Abstract
HLCS (holocarboxylase synthetase) is a nuclear protein that catalyses the binding of biotin to distinct lysine residues in chromatin proteins. HLCS-dependent epigenetic marks are overrepresented in repressed genomic loci, particularly in repeats. Evidence is mounting that HLCS is a member of a multi-protein gene repression complex, which determines its localization in chromatin. In the present study we tested the hypothesis that HLCS interacts physically with N-CoR (nuclear receptor co-repressor) and HDAC1 (histone deacetylase 1), thereby contributing toward the removal of H3K9ac (Lys9-acetylated histone H3) gene activation marks and the repression of repeats. Physical interactions between HLCS and N-CoR, HDAC1 and a novel splicing variant of HDAC1 were confirmed by co-immunoprecipitation, limited proteolysis and split luciferase complementation assays. When HLCS was overexpressed, the abundance of H3K9ac marks decreased by 50% and 68% in LTRs (long terminal repeats) 15 and 22 respectively in HEK (human embryonic kidney)-293 cells compared with the controls. This loss of H3K9ac marks was linked with an 83% decrease in mRNA coding for LTRs. Similar patterns were seen in pericentromeric alpha satellite repeats in chromosomes 1 and 4. We conclude that interactions of HLCS with N-CoR and HDACs contribute towards the transcriptional repression of repeats, presumably increasing genome stability.
Keywords: chromatin, epigenetics, gene repression, holocarboxylase synthetase (HLCS), histone deacetylase 1 (HDAC1), nuclear receptor co-repressor (N-CoR)
INTRODUCTION
HLCS (holocarboxylase synthetase) is the sole biotin protein ligase in the human genome [1]. Historically, HLCS was appreciated for its role in attaching biotin to distinct lysine residues in five human carboxylases, which catalyse essential steps in fatty acid metabolism, gluconeogenesis, and leucine metabolism in the cytoplasm and mitochondria [2]. Previously, it was demonstrated that full-length HLCS and a splicing variant also enter the nuclear compartment and that nuclear HLCS binds to chromosomes in a locus-specific punctate pattern [3,4]. These observations implicate HLCS in gene regulation. Unambiguous evidence suggests that HLCS has histone-biotin ligase activity [5,6], catalysing the binding of biotin to distinct lysine residues in histones [7–9]. HLCS-dependent histone biotinylation marks are enriched in repressed genomic loci [10–15], consistent with a role of HLCS in gene repression. Note that only less than 0.001% of histones H3 and H4 are biotinylated in human chromatin [16–18]. Because of the rarity of biotinylation marks, it is unlikely that histone biotinylation itself is responsible for causing the repression of genes and severe phenotypes of HLCS knockdown such as short lifespan and low heat-stress resistance in Drosophila melanogaster and chromosomal abnormalities in human cell cultures [3,14]. Moreover, HLCS does not contain a classical nuclear localization signal or a DNA-binding motif that would explain its nuclear localization and binding to chromosomes. Evidence suggests that the binding of HLCS to chromosomes and perhaps its nuclear entry might be facilitated by physical interactions with histone H3 [19], but these interactions do not explain the punctate distribution of HLCS in chromatin [3,4].
