Abstract
Objective:
To assess whether an online open-source tool would provide accurate calculations of T2* values for iron concentrations in the liver and heart compared with a standard reference software.
Methods:
An online open-source tool, written in pure HTML5/Javascript, was tested in 50 patients (age 26.0 ± 18.9 years, 46% males) who underwent T2* MRI of the liver and heart for iron overload assessment as part of their routine workup. Automated truncation correction was the default with optional manual adjustment provided if needed. The results were compared against a standard reference measurement using commercial software with manual truncation (CVI42® v. 5.1; Circle Cardiovascular Imaging; Calgary, AB).
Results:
The mean liver T2* values calculated with the automated tool was 4.3 ms [95% confidence interval (CI) 3.1 to 5.5 ms] vs 4.26 ms using the reference software (95% CI 3.1 to 5.4 ms) without any significant differences (p = 0.71). In the liver, the mean difference was 0.036 ms (95% CI −0.1609 to 0.2329 ms) with a regression correlation coefficient of 0.97. For the heart, the automated T2* value was 26.0 ms (95% CI 22.9 to 29.0 ms) vs 25.3 ms (95% CI 22.3 to 28.3 ms), p = 0.28. The mean difference was 0.72 ms (95% CI 0.08191 to 1.3621 ms) with a correlation coefficient of 0.96.
Conclusion:
The automated online tool provides similar T2* values for the liver and myocardial iron concentrations as compared with a standard reference software.
Advances in knowledge:
The online program provides an open-source tool for the calculation of T2* values, incorporating an automated correction algorithm in a simple and easy-to-use interface.
INTRODUCTION
Iron quantification using T2* MRI has significantly modified the management of diseases with chronic iron overload.1 Part of the limitation in widespread use of the technique occurs owing to restricted access to quantification software and difficulty in obtaining accurate numbers, especially in situations of severe iron overload where truncation or an offset model has to be applied.2,3 In order to facilitate the calculation of T2* values in these situations, He et al4 published an accurate algorithm for automated truncation and correction of the analysis of T2* decay curves, simplifying the method while maintaining excellent accuracy with a coefficient of variation of only 1.6%. Despite the significant results, the technique was implemented only in MATLAB® (MathWorks®, Natick, MA), limiting its access in most clinical centres worldwide. Because of this limitation, we sought to develop an online open-source tool incorporating the described algorithm in order to promote wider availability of the automated process to researchers and clinical physicians without the need for dedicated software.
METHODS AND MATERIALS
