Abstract
Background/Aim
Prostate cancer (PCa) shows disproportionately higher incidence and disease-associated mortality in African Americans. The human crystallin beta B2 (CRYBB2) gene has been reported as one tumor signature gene differentially expressed between African American and European American cancer patients. We investigated the role of CRYBB2 genetic variants in PCa in African Americans.
Materials and Methods
Subjects comprised of 233 PCa cases and 294 controls. Nine haplotype-tagged single nucleotide polymorphisms (SNPs) in and around the CRYBB2 gene were genotyped by pyrosequencing. Association analyses were performed for PCa with adjustment for age and prostate-specific antigen (PSA), under an additive genetic model.
Results
Out of the nine SNPs examined, rs9608380 was found to be nominally associated with PCa (odds ratio (OR)=2.619 (95% confidence interval (CI)=1.156–5.935), p=0.021). rs9306412 was in strong linkage disequilibrium with rs9608380 that showed an association p-value of 0.077. Using ENCODE data, we found rs9608380 mapped to a region annotated with regulatory motifs, such as DNase hypersensitive sites and histone modifications.
Conclusion
This is the first study to analyze the association between genetic variations in the CRYBB2 gene with PCa. rs9608380, associated with PCa, is a potentially functional variant.
Keywords: Prostate cancer, African Americans, single nucleotide polymorphism, CRYBB2, genetic association
Prostate cancer (PCa) is the most commonly diagnosed cancer in American men following only skin cancer and is the second leading cause of death from cancer in men of all ages in the US (1, 2). It is estimated that in the US in the year 2015, about 220,800 new cases of PCa will be diagnosed with about 27,540 projected deaths from this disease (2). The incidence and mortality rates of PCa vary substantially among different ethnic groups, with African Americans showing evidence of disease-associated disparity (2, 3). While the role of genetics in PCa is well-recognized (4) and disease susceptibility alleles have been shown to overlap between populations of different ethnic backgrounds (5), accumulating data point to genetic markers of risk for PCa that appear specific to men of African descent (6). Studies of the pathology and recurrence of tumors in African American and European American men have suggested that population differences in the biology of PCa tumors may explain observed differences in outcome (7).
The human crystallin, beta B2 (CRYBB2) gene is located on chromosomal locus 22q11.23. CRYBB2 encodes for the βB2-crystallin, the most abundant and water-soluble β-crystallin in the lens (8). Mutations in the CRYBB2 gene have been reported to be associated with cataracts in several independent studies (9, 10). Recent reports have observed a relationship of CRYBB2 expression with cancer. CRYBB2 expression was reported to be significantly up-regulated in African American vs. European American patients with colorectal cancer (11). CRYBB2 was also found to be differentially expressed between African American and European American breast cancer patients (12, 13). In a recent study examining the differences in tumor biology contributing to the disparity observed in the incidences and mortality from PCa by gene expression profiling, CRYBB2 was found significantly differently expressed in prostate tumors between African Americans and European Americans (14). The authors described CRYBB2 as one of the two tumor signature genes that accurately differentiated between African American and European American patients (14).
We, therefore, examined the association of CRYBB2 genetic variants with PCa in African Americans. Nine single nucleotide polymorphisms (SNPs) spanning the CRYBB2 gene were chosen for genotyping to examine their association with PCa in a cohort of African Americans by performing a case-control association study.
