Abstract
Motivation: DNA methylation patterns are well known to vary substantially across cell types or tissues. Hence, existing normalization methods may not be optimal if they do not take this into account. We therefore present a new R package for normalization of data from the Illumina Infinium Human Methylation450 BeadChip (Illumina 450 K) built on the concepts in the recently published funNorm method, and introducing cell-type or tissue-type flexibility.
Results: funtooNorm is relevant for data sets containing samples from two or more cell or tissue types. A visual display of cross-validated errors informs the choice of the optimal number of components in the normalization. Benefits of cell (tissue)-specific normalization are demonstrated in three data sets. Improvement can be substantial; it is strikingly better on chromosome X, where methylation patterns have unique inter-tissue variability.
Availability and Implementation: An R package is available at https://github.com/GreenwoodLab/funtooNorm, and has been submitted to Bioconductor at http://bioconductor.org.
Contact: celia.greenwood@mcgill.ca
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Recently, a normalization method was introduced by Fortin et al. (2014) specifically designed for the Illumina Infinium Human Methylation 450 BeadChip (Illumina 450 K) and implemented in Bioconductor’s minfi package(Aryee et al., 2014). The percentile-specific adjustments in funNorm are the key feature allowing batch effects and technical artefacts to have non-constant influence across the range of signal strengths.
However, since methylation patterns may differ substantially across cell types or tissues leading to cell- (or tissue)-type-specific quantiles, optimal normalization adjustments should capture this. Here we present an R package for normalization of Illumina 450 K data, funtooNorm (an extension of the ideas in funNorm) applicable to such heterogeneous data sets.
2 Methods
Key features of funtooNorm and funNorm are identical, i.e. normalization adjustments are estimated via regression models applied to a series of quantiles of the probe-type-specific signals in each sample. Covariates, derived from the control probes, capture variation not associated with the biological signals of interest. In funtooNorm, an augmented covariate matrix is constructed by including interactions between cell-type or tissue-type indicators and the average signal from each control probe type. Either principal component regression (PCR) or partial least squares regression (PLS) (Tenenhaus, 1998) can be fit (the type.fits option); as in funNorm, normalized methylation values are based on predictions from linear interpolations between the analyzed percentiles (see Supplemental Methods).
The function, funtoonorm, operates in two distinct modes:
Normalization mode: When validate = FALSE, normalization of the data is performed for a chosen number of components in the regressions. The model-fitting step requires only a set of quantiles for each sample, and hence is efficient both computationally and in memory usage. Calculations can be performed in a modular fashion; intermediary results can be saved by setting appropriate flags.
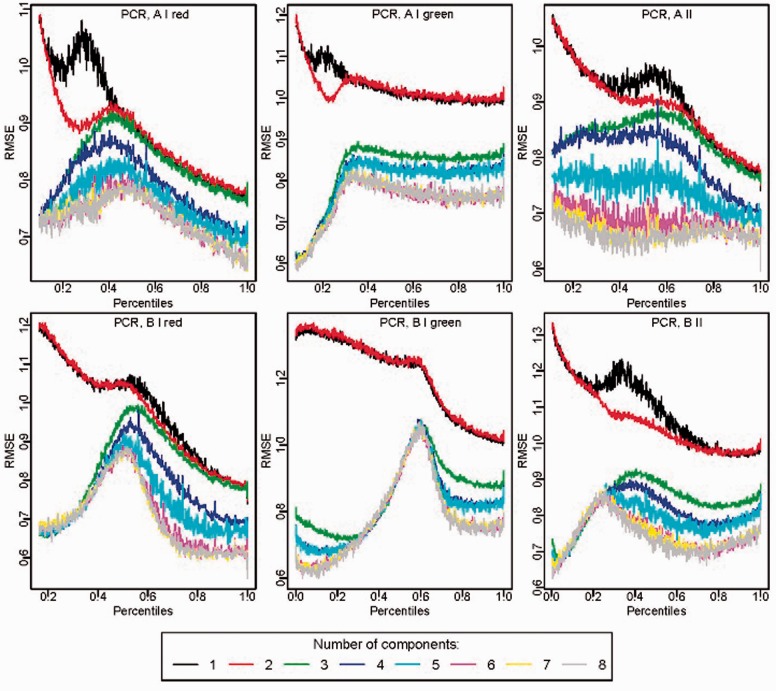
Cross-validation mode: When validate = TRUE, a graphical display of root mean squared errors (RMSE) obtained with cross-validation facilitates choice of an appropriate number of components (Fig. 1). Plots are provided for both PCR and PLS fits.
Three data sets are used to illustrate performance (Supplemental Table S1). In the Replication Data Set, methylation was measured in ten healthy individuals who contributed 2–3 samples of each of whole blood, buccal swab and dried blood spots, including a mixture of technical and biological replicates. In the Systemic Autoimmune Diseases Data (SARDS), monocytes and CD4 + T-cells from incident patients were separated from whole blood, with repeated samples drawn before and after 6 months of immunosuppressive treatment. For the Gestational Diabetes Data (GD), one technical replicate sample was available for each of fetal placenta and cord blood tissues. Agreement—within a tissue or cell type—is measured by the average (over probes) of the squared intra-replicate set differences, summed over distinct individuals.
Fig. 1.
Root mean square error from cross-validation comparing different numbers of components in funtooNorm on the Replication Data Set. Separate model fits are implemented for A and B signals, and for different probe types
3 Results
Figure 1 displays the cross-validation RMSE plot for the Replication Data set with PCR. The optimal number of components varies across the percentiles and signals; evidently there is substantial improvement in mean squared error from 2 to 3 components.
Technical replicate agreement was improved with funtooNorm compared to funNorm (Supplemental Figs S1 and S2, Supplemental Tables S2 and S3). Agreement improved by substantially for technical replicates of whole blood, blood spots, and fetal placenta tissues, although there was little difference between the methods for buccal swabs or cord blood. For biological replicates, we saw improvements of 10-20% in many tissues. Performance was particularly good for probes on the X chromosome. Supplemental Figure S3 shows that the distribution across probes of the differences between tissue types is distinct on the X chromosome; this is captured by our augmented covariate matrix. A similar argument explains enhanced performance for some probe annotations (Supplemental Fig. S4). Performance on the Y chromosome was poor, since with only 416 probes, a quantile-based model fit is overly complex; we recommend the simpler method implemented in funNorm for this chromosome.
4 Discussion
Most methylation studies today are designed to detect inter-individual differences, rather than inter-tissue differences. Improved normalization of datasets containing multiple tissues can be expected to translate into increased power to detect associations of interest, due to the inferred reduction in residual error; funNorm and this extension funtooNorm are designed with this goal in mind.
Supplementary Material
Acknowledgements
We thank Rani Damsteegt for assistance with data collection of the Replicate Data.
Funding
This work was supported by the Ludmer Center for Neuroinformatics & Mental Health, and by the Canadian Institutes of Health Research operating grant MOP-300545.
Conflict of Interest: none declared.
References
- Aryee M.J., et al. (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics, 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.-P., et al. (2014) Functional normalization of 450 K methylation array data improves replication in large cancer studies. Genome Biol., 15, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenhaus M. (1998) La Régression PLS: Théorie et Pratique. Paris: Éditions Technip. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.