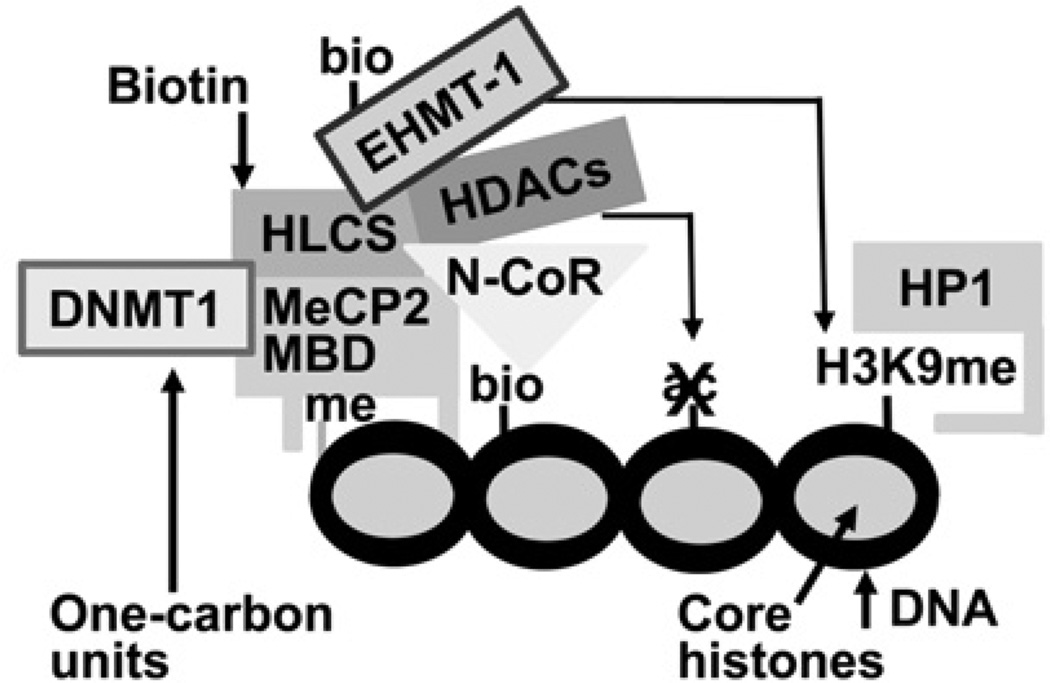
Recently, we have integrated the above reports into a coherent model that implicates HLCS in gene repression through epigenetic mechanisms (Figure 1). According to this model, HLCS is recruited to chromatin through physical interactions with the maintenance DNA methyltransferase DNMT1 (DNA methyltransferase 1) and the MeCP2 (methyl CpG-binding protein 2), [20], consistent with previous observations that erasure of DNA methylation marks impairs HLCS-dependent biotinylation events in chromatin [14]. Also according to this model, chromatin-bound HLCS recruits the eukaryotic histone H3 methyltransferase EHMT-1 (euchromatic histonelysine N-methyltransferase-1), which creates abundant H3K9me (Lys9-methylated histone H3) gene repression marks; the physical interaction between HLCS and EHMT-1 appears to be strengthened by HLCS-dependent biotinylation of Lys161 in EHMT-1 [21]. Consistent with a role of HLCS in the formation of a multi-protein gene repression complex, the abundance of H3K9me marks in repeats is severely reduced in HLCS-knockdown models [11,14]. Histone biotinylation marks are mere marks for HLCS-docking sites in chromatin and are caused by the close physical proximity between HLCS and histones [19].
Figure 1. HLCS-containing multi-protein gene repression complex.
The binding of HLCS to chromatin depends on physical interactions of HLCS with the maintenance DNA methyltransferase DNMT1 and MeCP2. Chromatin-bound HLCS recruits the eukaryotic histone H3 methyltransferase EHMT-1, N-CoR and HDAC. ac, acetylation; bio, biotinylation; HP1, heterochromatin protein 1; me, methylation.
The discovery of the interactions between HLCS and EHMT-1 was initiated by developing a protocol for predicting HLCS-binding proteins in silico [21]. Note that the protocol also predicted that HLCS interacts with N-CoR (nuclear receptor co-repressor), a protein known to facilitate the binding of HDACs (histone deacetylases) in chromatin [22–24]. HDACs play crucial roles in gene repression, mediated by HDAC-dependent removal of histone acetylation marks [25]. In the present study we tested the hypotheses that HLCS interacts with N-CoR and HDAC1 and that these interactions play roles in the repression of repeats.
MATERIALS AND METHODS
Prediction of HLCS-binding proteins
In previous studies, we have shown that HLCS-binding proteins in chromatin share the GGGG(K/R)G(I/M)R motif [21]. A BLAST search was conducted to identify proteins containing this motif, revealing N-CoR as a candidate for binding to HLCS (see the Results section). N-CoR ushers histone deacetylases HDAC1 and HDAC2 to N-CoR docking sites, thereby facilitating histone deacetylation and a repressive chromatin environment [22–24]. The amino acid sequences in HDAC1 and HDAC2 are 83% identical, and the sequences in the catalytic core domain and the C-terminal tail are nearly identical [26]. Subsequent experiments used HDAC1 as a model for deacetylases.