Materials and Methods
Study sample
Individuals studied were unrelated, self-reported African American men from the Washington, DC area. Participants were between 40–85 years old and recruited either from the Urology Clinic at Howard University Hospital or from a PCa screening program conducted at the Howard University Cancer Center. The screening program was demographically similar to the patient population seen in the Urology Clinic. Ethical approval for the study was obtained from the Howard University’s Institutional Review Board. All individuals provided their written informed consent for the collection of data and samples, as well as subsequent analyses. The participants for this study comprised of 233 PCa cases and 294 controls. PCa cases were diagnosed by an urologist, initially by clinical examination followed by transrectal ultrasound-guided biopsy using standard saturation technique (15). Biopsy cores were reviewed by members of the Department of Pathology of Howard University Hospital. PCa cases were classified according to the well-established parameters of the Gleason Scoring System (16). Controls included men of the same age group with a prostate-specific antigen (PSA) value of ≤4.0 ng/ml and a normal finding by digital rectal examination (DRE) and, in selected cases, by biopsies. Individuals who were ever diagnosed with benign prostatic hyperplasia (BPH) were not included as controls. During a clinical examination, demographic and medical information were collected by interview. Blood samples were collected from each subject from which genomic DNA was obtained. Clinical characteristics, including Gleason grade, PSA, age at diagnosis and relevant clinical data were obtained from medical records.
Genotyping and quality control
Using the International HapMap Project YRI data as a reference, nine tag SNPs spanning the CRYBB2 genomic locus at a pairwise r2 value of at least 0.8 were selected for genotyping. Genotyping was performed by pyrosequencing (Qiagen, Germantown, MD, USA) according to the manufacturer’s recommendations. Briefly, primers were designed using the PSQ Assay Design Software (Qiagen, Germantown, MD, USA) and polymerase chain reaction (PCR) amplification was performed using 20 pico moles of forward unlabeled and a reverse biotin labeled primer (or vice versa), MgCl2, deoxynucleotide triphosphates and platinum Taq DNA polymerase (Life Tech, Carlsbad, CA, USA). Biotinylated single-stranded DNA fragments were separated using Streptavidin Sepharose™ High Performance (GE Healthcare, Piscataway, NJ, USA) and pyrosequenced. Results were analyzed with the PyroMark Q24 software (Qiagen, Germantown, MD, USA). Duplicate test samples and negative controls were included in each of the 96-well plates.
Statistical analyses
Power calculations were performed using QUANTO (17). Assuming an additive genetic model and with a minor allele frequency (MAF) of 0.05, this study had a power of 43% to 78% to detect associations with a genetic relative risk of 1.6 to 2.0, respectively. The Fisher’s exact test was performed to determine deviations of genotype distributions from the expected Hardy-Weinberg equilibrium (HWE). Linkage disequilibrium (LD) was visualized using Haploview (18). PCa was analyzed as a binary trait (cases versus controls) and odds ratios (OR), 95% confidence intervals (CI) and p-values were calculated using logistic regression under an additive genetic model. Genetic effects were adjusted for age (at time of diagnosis for cases and at time of ascertainment for controls) and PSA value. Two-sided p-values ≤0.05 were considered as statistically significant. All association analyses were performed using the SPSS statistical package, version 22.0 (IBM Corporation, Armonk, NY, USA).
Expression analysis and functional annotation
To search for prostate tissue-specific expression of CRYBB2, we queried the Gene Expression Atlas (19). We examined the regulatory role of the disease-associated non-coding SNPs by querying the ENCODE data using HaploReg version 2 (20) and RegulomeDB (21).
Results
The frequencies of clinical characteristics of our study population are shown in Table I. The mean age (±SD) was 64.88±9.18 years for the cancer patients and 58.28±10.72 years for the controls, with significant difference between the two groups (p<0.0001). Similarly, PSA level was found significantly higher in the case group (11.12±14.64) compared to the controls (3.35±7.59). The cancer group had a higher percent of family history of PCa (18.45%) compared to the controls (11.56%).
Table I.
Clinical characteristics of the study population (n=527).
| Characteristic | Cases | Controls | p-Value |
|---|---|---|---|
| No. of participants | 233 | 294 | |
| Age, years | 64.88 (9.18) | 58.28 (10.72) | <0.0001 |
| Age group, number (%) | |||
| <50 years | 12 (5.15) | 64 (21.77) | |
| 50–59 | 59 (25.32) | 99 (33.67) | |
| 60–69 | 88 (37.77) | 75 (25.51) | |
| ≥70 | 74 (31.76) | 56 (19.05) | |
| PSA (ng/ml) | 11.12 (14.64) | 3.35 (7.59) | < 0.0001 |
| Family history of PCa (%) | 43 (18.45) | 34 (11.56) | |
PSA, Prostate-specific antigen; PCa, prostate cancer. Data are shown as number (percent) or mean (SD).
