol consumption.
Acknowledgments
This work was supported by the Department of Veterans Affairs [Merit Review Grant Career Development Award (CDA-2) (IK2BX001650) to (D.P.G.) and (Merit Review Grant (I01BX000143); Senior Research Career Scientist award) to (S.C.P.)], NARSAD Young Investigator Award donation from The Family of Joseph M. Evans (D.P.G.), National Institute on Alcohol Abuse and Alcoholism Grants (AA-010005; AA-0212662) (S.C.P), and Center for Alcohol Research in Epigenetics (AA022538) (S.C.P).
Footnotes
The authors have no conflicts of interest to disclose.
References
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Belknap ND, Berg JH, Coleman R. Preabsorptive vs. postabsorptive control of ethanol intake in C57BL/6J and DBA/2J mice. Behav Genet. 1977;7:413–425. doi: 10.1007/BF01066776. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Crabbe JC, Young ER. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology (Berl) 1993;112:503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- Blednov YA, Ozburn AR, Walker D, Ahmed S, Belknap JK, Harris RA. Hybrid mice as genetic models of high alcohol consumption. Behav Genet. 2010;40:93–110. doi: 10.1007/s10519-009-9298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botia B, Legastelois R, Alaux-Cantin S, Naassila M. Expression of ethanol-induced behavioral sensitization is associated with alteration of chromatin remodeling in mice. PLoS One. 2012;7:e47527. doi: 10.1371/journal.pone.0047527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai YQ, Cai GQ, Liu GX, Cai Q, Shi JH, Shi J, Ma SK, Sun X, Sheng ZJ, Mei ZT, Cui D, Guo L, Wang Z, Fei J. Mice with genetically altered GABA transporter subtype I (GAT1) expression show altered behavioral responses to ethanol. J Neurosci Res. 2006;84:255–267. doi: 10.1002/jnr.20884. [DOI] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MI. Ethanol-BDNF interactions: still more questions than answers. Pharmacol Ther. 2008;118:36–57. doi: 10.1016/j.pharmthera.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detich N, Bovenzi V, Szyf M. Valproate induces replication-independent active DNA demethylation. J Biol Chem. 2003;278:27586–27592. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- Dong E, Guidotti A, Grayson DR, Costa E. Histone hyperacetylation induces demethylation of reelin and 67-kDa glutamic acid decarboxylase promoters. Proc Natl Acad Sci U S A. 2007;104:4676–4681. doi: 10.1073/pnas.0700529104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Floreani C. Epigenetics of schizophrenia: an open and shut case. Int Rev Neurobiol. 2014;115:155–201. doi: 10.1016/B978-0-12-801311-3.00005-6. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Kartan S, Chase K, Jayaraman S, Sharma RP. Histone deacetylase inhibitors and candidate gene expression: An in vivo and in vitro approach to studying chromatin remodeling in a clinical population. J Psychiatr Res. 2009;43:870–876. doi: 10.1016/j.jpsychires.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Gavin DP, Kusumo H, Sharma RP, Guizzetti M, Guidotti A, Pandey SC. Gadd45b and N-methyl-d-aspartate induced DNA demethylation in postmitotic neurons. Epigenomics. 2015;7:567–579. doi: 10.2217/epi.15.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth Arrest and DNA-Damage-Inducible, Beta (GADD45b)-Mediated DNA Demethylation in Major Psychosis. Neuropsychopharmacology. 2012;37:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Gavin DP, Veldic M, Zhao W, Bhaumik DK, Pandey SC, Grayson DR. DNA Methylation/Demethylation Network Expression in Psychotic Patients with a History of Alcohol Abuse. Alcohol Clin Exp Res. 2012 doi: 10.1111/j.1530-0277.2012.01947.x. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M, Gupta SK, Balliet AG, Hollander MC, Fornace AJ, Hoffman B, Liebermann DA. Hematopoietic cells from Gadd45a- and Gadd45b-deficient mice are sensitized to genotoxic-stress-induced apoptosis. Oncogene. 2005;24:7170–7179. doi: 10.1038/sj.onc.1208847. [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensler JG, Ladenheim EE, Lyons WE. Ethanol consumption and serotonin-1A (5-HT1A) receptor function in heterozygous BDNF (+/−) mice. J Neurochem. 2003;85:1139–1147. doi: 10.1046/j.1471-4159.2003.01748.x. [DOI] [PubMed] [Google Scholar]
- Huang MC, Chen CH, Liu HC, Chen CC, Ho CC, Leu SJ. Differential patterns of serum brain-derived neurotrophic factor levels in alcoholic patients with and without delirium tremens during acute withdrawal. Alcohol Clin Exp Res. 2011;35:126–131. doi: 10.1111/j.1530-0277.2010.01329.x. [DOI] [PubMed] [Google Scholar]