Cell lines and HLCS overexpression
HEK (human embryonic kidney)-293 cells (A.T.C.C.) were cultured in DMEM (Dulbecco’s modified Eagle’s medium; Thermo Scientific) containing 10% FBS, 1% l-glutamine, 1% penicillin/streptomycin and 0.1% sodium pyruvate. HLCS-overexpression cells were created by transfecting HEK-293 cells with the plasmid FLAG/Myc-HLCS using electroporation as described previously [20]. The expression of HLCS was assessed by Western blot analysis and qPCR (quantitative real-time PCR) as described previously using the PCR primer pair denoted ‘25’ given in Supplementary Table S1 (http://www.biochemj.org/bj/461/bj4610477add.htm) [7].
Plasmids
Full-length human HLCS was subcloned from plasmid pGBKT7-HLCS [27] into vectors pCMV-Myc and pCMV-HA (Clontech) using SfiI and SalI, thereby creating the plasmids pCMV-Myc-HLCS and pCMV-HA-HLCS. Previous studies suggest the existence of four distinct domains in human HLCS [27]. Myc-tagged overexpression plasmids coding for the individual domains were created as described previously [27], using plasmid pGBKT7-HLCS as a template, PCR primer pairs 1, 2, 3 and 4, and vector pCMV-Myc as acceptor, thereby creating the plasmids pCMV-Myc-HLCS-NT (N-terminus, Ser2–Phe446), pCMV-Myc-HLCS-CD (central domain, Phe471–S575), pCMV-Myc-HLCS-L (linker domain, Thr610–Val668) and pCMV-Myc-HLCS-CT (C-terminus, His669–Arg718) respectively. Plasmid pET41a-HLCS codes for GST (glutathione transferase)-tagged recombinant HLCS and was prepared as described previously using the PCR primer pair 5 [19].
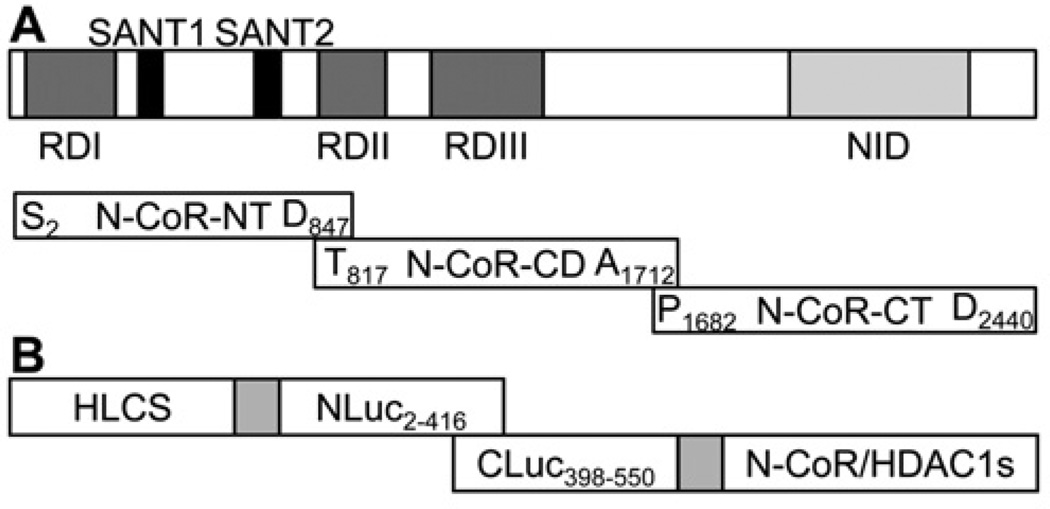
N-CoR is a 270-kDa protein, containing three autonomous repression domains and a conserved bipartite nuclear receptor interaction domain [28,29]. In order to achieve reasonable transfection efficiencies (through keeping plasmids reasonably small) and to assign putative interactions with HLCS to distinct domains in N-CoR, the following three overlapping fragments of N-CoR were cloned using HEK-293 cell cDNA as template (Figure 2A): NT (N-terminal domain), CD (central domain) and CT (C-terminal domain). PCRs were conducted using primers 6 (NT), 7 (CD), and 8 (CT); PCR products were digested using EcoRI and KpnI and cloned into the vectors pCMV-Myc and pCMV-HA, thereby creating the plasmids pCMV-Myc-N-CoR-NT, pCMV-Myc-N-CoR-CD, pCMV-Myc-N-CoR-CT, pCMV-HA-N-CoR-NT, pCMV-HA-N-CoR-CD