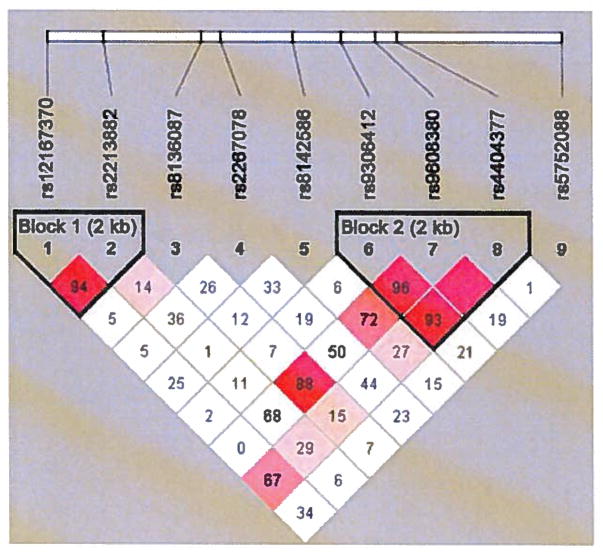
Nine SNPs from about 20 kb genomic sequences spanning the CRYBB2 gene were selected for examination. Table II shows the list of the SNPs with their corresponding chromosomal location. Genotype distribution of the examined SNPs conformed to HWE (p>0.001). MAFs of the examined SNPs ranged from 5–46% in our population sample. Detailed results of association testing are presented in Table II. Out of the nine SNPs tested, rs9608380 was found associated with PCa at nominal significance (p=0.021) under an additive genetic model. The A allele of the SNP had a frequency of about 6% and was found to increase PCa risk (OR=2.619) (CI=1.156–5.935) (Table II). rs9608380 is located in the intronic region of CRYBB2 gene. Figure 1 shows the pattern of LD of the genomic region of the CRYBB2 gene spanning the examined SNPs in our tested samples. rs9306412, another intronic SNP, showed an association p-value of 0.077 and was in strong LD with rs9608380 (Figure 1).
Table II.
Association results of CRYBB2 SNPs.
| SNP | Position | Type | Minor allele | MAF | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| rs12167370 | 25609680 | Upstream | T | 0.0788 | 1.527 (0.774–3.01) | 0.222 |
| rs2213882 | 25611881 | Upstream | T | 0.4665 | 0.855 (0.587–1.245) | 0.414 |
| rs8136087 | 25615710 | Intronic | A | 0.4655 | 1.062 (0.79–1.427) | 0.69 |
| rs2267078 | 25616451 | Intronic | A | 0.0601 | 0.919 (0.461–1.832) | 0.81 |
| rs8142586 | 25619267 | Intronic | A | 0.4464 | 0.839 (0.605–1.165) | 0.295 |
| rs9306412 | 25621123 | Intronic | A | 0.2319 | 1.432 (0.962–2.133) | 0.077 |
| rs9608380 | 25622442 | Intronic | A | 0.0578 | 2.619 (1.156–5.935) | 0.021 |
| rs4404377 | 25623323 | Intronic | A | 0.3396 | 1.2 (0.874–1.649) | 0.26 |
| rs5752088 | 25629682 | Downstream | G | 0.4605 | 1.244 (0.872–1.774) | 0.228 |
Position based on NCBI genome build 105; SNP, single nucleotide polymorphism; MAF, minor allele frequency; OR, odds ratio; CI, confidence interval.
Figure 1.
Schematic diagram of the location of 9 tag SNPs in the CRYBB2 gene and patterns of pairwise-linkage disequilibrium (LD) between the SNPs. Strength of LD in the samples is represented as red squares for strong LD and white squares for little or no LD.