In order to provide amplified access to the automated algorithm, a tool was written in pure HTML5/Javascript, incorporating the previously established rules while running in most modern web browsers (http://www.isodense.com/ic) (Figure 1). To test the tool, we selected 50 patients who underwent T2* MRI of the liver and heart for iron overload assessment as part of their routine work-up (mean age 26.0 ± 18.9 years, 46% males, 80% with thalassaemia major as the primary haematological disorder). T2* acquisition was performed after patients completed the institutional review board-approved informed consent in a 1.5-T scanner (MAGNETOM® Aera; Siemens, Erlangen, Germany). A short-axis single-slice black-blood image of the heart in the mid-ventricular level as well as an axial slice of the liver using multiecho gradient-echo images was obtained according to previous published methods.5,6 For each image, we identified the individual echo times (TEs) and region-of-interest-based signal intensities for each organ in the scanner console itself. The operator would draw the region of interest on each T2* image and then manually insert these values in the online webpage. Automated truncation was set as the default, but the user could manually adjust the results if truncation failed (for example, in cases with fewer than three data sets). For comparison against a standard reference, the same data sets were loaded in a commercial software (CVI42® v. 5.1; Circle Cardiovascular Imaging) and regions of interest drawn as close as possible to the original drawing on the console in the T2* module of the software with manual truncation for correction.2
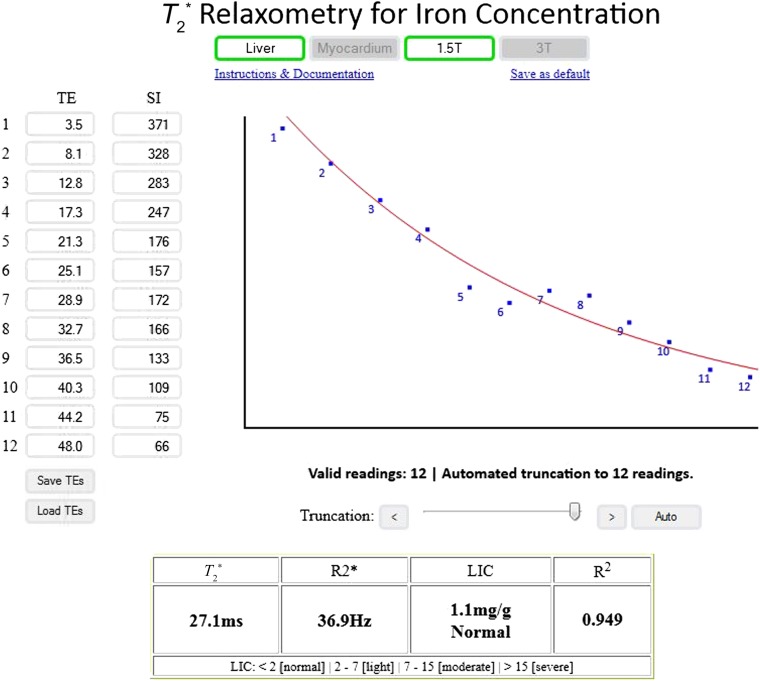
Figure 1.
A sample calculation performed with the online tool that allows for automated truncation and provides the results in T2*, R2* and final liver and myocardial iron concentrations. LIC, liver iron concentration; SI, signal intensity; TE, echo time.
Values obtained with both tools were compared using paired t-test, Bland–Altman plots and correlation coefficients (MedCalc v. 15.2; MedCalc Software, Ostend, Belgium).
RESULTS
14 patients (28%) had myocardial T2* values below the normal 20 ms cut-off while only 2 patients (4%) had normal liver iron levels with 22 patients (44%) with severe iron overload. The mean myocardial T2* values calculated by the reference software was 25.3 ms [95% confidence interval (CI) 22.3 to 28.3 ms] corresponding to a myocardial iron concentration of 0.87 mg g−1 (95% CI 0.76 to 1.02 mg g−1).7 No significant differences were observed comparing these values to the myocardial T2* values calculated using the online tool of 26.0 ms (95% CI 22.9 to 29.0 ms; p = 0.28). For the liver, the mean T2* value measured using the reference software was 4.26 ms (95% CI 3.1 to 5.4 ms) corresponding to a liver iron concentration of 7.4 mg g−1 (95% CI 5.7 to 10.1 mg g−1).8 No significant differences were observed compared with the online calculation of 4.30 ms (95% CI 3.1 to 5.5 ms; p = 0.71).
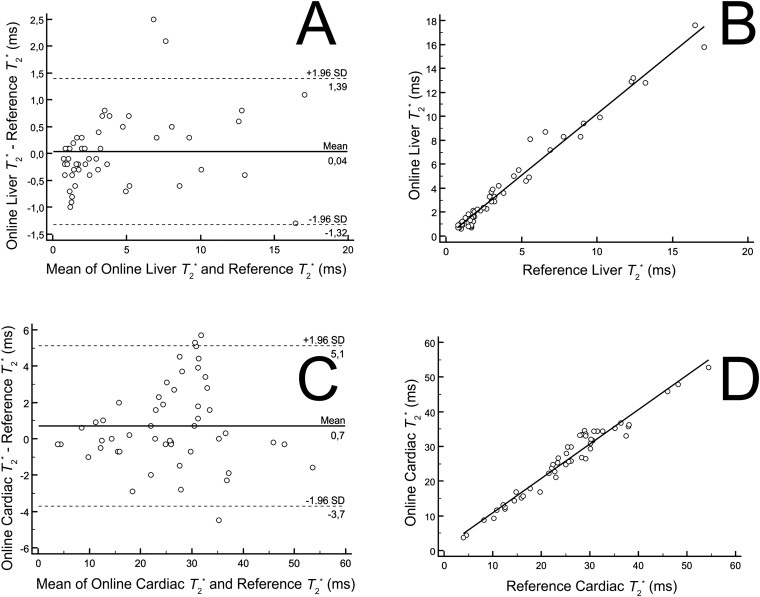
The results for the liver and heart demonstrated excellent agreement with the reference method (Figure 2). In the liver, the mean difference was 0.036 ms (95% CI −0.1609 to 0.2329 ms) with a regression correlation coefficient of 0.97; in the heart, the mean difference was 0.72 ms (95% CI 0.08191 to 1.3621 ms) with a regression correlation coefficient of 0.96.