- Jeanblanc J, He DY, Carnicella S, Kharazia V, Janak PH, Ron D. Endogenous BDNF in the dorsolateral striatum gates alcohol drinking. J Neurosci. 2009;29:13494–13502. doi: 10.1523/JNEUROSCI.2243-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanblanc J, He DY, McGough NN, Logrip ML, Phamluong K, Janak PH, Ron D. The dopamine D3 receptor is part of a homeostatic pathway regulating ethanol consumption. J Neurosci. 2006;26:1457–1464. doi: 10.1523/JNEUROSCI.3786-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe KH, Kim YK, Kim TS, Roh SW, Choi SW, Kim YB, Lee HJ, Kim DJ. Decreased plasma brain-derived neurotrophic factor levels in patients with alcohol dependence. Alcohol Clin Exp Res. 2007;31:1833–1838. doi: 10.1111/j.1530-0277.2007.00507.x. [DOI] [PubMed] [Google Scholar]
- Jung HJ, Kim EH, Mun JY, Park S, Smith ML, Han SS, Seo YR. Base excision DNA repair defect in Gadd45a-deficient cells. Oncogene. 2007;26:7517–7525. doi: 10.1038/sj.onc.1210557. [DOI] [PubMed] [Google Scholar]
- Kapfhamer D, Bettinger JC, Davies AG, Eastman CL, Smail EA, Heberlein U, McIntire SL. Loss of RAB-3/A in Caenorhabditis elegans and the mouse affects behavioral response to ethanol. Genes Brain Behav. 2008;7:669–676. doi: 10.1111/j.1601-183X.2008.00404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde B, Gabel HW, Gilbert CS, Griffith EC, Greenberg ME. Reading the unique DNA methylation landscape of the brain: Non-CpG methylation, hydroxymethylation, and MeCP2. Proc Natl Acad Sci U S A. 2015;112:6800–6806. doi: 10.1073/pnas.1411269112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan HR, Sakharkar AJ, Teppen TL, Berkel TD, Pandey SC. The epigenetic landscape of alcoholism. Int Rev Neurobiol. 2014;115:75–116. doi: 10.1016/B978-0-12-801311-3.00003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasek AW, Giorgetti F, Berger KH, Tayor S, Heberlein U. Lmo genes regulate behavioral responses to ethanol in Drosophila melanogaster and the mouse. Alcohol Clin Exp Res. 2011;35:1600–1606. doi: 10.1111/j.1530-0277.2011.01506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach PT, Poplawski SG, Kenney JW, Hoffman B, Liebermann DA, Abel T, Gould TJ. Gadd45b knockout mice exhibit selective deficits in hippocampus-dependent long-term memory. Learn Mem. 2012;19:319–324. doi: 10.1101/lm.024984.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- McGough NN, He DY, Logrip ML, Jeanblanc J, Phamluong K, Luong K, Kharazia V, Janak PH, Ron D. RACK1 and brain-derived neurotrophic factor: a homeostatic pathway that regulates alcohol addiction. J Neurosci. 2004;24:10542–10552. doi: 10.1523/JNEUROSCI.3714-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra K, Pandey SC. Differences in basal levels of CREB and NPY in nucleus accumbens regions between C57BL/6 and DBA/2 mice differing in inborn alcohol drinking behavior. J Neurosci Res. 2003;74:967–975. doi: 10.1002/jnr.10831. [DOI] [PubMed] [Google Scholar]
- Moonat S, Sakharkar AJ, Zhang H, Pandey SC. The role of amygdaloid brain-derived neurotrophic factor, activity-regulated cytoskeleton-associated protein and dendritic spines in anxiety and alcoholism. Addict Biol. 2011;16:238–250. doi: 10.1111/j.1369-1600.2010.00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonat S, Sakharkar AJ, Zhang H, Tang L, Pandey SC. Aberrant Histone Deacetylase2-Mediated Histone Modifications and Synaptic Plasticity in the Amygdala Predisposes to Anxiety and Alcoholism. Biol Psychiatry. 2013 doi: 10.1016/j.biopsych.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Roy A, Zhang H, Xu T. Partial deletion of the cAMP response element-binding protein gene promotes alcohol-drinking behaviors. J Neurosci. 2004;24:5022–5030. doi: 10.1523/JNEUROSCI.5557-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008a;28:3729–3737. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Roy A, Misra K. Central and medial amygdaloid brain-derived neurotrophic factor signaling plays a critical role in alcohol-drinking and anxiety-like behaviors. J Neurosci. 2006;26:8320–8331. doi: 10.1523/JNEUROSCI.4988-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SC, Zhang H, Ugale R, Prakash A, Xu T, Misra K. Effector immediate-early gene arc in the amygdala plays a critical role in alcoholism. J Neurosci. 2008b;28:2589–2600. doi: 10.1523/JNEUROSCI.4752-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash A, Zhang H, Pandey SC. Innate differences in the expression of brain-derived neurotrophic factor in the regions within the extended amygdala between alcohol preferring and nonpreferring rats. Alcohol Clin Exp Res. 2008;32:909–920. doi: 10.1111/j.1530-0277.2008.00650.x. [DOI] [PubMed] [Google Scholar]