We queried the Gene Expression Atlas for tissue-specific baseline expression of CRYBB2 and found it to be expressed at a low level in normal prostate tissue. We also performed in silico lookup for DNA feature and potential regulatory elements spanning the associated locus. rs9608380 was predicted from ENCODE data as a functional SNP with a regulatory role. The rs9608380 locus maps to several regulatory motifs, including Barx1 in the LNCaP cell line as revealed by DNase footprinting, and Lhx4 as well.
As determined by ENCODE ChIP-seq data, the variant locus was annotated with two histone marks, H3k79me2 and H3k27me3, localized in a short chromosomal region (chromosomal location: 25622304-25622496 and 25622330-25622518, respectively).
Discussion
In the present study, we examined the association of genetic variants in the CRYBB2 gene with PCa in an African American population. We chose the CRYBB2 gene based on recent reports regarding its differential expression between African American and European American men in different cancers (11–13), including PCa patients (14). Regional deletion distal to CRYBB2 on 22q has been reported in a retroperitoneal rhabdoid malignant tumor (22). At least eleven missense, one splicing and one insertion/deletion mutation in the CRYBB2 gene have been associated with congenital cataracts (23, 24). To our knowledge, this is the first report examining the association of genetic variants in CRYBB2 with PCa. As our study included only self-reported African Americans, population stratification is not expected to be an issue. Many candidate gene studies rely on self-reported ancestry to control for population stratification, an approach that has been shown to be reliable in genetic studies (25, 26).
Using nominal significance (p<0.05) for association, we found one SNP, rs9608380, associated with PCa. However, the significance was lost when corrected for multiple testing using the Bonferroni method. A limitation of genetic studies, such as ours, which evaluate many SNPs, is the potential for false positive findings as a result of multiple testing (type I error). Due to the multiple testing burden associated with the number of SNPs tested, it is possible that this study did not have enough power to see an association after Bonferroni correction. Our observed frequency of the associated allele A (5.78%) is somewhat less than what was found in the HapMap-ASW data (10.2%). We did not find any report of genetic association of rs9608380 with any other phenotype.
An important issue in many association studies, including ours, is that significantly associated variants are often located in non-coding regions, thus raising the question of their functionality and possible role in disease predisposition. An intronic SNP may alter the function of a nearby regulatory element or be in LD with another causative variant that is directly involved in PCa susceptibility. Introns also contain several short sequences of cis-splicing motifs that are important for efficient splicing, such as acceptor and donor sites at either end of the intron and a branch point site, which are required for proper splicing by the spliceosome. Additionally, about 40% of the miRNA genes that are short non-coding RNAs and known to regulate gene expression by binding to sequences on the target mRNA (27) are often embedded within introns (28). Another possible mechanism of action is thought to be via the influence of SNPs on distal enhancer elements that regulate the expression of critical target genes involved in the etiopathogenesis of disease (29, 30).
The potential binding motifs spanning the rs9608380 locus warrant special mention. BARX1 is a homeobox transcription factor whose promoter region has been reported to be hypermethylated in colorectal cancer (31) and in gastric cancer (GC) cell lines and patient samples, with BARX1 mRNA expression in GC tissues and cell lines being reduced (32). A genome-wide association study has identified rs11789015, an intronic SNP in BARX1 associated with esophageal adenocarcinoma and Barrett’s esophagus (33). LHX4 is abundantly expressed in cancers (34, 35). Treatment with simvastatin, a known anticancer agent that inhibits prostate cancer cell functions and tumor growth (36), resulted in reduced LHX4 protein level in PC3 cells (37). Further studies are, thus, warranted to explore the role of these regulatory molecules in CRYBB2 expression in the context of PCa development and progression.
