Abstract
Summary: Omokage search is a service to search the global shape similarity of biological macromolecules and their assemblies, in both the Protein Data Bank (PDB) and Electron Microscopy Data Bank (EMDB). The server compares global shapes of assemblies independent of sequence order and number of subunits. As a search query, the user inputs a structure ID (PDB ID or EMDB ID) or uploads an atomic model or 3D density map to the server. The search is performed usually within 1 min, using one-dimensional profiles (incremental distance rank profiles) to characterize the shapes. Using the gmfit (Gaussian mixture model fitting) program, the found structures are fitted onto the query structure and their superimposed structures are displayed on the Web browser. Our service provides new structural perspectives to life science researchers.
Availability and implementation: Omokage search is freely accessible at http://pdbj.org/omokage/.
Contact: hirofumi@protein.osaka-u.ac.jp
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Electron microscopy of cellular macromolecules provides 3D density maps of many important molecular machines. More than 3000 density maps are now stored in the Electron Microscopy Data Bank (EMDB) (Lawson et al., 2010). Atomic models, obtained by X-ray crystallography and the current hybrid method, are also available in the Protein Data Bank (PDB) (Berman et al., 2007). We have been providing Web-based services, EM Navigator and Yorodumi, for both databanks (Kinjo et al., 2012). Shape comparisons among these 3D density maps and atomic models facilitate the elucidation of structural differences and conformational changes, and the generation of atomic models from the density maps. However, very few Web services look for shapes represented as 3D density maps that are similar to the atomic models. The Web server EM-SURFER (Esquivel-Rodriguez et al., 2015) was recently developed for searching 3D density maps. However, it only handles 3D density maps in the EMDB, and not in the PDB, and does not provide 3D superimpositions.
Here, we describe our new search service, Omokage search, based on the global shape similarity of the structure data, for both the PDB and EMDB. Our server provides superimposed structures, using the program gmfit. Users can visually assess the similarities by the 3D superimpositions.
2 Methods
2.1 Similarity search
For a fast search through the large dataset, a comparison is performed using one-dimensional (1D) profiles generated from 3D point models. We employ the vector quantization method to convert both density maps and atomic models into 3D point models, and use the Situs software for the conversion (Wriggers et al., 1998). Four 1D profiles are generated from the 3D point model. Three of them are generated based on the distances of the 3D point pairs (incremental distance rank profiles). The other is based on the principal components analysis (PCA) of the 3D point model. Details of the procedure are described in the Supplementary Data.
2.2 3D superimposition of assemblies
A superimposition of assemblies is performed using the gmfit program (Kawabata, 2008), which employs the Gaussian mixture model (GMM) algorithm to represent both the 3D density maps and atomic models. The GMM representation considerably reduces the computational cost for superimposition. We calculate the ‘one-to-one’ fitting, where a single density map or an atomic model is superimposed onto another fixed density map or atomic model. We employ the principal component axes alignment to generate the initial configurations, and the steepest descent method to refine the initial configurations. The computation time for the one-to-one superimposition is less than one second. Details of the procedure are described in the Supplementary Data.
3 Server description
3.1 Dataset
In this service, density maps from the EMDB, the asymmetric unit (AU) and a biological unit (BU) from the PDB are stored in the dataset. Approximately 2800 EMDB map data, 100 000 PDB AU models and 100 000 PDB BU models are presently available, and they are updated weekly.
3.2 Input
Input query structure data can be submitted by specifying the ID of the data in the search dataset or by uploading one’s own data file. As trials, some sample query data are shown. For a PDB entry, the user can specify an assembly ID by adding a number to the four component PDB ID (e.g. ‘1oel-1’) or by selecting one from the AU or BU images. PDB format files are acceptable for uploading an atomic model or a dummy atom model by small angle scattering. CCP4/MRC format files are acceptable for uploading a 3D density map, and the surface level should be specified.