and pCMV-HA-N-CoR-CT. Lysine to arginine mutations and GGGG(K/R)G(I/M)R motif deletion were created using pCMV-Myc-N-CoR-CT as the template and the GENEART Site-Directed Mutagenesis System (Invitrogen), thereby creating the plasmids pCMV-Myc-N-CoRCTK2323R (primer pair 9), pCMV-Myc-N-CoR-CTK2325R (primer pair 10) and pCMV-Myc-N-CoR-CTdel (primer pair 11). For the preparation of recombinant proteins, pCMV-Myc-N-CoR-NT, pCMV-Myc-N-CoR-CD and pCMV-Myc-N-CoR-CT were PCR amplified using primer pairs 12, 13 and 14, and the three N-CoR domains were subcloned into the pET41a vector using EcoRI and SacI, thereby creating the plasmids pET41a-N-CoR-NT, pET41a-N-CoR-CD and pET41a-N-CoR-CT respectively.
Figure 2. Schematic diagrams of domains in N-CoR and luciferase constructs used in the split luciferase complementation assays.
(A) The N-terminus of N-CoR contains transcriptional repression domains (RDs) responsible for the recruitment of additional components of the co-repressor complex such as HDAC, mSin3 and GPS2 (G-protein-pathway suppressor 2). A pair of potent repressor motifs known as SANT motifs (SWI3, ADA2, N-CoR and TFIIIB) is positioned between the repression domains. SANT motifs recruit HDAC3 and histones to the repressor complex in order to enhance HDAC3 activity. The C-terminus of N-CoR includes a nuclear receptor interaction domain (NID), which binds ligand-free nuclear receptors. In order to assign putative interactions with HLCS to distinct domains in N-CoR, three overlapping fragments of N-CoR were cloned, N-terminal domain (NT), central domain (CD) and C-terminal domain (CT). (B) In split luciferase complementation assays, N-terminal and C-terminal fragments are fused to interacting proteins. Physical interactions between the two proteins reconstitute luciferase activity.
HDAC1 and its novel splicing variant HDAC1Δ31, which lacks the entire 31 amino acids encoded by exon 7 (D. Liu, and J. Zempleni, unpublished work), were cloned from HEK-293 cell cDNA into the vectors pCMV-Myc and pCMV-HA using PCR primer pair 15 and EcoRI and KpnI, thereby creating the plasmids pCMV-Myc-HDAC1, pCMV-Myc-HDAC1Δ31, pCMV-HA-HDAC1 and pCMV-HA-HDAC1Δ31. HDAC ORFs were subcloned into the pET41a vector using PCR primer pair 16, and EcoRI and SacI to create the plasmids pET41a-HDAC1 and pET41a-HDAC1Δ31 for preparing recombinant HDAC1 and HDAC1Δ31 respectively.