Several limitations should be kept in mind when interpreting the results of this study. First, the sample size was relatively small, which can result in insufficient power to detect minor contributions of associated alleles and contributions of those with low MAF. Similarly, small sample sizes can provide imprecise or incorrect estimates of the magnitude of the observed effects. Second, like other cancers, PCa is a complex disease associated with both genetic and environmental factors. Socio-environmental factors were not included in the present analysis, which limited the interpretation of these results and evaluation of the gene-environment interaction. We do, however, envision that since the participants are recruited from a defined demographic region, there will be some shared environmental factors, thereby minimizing the bias imposed by them. Thirdly, the incidence of PCa increases with age. Therefore, even though the effect of age was adjusted for by statistical methods, some of the younger control subjects may yet develop PCa in the future. The third limitation is not specific to our study design but, in fact, is applicable for many studies exploring germ-line genetics of PCa.
In conclusion, we have, for the first time, demonstrated that rs9608380, an intronic SNP in the CRYBB2 gene, is associated with the risk of PCa in an African American population. We also found that rs9608380 maps to a locus potentially containing regulatory motifs, as predicted by in silico analyses. While we acknowledge both the strengths and limitations of our study, further replication studies with larger sample sizes and deep sequencing are required to confirm the genetic association of CRYBB2 gene with PCa and to pin-point the causal variant(s) as well. The functional role of CRYBB2 in prostate tissue and how it contributes to PCa remains speculative. CRYBB2 is among the major proteins of the vertebrate eye lens. Although it is not clear how a lens protein would contribute to prostate tumorigenesis, in a rat model, the expression of Crybb2 increased in damaged optic nerves promoting regrowth of axons (38). Thus, it is possible that over-expression of CRYBB2 in African American men may promote or enhance tumor growth. Additional functional studies are warranted to decipher the role of CRYBB2 gene in PCa development and progression.
Acknowledgments
This research was supported in part by grant S06 GM08016 from the MBRS/SCORE Program of the NIGMS and by grant G12 RR003048 to the RCMI Program at Howard University by Division of Research Infrastructure, NCRR, NIH. Part of the work was also supported by an internal award from the Health Sciences Faculty Seed Grant Program at Howard University. The Authors would like to thank participants of the study.
Footnotes
Conflicts of Interest
The Authors declare no conflicts of interest.
References
- 1.U.S. Cancer Statistics Working Group. United States Cancer Statistics: 1999–2006 Incidence and Mortality Web-Based Report. Atlanta, Ga, USA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute; 2010. [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Brawley OW, Jani AB, Master V. cancer and race. Curr Probl Cancer. 2007;31:211–225. doi: 10.1016/j.currproblcancer.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Langeberg WJ, Isaacs WB, Stanford JL. Genetic etiology of hereditary prostate cancer. Front Biosci. 2007;12:4101–4110. doi: 10.2741/2374. [DOI] [PubMed] [Google Scholar]