Figure 2.
Bland–Altman plots and scatter diagrams with regression line demonstrating the comparison of T2* values in the liver (a, b) and heart (c, d) using the online tool vs the standard offline calculations. SD, standard deviation.
DISCUSSION
One of the main limitations to the implementation of routine iron overload assessment in many centres in the world is the inability to accurately calculate T2* values with currently available tools. Not only that, training and understanding the main limitations of these calculations are especially important in order to avoid making significant mistakes in the final iron values by underestimating the true iron concentrations in cases where severe iron overload exists.9 Although some authors suggest that using an offset model might solve part of the problems with noise in the current gradient-echo sequences, the true T2* decay curve appears to be best reflected by a monoexponential formula,10 especially when analysing the data using region-of-interest-based methods as is most commonly performed by routine commercial software.11 While CVI42 provides an option to calculate the T2* values using baseline correction, we chose not to include this function in the online tool so as not to add extra variability to the results provided, especially considering the possible use of this tool in less experienced centres. While we recognize that the choice of methods merits scientific discussion, in clinical practice it appears that the differences are not specifically appreciable and so we opted for a more simple unique approach.12
With the monoexponential model, the application of truncation of the last data points is fundamental in order to obtain accurate results in cases with severe iron concentration. Lack of application of this principle accounts for most of the errors in the interpretation of the examination in less experienced centres.3 Therefore, the development of an automated algorithm by He et al4 reducing the need for user manipulation of the data with correction for imperfections in the original T2* signal was greatly sought. This algorithm incorporates the following rules in order to correct for the apparent offset in the decay curve: if the original correlation coefficient is >0.995 for all data sets, it accepts the original T2*; if not, it automatically eliminates the last data points until the correlation coefficient exceeds that threshold; the elimination proceeds until the T2* values drop to <2.5%. While the method is very effective in providing corrected numbers, the need to use the original implementation in MATLAB significantly limits its availability, as most clinical centres do not routinely use that software. With the incorporation of the same rules previously described in an online platform with an easy-to-use interface and graphical visualization of the original fitting and corrected curves, we believe that more centres might be able to perform the accurate interpretation of T2* examinations.
While the algorithm corrects for most cases of noise in the images, in liver cases with very severe iron overload (T2* < 1.0 ms) or when the initial TE is higher than 1.3 ms, the automated truncation cannot always be performed. In these cases, users will have to choose to use a maximum of three or even two data points manually, increasing the overall spread of CIs of the calculated iron concentrations. While this can become a limitation, it has more to do with the restraints of the original multiecho gradient-echo technique than with the software used to analyse the data per se.3 Added to this limitation, the results presented in this article were produced in a single-centre setting with experienced users in T2* analysis and there is uncertainty as to how the online tool might perform in less proficient centres where other difficulties might appear. Finally, while one might expect the exact same results in both platforms, as the exponential equation is the same, slightly different regions of interest used for each software may explain part of the variation observed. This can also be explained by the fact that many of the patients studied presented with only mildly abnormal values specifically in the heart where other factors besides iron concentration affect the accuracy of the measurements at such levels.13
CONCLUSION
In conclusion, the online open-source tool provides accurate T2* values, incorporating a previously published automated algorithm in an accessible platform for clinical centres worldwide without the need for additional software while correcting for the most common source of error in the interpretation of these examinations.
FUNDING
This work was supported by the Fundacao de Amparo a Pesquisa do Estado de São Paulo (FAPESP)—Public Agency and Sultan Bin Khalifa International Thalassemia Award.