Two fragments of Photinus pyralis firefly luciferase (N-terminus, Glu2–Gly416, and C-terminus, Met398–Cys550) were cloned and used to create split luciferase reporter plasmids. The N-terminal fragment was cloned using plasmid pGL3-Control (Promega) as a template (primer pair 17) and inserted into pCMV-Myc, pCMV-Myc-HLCS and pCMV-Myc-TP53 using KpnI and NotI, thereby creating the plasmids pCMV-Myc-NLuc, pCMV-Myc-HLCS-NLuc and pCMV-Myc-TP53-NLuc. The C-terminal fragment was also cloned using plasmid pGL3-Control as a template (primer pair 18) and inserted into pCMV-Myc, pCMV-Myc-N-CoR-NT, pCMV-Myc-N-CoR-CD,pCMV-Myc-N-CoR-CT,pCMV-Myc-HDAC1, pCMV-Myc-HDAC1Δ31, pCMV-Myc-MDM2 and pCMV-Myc-CDK3 using SfiI and EcoRI, thereby creating the plasmids pCMV-Myc-CLuc, pCMV-Myc-N-CoR-NT-CLuc, pCMV-Myc-N-CoR-CD-CLuc, pCMV-Myc-N-CoR-CT-CLuc, pCMV-Myc-HDAC1-CLuc, pCMV-Myc-HDAC1Δ31-CLuc, pCMV-Myc-MDM2-CLuc and pCMV-Myc-CDK3-CLuc respectively (Figure 2B).
The following control plasmids were created for the split luciferase complementation assay. The human tumour suppressor TP53 (tumour protein p53) was cloned from HEK-293 cell cDNA into vector pCMV-Myc using primer pair 19 and SfiI and SalI, thereby creating the plasmid pCMV-Myc-TP53. The E3 ubiquitin-protein ligase MDM2 (murine double minute 2) and CDK3 (cyclin-dependent kinase 3) were cloned from HEK-293 cell cDNA into the vector pCMV-Myc using EcoRI and XhoI, thereby creating the plasmids pCMV-Myc-MDM2 (primer pair 20) and pCMV-Myc-CDK3 (primer pair 21).
Co-immunoprecipitation assays
HEK-293 cells were co-transfected with equal amounts of overexpression and control vectors in the permutations as described in the Results section, using TurboFect™ reagent (Thermo Scientific) according to the manufacturer’s instructions. A total of 4×106 cells were collected 24 h after transfection and lysed in 20 mM Tris/HCl buffer (pH 8.0), containing 137 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA, PMSF and protease inhibitor cocktail (Sigma–Aldrich). Target proteins were precipitated with mouse anti-Myc antibody (Origene), rabbit anti-HA (haemagglutinin) antibody (Abcam) and settled Protein A beads (Pierce). Precipitated proteins were analysed by Western blotting with the antibodies listed in the Results section. Non-transfected HEK-293 cells were used as negative controls.
Interactions between endogenous proteins were assessed as follows. HEK-293 cells (8×106) were suspended in 3 ml of PBS and proteins were cross-linked with 37% formaldehyde (final concentration 1.8%). After lysis with RIPA buffer (Boston Bioproducts) containing protease inhibitor cocktail for 1 h, samples were incubated with rabbit anti-(human HLCS) antibody [7] or anti-(rabbit IgG) antibody (Santa Cruz Biotechnology) at 4°C overnight, followed by incubation with about 50 µl of settled Protein A/G–agarose beads (Thermo Scientific) at 4°C for 3 h. Beads were washed with RIPA buffer to remove non-specifically bound proteins before antibody-bound proteins were released through boiling for subsequent analysis by immune blotting.
Limited proteolysis assays
This assay is based on the principle that the proteolytic digestion of proteins by dilute proteases is slowed if two or more proteins interact physically (http://www.ihcworld.com/_protocols/lab_protocols/chazin-lab-protocols.htm). Recombinant proteins were overexpressed in Arctic Express (DE3) competent cells (Stratagene) and purified using GSTrap HP Columns (GE Healthcare) according to the manufacturer’s instructions. The GST tag was removed using thrombin, and the identity of recombinant proteins was assessed by Western blot analysis. Limited proteolysis assays were performed using recombinant HLCS and equal amounts of recombinant N-CoR fragments, HDAC1 or HDAC1Δ31 in 150 mM Tris/HCl acetate buffer (pH 7.5), containing 0.6 mM DTT and 90 mM MgCl2. After 1 h of pre-incubation at 37°C, aliquots were collected before the addition of trypsin (10 ng of trypsin/µg of recombinant protein; Sigma–Aldrich), and equal volumes of aliquots were collected at timed intervals after initiation of digestion. Digestion of proteins was visualized by gel electrophoresis and staining with Coomassie Blue.