3.3 Output
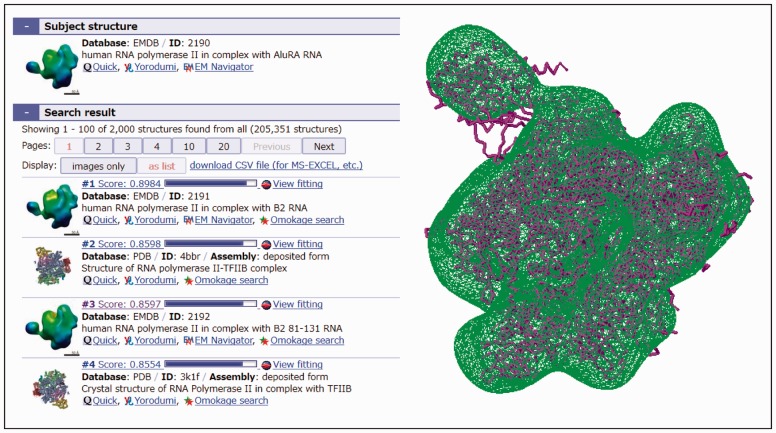
The search usually finishes within 1 min, and a list of similar shaped structure data (at most, 2000) to the input query data is shown, in the order of the similarities (Fig. 1, left). The users can open the page with the interactive viewer, Jmol/JSmol, which will show the found model superimposed onto the query model by the program gmfit (Kawabata, 2008; Fig. 1, right).
Fig. 1.
The search results using the map data RNA polymerase II (EMDB 2190) as the search input query (left), and the fitting of the atomic data (PDB 4BBR) onto the map data (right)
3.4 Evaluation
We evaluated the performance to detect biological similarities for both Omokage and EM-SURFER, and concluded that our Omokage is more powerful to detect biological similarities among various density maps with different resolutions and volumes. Details about the performance comparison using the ClpP-ClpB and 70S-ribosome datasets are described in the Supplementary Data.
4 Case studies and outlook
We emphasize that the advantage of our server is its ability to rapidly compare global shapes independent of sequence-order, subunit number and type of structural data (atomic model and density map). We introduce three types of case studies. The first is a search for low resolution structures. For the query of the 3D map data of RNA polymerase II (EMDB 2190; 25 Å resolution), 100 RNA polymerase structures were found in both databanks, independent of their resolutions. The second is a search for similar assembly forms with different subunits. A search with the PCNA clamp in the trimer ring form (AU of PDB 3IFV) yielded 100 clamps, including dimer beta clamps. The third is finding unexpected similar shapes, and implying some functional similarity (‘molecular mimicry’). The shape similarity of the tRNA-EF-Tu complex (RNA and protein) and EF-G (single protein) is a famous example (Nissen et al., 1995). The search using the tRNA-EF-Tu complex structure (AU of PDB 1OB2) yielded some EF-G structures. The current case studies and the performance are described in detail in the Supplementary Data. Our server is not meant for sequence-order comparisons of atomic models of single protein chains, which can be easily performed by BLAST and DALI. The detection of local substructure similarity has not been incorporated in the current algorithm. For the hybrid and integrative methods (Sali et al., 2015), such an algorithm should be developed in the future.
Supplementary Material
Funding
This work was supported by National Bioscience Database Center (NBDC) of Japan Science and Technology Agency (JST), and T. K. and H. S. were supported by JSPS KAKENHI Grant Number 26440078.
Conflict of Interest: none declared.
References
- Berman H., et al. (2007) The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res., 35, D301–D303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquivel-Rodriguez J., et al. (2015) Navigating 3D electron microscopy maps with EM-SURFER. BMC Bioinformatics, 16, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata T. (2008) Multiple subunit fitting into a low-resolution density map of a macromolecular complex using Gaussian mixture model. Biophys.J., 95, 4643–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo A.R., et al. (2012) Protein Data Bank Japan (PDBj): maintaining a structural data archive and resource description framework format. Nucleic Acids Res., 40, D453–D460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson C.L., et al. (2010) EMDataBank.org: unified data resource for CryoEM. Nucleic Acids Res., 39, D456–D464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P., et al. (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science, 5241, 1464–1472. [DOI] [PubMed] [Google Scholar]
- Wriggers W., et al. (1998) Self-organizing neural networks bridge the biomolecular resolution gap. J. Mol. Biol., 284, 1247–1254. [DOI] [PubMed] [Google Scholar]
- Sali A., et al. (2015) Outcome of the first wwPDB hybrid/integrative methods task force. Structure, 23, 1156–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.