Contributor Information
K-A Git, Email: emlscs@yahoo.com.
L A B Fioravante, Email: rcccardio@mpc.com.br.
J L Fernandes, Email: jlaraf@terra.com.br.
REFERENCES
- 1.Voskaridou E, Ladis V, Kattamis A, Hassapopoulou E, Economou M, Kourakli A, et al. A national registry of haemoglobinopathies in Greece: deducted demographics, trends in mortality and affected births. Ann Hematol 2012; 91: 1451–8. doi: 10.1007/s00277-012-1465-7 [DOI] [PubMed] [Google Scholar]
- 2.Fernandes JL, Sampaio EF, Verissimo M, Pereira FB, da Silva JA, de Figueiredo GS, et al. Heart and liver T2 assessment for iron overload using different software programs. Eur Radiol 2011; 21: 2503–10. doi: 10.1007/s00330-011-2208-1 [DOI] [PubMed] [Google Scholar]
- 3.Feng Y, He T, Gatehouse PD, Li X, Harith Alam M, Pennell DJ, et al. Improved MRI R2* relaxometry of iron-loaded liver with noise correction. Magn Reson Med 2013; 70: 1765–74. doi: 10.1002/mrm.24607 [DOI] [PubMed] [Google Scholar]
- 4.He T, Zhang J, Carpenter JP, Feng Y, Smith GC, Pennell DJ, et al. Automated truncation method for myocardial T2* measurement in thalassemia. J Magn Reson Imaging 2013; 37: 479–83. doi: 10.1002/jmri.23780 [DOI] [PubMed] [Google Scholar]
- 5.Hankins JS, McCarville MB, Loeffler RB, Smeltzer MP, Onciu M, Hoffer FA, et al. R2* magnetic resonance imaging of the liver in patients with iron overload. Blood 2009; 113: 4853–5. doi: 10.1182/blood-2008-12-191643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He T, Gatehouse PD, Kirk P, Tanner MA, Smith GC, Keegan J, et al. Black-blood T2* technique for myocardial iron measurement in thalassemia. J Magn Reson Imaging 2007; 25: 1205–9. doi: 10.1002/jmri.20929 [DOI] [PubMed] [Google Scholar]
- 7.Carpenter JP, He T, Kirk P, Roughton M, Anderson LJ, de Noronha SV, et al. On T2* magnetic resonance and cardiac iron. Circulation 2011; 123: 1519–28. doi: 10.1161/CIRCULATIONAHA.110.007641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garbowski MW, Carpenter JP, Smith G, Roughton M, Alam MH, He T, et al. Biopsy-based calibration of T2* magnetic resonance for estimation of liver iron concentration and comparison with R2 Ferriscan. J Cardiovasc Magn Reson 2014; 16: 40. doi: 10.1186/1532-429X-16-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghugre NR, Enriquez CM, Coates TD, Nelson MD, Jr, Wood JC. Improved R2* measurements in myocardial iron overload. J Magn Reson Imaging 2006; 23: 9–16. doi: 10.1002/jmri.20467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghugre NR, Enriquez CM, Coates TD, Nelson Jr MD, Wood JC. Improved R2* measurements in myocardial iron overload. J Magn Reson Imaging 2006; 23: 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He T, Gatehouse PD, Smith GC, Mohiaddin RH, Pennell DJ, Firmin DN. Myocardial T2* measurements in iron-overloaded thalassemia: an in vivo study to investigate optimal methods of quantification. Magn Reson Med 2008; 60: 1082–9. doi: 10.1002/mrm.21744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meloni A, Rienhoff HY, Jr, Jones A, Pepe A, Lombardi M, Wood JC. The use of appropriate calibration curves corrects for systematic differences in liver R2* values measured using different software packages. Br J Haematol 2013; 161: 888–91. doi: 10.1111/bjh.12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ou P, Zhao Y, Fawal SE, Banka P, Powell AJ. Cardiac T2 measurements in patients with iron overload: a comparison of imaging parameters and analysis techniques. Magn Reson Imaging 2012; 30: 641–8. doi: 10.1016/j.mri.2011.12.020 [DOI] [PubMed] [Google Scholar]