- 5.Xu Z, Bensen JT, Smith GJ, Mohler JL, Taylor JA. GWAS SNP Replication among African American and European American men in the North Carolina-Louisiana prostate cancer project (PCaP) Prostate. 2011;71:881–891. doi: 10.1002/pros.21304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, Rybicki BA, Isaacs WB, Ingles SA, Stanford JL, Diver WR, Witte JS, Hsing AW, Nemesure B, Rebbeck TR, Cooney KA, Xu J, Kibel AS, Hu JJ, John EM, Gueye SM, Watya S, Signorello LB, Hayes RB, Wang Z, Yeboah E, Tettey Y, Cai Q, Kolb S, Ostrander EA, Zeigler-Johnson C, Yamamura Y, Neslund-Dudas C, Haslag-Minoff J, Wu W, Thomas V, Allen GO, Murphy A, Chang BL, Zheng SL, Leske MC, Wu SY, Ray AM, Hennis AJ, Thun MJ, Carpten J, Casey G, Carter EN, Duarte ER, Xia LY, Sheng X, Wan P, Pooler LC, Cheng I, Monroe KR, Schumacher F, Le Marchand L, Kolonel LN, Chanock SJ, Van Den Berg D, Stram DO, Henderson BE. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43:570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powell IJ. Prostate cancer in the African American: is this a different disease? Semin Urol Oncol. 1998;16:221–226. [PubMed] [Google Scholar]
- 8.Huang B, He W. Molecular characteristics of inherited congenital cataracts. Eur J Med Genet. 2010;53:347–357. doi: 10.1016/j.ejmg.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Litt M, Carrero-Valenzuela R, LaMorticella DM, Schultz DW, Mitchell TN, Kramer P, Maumenee IH. Autosomal dominant cerulean cataract is associated with a chain termination mutation in the human beta-crystallin gene CRYBB2. Hum Mol Genet. 1997;6:665–668. doi: 10.1093/hmg/6.5.665. [DOI] [PubMed] [Google Scholar]
- 10.Gill D, Klose R, Munier FL, McFadden M, Priston M, Billingsley G, Ducrey N, Schorderet DF, Héon E. Genetic heterogeneity of the Coppock-like cataract: a mutation in CRYBB2 on chromosome 22q11.2. Invest Ophthal Vis Sci. 2000;41:159–165. [PubMed] [Google Scholar]
- 11.Jovov B, Araujo-Perez F, Sigel CS, Stratford JK, McCoy AN, Yeh JJ, Keku T. Differential gene expression between African American and European American colorectal cancer patients. PLoS One. 2012;7:e30168. doi: 10.1371/journal.pone.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin DN, Boersma BJ, Yi M, Reimers M, Howe TM, Yfantis HG, Tsai YC, Williams EH, Lee DH, Stephens RM, Weissman AM, Ambs S. Differences in the tumor microenvironment between African-American and European-American breast cancer patients. PLoS One. 2009;4:e4531. doi: 10.1371/journal.pone.0004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Field LA, Love B, Deyarmin B, Hooke JA, Shriver CD, Ellsworth RE. Identification of differentially expressed genes in breast tumors from African American compared with Caucasian women. Cancer. 2012;118:1334–1344. doi: 10.1002/cncr.26405. [DOI] [PubMed] [Google Scholar]
- 14.Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, Stephens RM, Caporaso NE, Loffredo CA, Ambs S. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–936. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 15.Raja J, Ramachandran N, Munneke G, Patel U. Current status of transrectal ultrasound-guided prostate biopsy in the diagnosis of prostate cancer. Clin Radiol. 2006;61:142–153. doi: 10.1016/j.crad.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Gleason DF. Classification of prostatic carcinomas. Cancer Chemother Rep. 1966;50:125–128. [PubMed] [Google Scholar]
- 17.Gauderman W, Morrison J. QUANTO 1.1: A computer program for power and sample size calculations for genetic-epidemiology studies. 2006 ( http://hydra.usc.edu/gxe)
- 18.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 19.Lukk M, Kapushesky M, Nikkilä J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A. A global map of human gene expression. Nat Biotechnol. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Besnard-Guérin C, Cavenee W, Newsham I. The t(11;22) (p15.5;q11.23) in a retroperitoneal rhabdoid tumor also includes a regional deletion distal to CRYBB2 on 22q. Genes Chromosomes Cancer. 1995;13:145–150. doi: 10.1002/gcc.2870130302. [DOI] [PubMed] [Google Scholar]
- 23.Faletra F, d’Adamo AP, Pensiero S, Athanasakis E, Catalano D, Bruno I, Gasparini P. A novel CRYBB2 missense mutation causing congenital autosomal dominant cataract in an Italian family. Ophthalmic Genet. 2013;34:115–117. doi: 10.3109/13816810.2012.707273. [DOI] [PubMed] [Google Scholar]
- 24.BIOBASE HGMD®. http://www.biobase-international.com/product/hgmd.