Split luciferase complementation assays
The split luciferase complementation assay is based on the principle that luciferase produces chemiluminescence only if its N- and C-termini are in close physical proximity [31]. If the N- and C-terminal fragments of luciferase are fused to proteins, chemiluminescence will be produced only if the two fusion proteins interact physically, thereby bringing the luciferase termini in close physical proximity. HEK-293 cells were co-transfected with equal amounts of luciferase fusion constructs using TurboFect™ reagent. Cells were collected 48 h after transfection and the cell suspension was mixed with an equal volume of LucLite substrate (Promega). Luminescence was quantified using a microplate scintillation and luminescence counter (Packard) and normalized by β-gal activity as described previously [32]. Fusion constructs of TP53 andMDM2 were used as positive controls [33] and fusion constructs of TP53 and CDK3 were used as negative controls [34].
Biotinylation of HLCS-binding proteins by recombinant HLCS
There is precedent for the HLCS-dependent covalent binding of biotin to chromatin proteins such as EHMT-1 and histones [19,21]. We determined whether putative HLCS-interacting proteins are potential targets for biotinylation by HLCS. Briefly, recombinant N-CoR fragments, HDAC1 and HDAC1Δ31 were incubated with recombinant HLCS in biotinylation buffer and protein-bound biotin was probed with an anti-biotin antibody (Abcam) as described previously [35]. Negative controls were created by omitting HLCS, N-CoR or HDAC1. Equal loading was confirmed using Coomassie Blue.
ChIP assay
The enrichment of H3K9ac (Lys9-acetylated histone H3) marks in LTRs (long terminal repeats) 15 and 22 and pericentromeric alpha satellite repeats in chromosomes 1 (Chr1alpha) and 4 (Chr4alpha) in HLCS-overexpressing cells was assessed by ChIP as described in [11,36]. ChIP-grade anti-H3K9ac (Abcam) and anti-H3 (Abcam) antibodies were used to precipitate chromatin associated with H3K9ac marks and to normalize for nucleosomal occupancy respectively. The promoter regulating the expression of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) localizes in euchromatin and was used as the control locus. Data are expressed as a percentage of input DNA.
qPCR
The abundance of mRNA coding for LTRs 15 and 22 and Chr1alpha and Chr4alpha was quantified by qPCR using Power SYBR Green PCR Master Mix (Applied Biosystems). PerfeCTa SYBR Green FastMix (Quanta Biosciences) was used to quantify the abundance of amplicons in immunoprecipitated chromatin [11]. The PCR primer pairs used are listed in Supplementary Table S1 [37,38].
Statistics
Data were tested for normality of distribution by Komolgorov–Smirnov normality test. Data from split luciferase complementation assays were tested for homogeneity of variances by Bartlett’s test and analysed for significance of difference by one-way ANOVA. Data from ChIP and qPCR were tested for homogeneity of variances by F-test and analysed by Student’s t test (http://support.sas.com/documentation/cdl/en/statug/63033/HTML/default/viewer.htm). Variances of data from qPCR testing HLCS expression levels were heterogeneous and therefore the conservative Mann–Whitney U test was used for analysis. All analyses and data points are based on three biologically independent repeats. StatView 5.0.1 (SAS Institute) was used to perform all calculations. Differences were considered significant if P < 0.05. Data are expressed as means ± S.D.
RESULTS
In silico predictions
In silico predictions suggested that the GGGG(K/R)G(I/M)R sequence in N-CoR is a candidate for mediating physical interactions with HLCS and that, because of the known interactions between N-CoR and HDAC1, the latter might also be a candidate for interacting physically with HLCS. N-CoR had a score of 17.2 bits and an E value of 393, compared with scores of 20.2 bits and 16.3 bits and E values of 57 and 634 for the HLCS-interacting proteins propionyl-CoA carboxylase and EHMT-1 respectively.