- Renthal W, Nestler EJ. Chromatin regulation in drug addiction and depression. Dialogues Clin Neurosci. 2009;11:257–268. doi: 10.31887/DCNS.2009.11.3/wrenthal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JS, Best K, Belknap JK, Finn DA, Crabbe JC. Evaluation of a simple model of ethanol drinking to intoxication in C57BL/6J mice. Physiol Behav. 2005;84:53–63. doi: 10.1016/j.physbeh.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Russo SJ, Mazei-Robison MS, Ables JL, Nestler EJ. Neurotrophic factors and structural plasticity in addiction. Neuropharmacology. 2009;56(Suppl 1):73–82. doi: 10.1016/j.neuropharm.2008.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar AJ, Zhang H, Tang L, Shi G, Pandey SC. Histone deacetylases (HDAC)-induced histone modifications in the amygdala: a role in rapid tolerance to the anxiolytic effects of ethanol. Alcohol Clin Exp Res. 2012;36:61–71. doi: 10.1111/j.1530-0277.2011.01581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarese A, Zou ME, Kharazia V, Maiya R, Lasek AW. Increased behavioral responses to ethanol in Lmo3 knockout mice. Genes Brain Behav. 2014;13:777–783. doi: 10.1111/gbb.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelieva KV, Caudle WM, Findlay GS, Caron MG, Miller GW. Decreased ethanol preference and consumption in dopamine transporter female knock-out mice. Alcohol Clin Exp Res. 2002;26:758–764. [PubMed] [Google Scholar]
- Shelton KL, Grant KA. Discriminative stimulus effects of ethanol in C57BL/6J and DBA/2J inbred mice. Alcohol Clin Exp Res. 2002;26:747–757. [PubMed] [Google Scholar]
- Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD. Genetic Deletion of gadd45b, a Regulator of Active DNA demethylation, Enhances Long-Term Memory and Synaptic Plasticity. J Neurosci. 2012;32:17059–17066. doi: 10.1523/JNEUROSCI.1747-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. A dynamic methylome; implications of non-CG methylation/demethylation. Cell Cycle. 2010;9:3846–3847. doi: 10.4161/cc.9.19.13563. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Cubero I, van Dijk G, Mediavilla C, Bernstein IL. Ethanol-induced c-fos expression in catecholamine- and neuropeptide Y-producing neurons in rat brainstem. Alcohol Clin Exp Res. 2000;24:802–809. [PubMed] [Google Scholar]
- Warnault V, Darcq E, Levine A, Barak S, Ron D. Chromatin remodeling - a novel strategy to control excessive alcohol drinking. Transl Psychiatry. 2013;3:e231. doi: 10.1038/tp.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme JT, Warner JA, Capparuccini MI, Archer KJ, Shelton KL, Miles MF. Genomic analysis of individual differences in ethanol drinking: evidence for non-genetic factors in C57BL/6 mice. PLoS One. 2011;6:e21100. doi: 10.1371/journal.pone.0021100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan QS, Feng MJ, Yan SE. Different expression of brain-derived neurotrophic factor in the nucleus accumbens of alcohol-preferring (P) and -nonpreferring (NP) rats. Brain Res. 2005;1035:215–218. doi: 10.1016/j.brainres.2004.12.039. [DOI] [PubMed] [Google Scholar]
- Yoneyama N, Crabbe JC, Ford MM, Murillo A, Finn DA. Voluntary ethanol consumption in 22 inbred mouse strains. Alcohol. 2008;42:149–160. doi: 10.1016/j.alcohol.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanardini R, Fontana A, Pagano R, Mazzaro E, Bergamasco F, Romagnosi G, Gennarelli M, Bocchio-Chiavetto L. Alterations of brain-derived neurotrophic factor serum levels in patients with alcohol dependence. Alcohol Clin Exp Res. 2011;35:1529–1533. doi: 10.1111/j.1530-0277.2011.01489.x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Kusumo H, Sakharkar AJ, Pandey SC, Guizzetti M. Regulation of DNA methylation by ethanol induces tissue plasminogen activator expression in astrocytes. J Neurochem. 2014;128:344–349. doi: 10.1111/jnc.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Zhu J, Lv T, Chen G, Sun H, Yang X, Huang X, Tian J. Ethanol and its metabolites induce histone lysine 9 acetylation and an alteration of the expression of heart development-related genes in cardiac progenitor cells. Cardiovasc Toxicol. 2010;10:268–274. doi: 10.1007/s12012-010-9081-z. [DOI] [PubMed] [Google Scholar]