- 25.Tang H, Quertermous T, Rodriguez B, Kardia SL, Zhu X, Brown A, Pankow JS, Province MA, Hunt SC, Boerwinkle E, Schork NJ, Risch NJ. Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. Am J Hum Genet. 2005;76:268–275. doi: 10.1086/427888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Haiman CA, Kolonel LN, Henderson BE, Wilkens LR, Le Marchand L, Stram DO. Self-reported ethnicity, genetic structure and the impact of population stratification in a multiethnic study. Hum Genet. 2010;128:165–177. doi: 10.1007/s00439-010-0841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Björklund M, Wei G, Yan J, Niittymäki I, Mecklin JP, Järvinen H, Ristimäki A, Di-Bernardo M, East P, Carvajal-Carmona L, Houlston RS, Tomlinson I, Palin K, Ukkonen E, Karhu A, Taipale J, Aaltonen LA. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 31.Kober P, Bujko M, Olędzki J, Tysarowski A, Siedlecki JA. Methyl-CpG binding column-based identification of nine genes hypermethylated in colorectal cancer. Mol Carcinog. 2011;50:846–856. doi: 10.1002/mc.20763. [DOI] [PubMed] [Google Scholar]
- 32.Agarwal R, Mori Y, Cheng Y, Abraham JM, Montgomery E, Meltzer SJ. BARX1, CYGB, and other epigenetic targets of gastric cancer. Cancer Res. 2012;72(Suppl 1):abstract LB-384. [Google Scholar]
- 33.Levine DM, Ek WE, Zhang R, Liu X, Onstad L, Sather C, Lao-Sirieix P, Gammon MD, Corley DA, Shaheen NJ, Bird NC, Hardie LJ, Murray LJ, Reid BJ, Chow WH, Risch HA, Nyrén O, Ye W, Liu G, Romero Y, Bernstein L, Wu AH, Casson AG, Chanock SJ, Harrington P, Caldas I, Debiram-Beecham I, Caldas C, Hayward NK, Pharoah PD, Fitzgerald RC, Macgregor S, Whiteman DC, Vaughan TL. A genome-wide association study identifies new susceptibility loci for esophageal adenocarcinoma and Barrett’s esophagus. Nat Genet. 2013;45:1487–1493. doi: 10.1038/ng.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawamata N, Sakajiri S, Sugimoto KJ, Isobe Y, Kobayashi H, Oshimi K. A novel chromosomal translocation t(1;14) (q25;q32) in pre-B acute lymphoblastic leukemia involves the LIM homeodomain protein gene, Lhx4. Oncogene. 2002;21:4983–4991. doi: 10.1038/sj.onc.1205628. [DOI] [PubMed] [Google Scholar]
- 35.Hung TM, Hu RH, Ho CM, Chiu YL, Lee JL, Jeng YM, Shih DT, Lee PH. Downregulation of alpha-fetoprotein expression by LHX4: a critical role in hepatocarcinogenesis. Carcinogenesis. 2011;32:1815–1823. doi: 10.1093/carcin/bgr219. [DOI] [PubMed] [Google Scholar]
- 36.Kochuparambil ST, Al-Husein B, Goc A, Soliman S, Somanath PR. Anticancer efficacy of simvastatin on prostate cancer cells and tumor xenografts is associated with inhibition of Akt and reduced prostate-specific antigen expression. J Pharmacol Exp Ther. 2011;336:496–505. doi: 10.1124/jpet.110.174870. [DOI] [PubMed] [Google Scholar]
- 37.Goc A, Kochuparambil ST, Al-Husein B, Al-Azayzih A, Mohammad S, Somanath PR. Simultaneous modulation of the intrinsic and extrinsic pathways by simvastatin in mediating prostate cancer cell apoptosis. BMC Cancer. 2012;12:409. doi: 10.1186/1471-2407-12-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liedtke T, Schwamborn JC, Schroer U, Thanos S. Elongation of axons during regeneration involves retinal crystalline beta b2 (crybb2) Mol Cell Proteomics. 2007;6:895–907. doi: 10.1074/mcp.M600245-MCP200. [DOI] [PubMed] [Google Scholar]