Interactions between HLCS and N-CoR
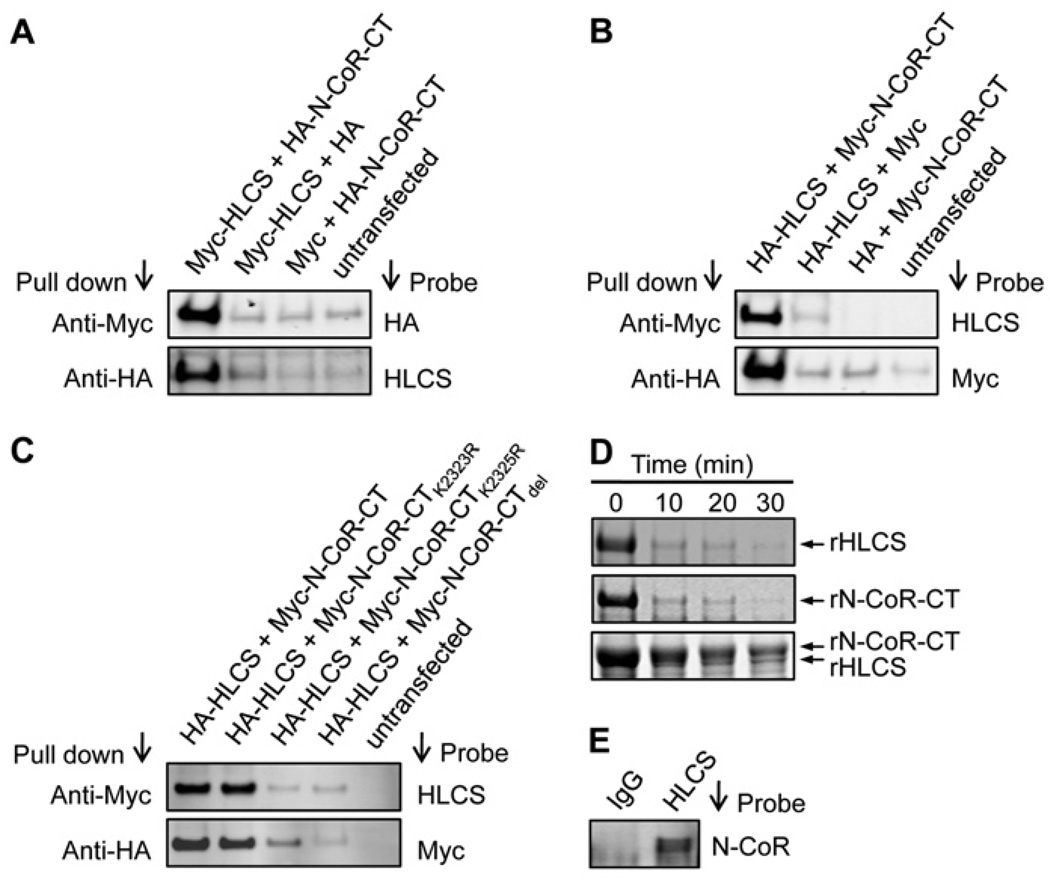
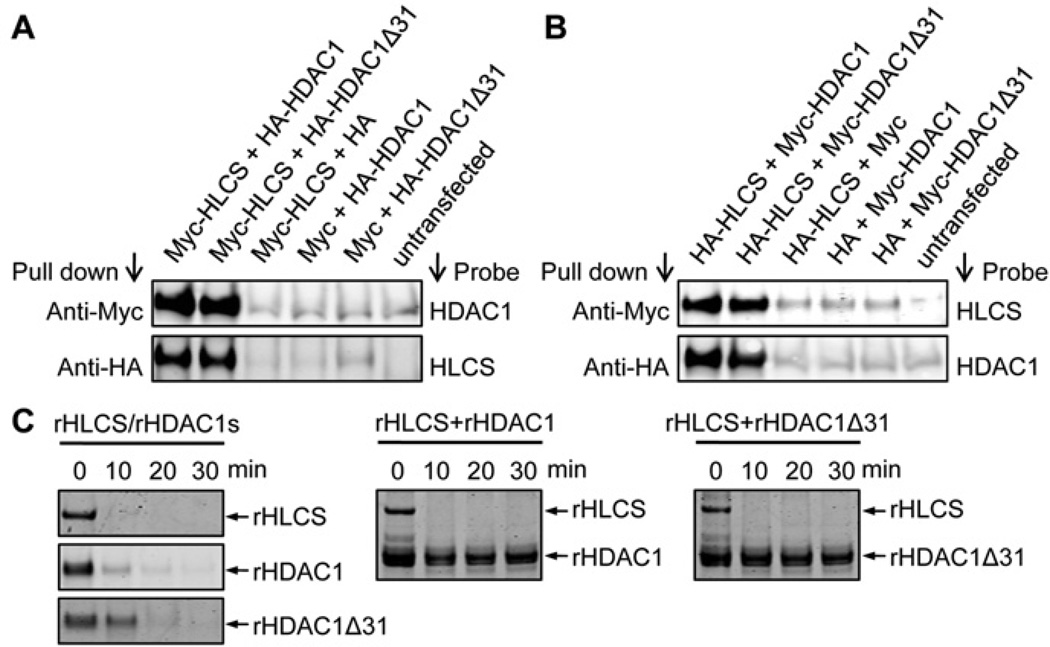
HLCS interacts with the C-terminus in N-CoR, based on the following lines of evidence. HEK-293 cells were co-transfected with plasmid pCMV-Myc-HLCS and plasmids coding for the three N-CoR fragments, i.e. pCMV-HA-N-CoR-NT, pCMV-HA-N-CoR-CD or pCMV-HA-N-CoR-CT. When cell lysates were precipitated with an anti-Myc antibody and probed with an anti-HA antibody, a distinct signal was obtained only for cells transfected with the plasmid coding for the N-CoR C-terminus, pCMV-HA-N-CoR-CT (Figure 3A, upper panel), but not for those transfected with pCMV-HA-N-CoR-NT and pCMV-HA-N-CoR-CD (results not shown). The same pattern was seen when cell lysates were precipitated with an anti-HA antibody and probed with an anti-HLCS antibody (Figure 3A, lower panel, and results not shown). Empty vectors in various permutations and non-transfected cells produced no detectable signal (negative controls). Next, tags were swapped and HEK-293 cells were co-transfected with pCMV-HA-HLCS and either pCMV-Myc-N-CoR-NT, pCMV-Myc-N-CoR-CD or pCMV-Myc-N-CoR-CT. Again, only cells co-transfected with HLCS and the plasmid coding for the N-CoR C-terminus, pCMV-HA-N-CoR-CT, produced distinct signals in co-immunoprecipitation assays. No signal was produced by the negative controls (Figure 3B) and by cells co-transfected with HLCS and plasmids coding for the N-terminus and central domain in N-CoR (results not shown). In addition, interactions between endogenous HLCS and N-CoR were verified by co-immunoprecipitation. When cell lysates were precipitated with an anti-HLCS antibody and probed with an anti-N-CoR antibody, the signal produced was clearly distinct from the IgG controls (Figure 3E).
Figure 3. HLCS interacts physically with N-CoR.
(A) In co-immunoprecipitation assays, Myc–HLCS and HA–N-CoR-CT were overexpressed in HEK-293 cells and cell lysates were precipitated with anti-Myc or anti-HA antibodies. Proteins were resolved by electrophoresis and probed with anti-HA or anti-HLCS antibodies. Empty vectors in various permutations and non-transfected cells were used as negative controls. (B) As for (A), but tags were swapped. (C) HA-tagged HLCS and Myc-tagged wild-type or mutant N-CoR C-terminus were overexpressed in HEK-293 cells and cell lysates were precipitated with anti-Myc or anti-HA antibodies. Proteins were resolved by electrophoresis and probed with anti-HLCS or anti-Myc antibodies. Non-transfected cells were used as negative controls. (D) Limited proteolysis assays: recombinant (r) HLCS and N-CoR C-terminus were pre-incubated before trypsin digestion (bottom panel). Negative controls were generated by omission of HLCS or N-CoR C-terminus (top and middle panels). (E) Extracts from normal non-transfected HEK-293 cells were precipitated using anti-HLCS antibody and probed using anti-N-CoR antibody. An anti-IgG antibody was used as a control for the anti-HLCS antibody. Gels depict representative examples from three biological repeats.
Previous studies have suggested that lysine residues in HLCS-binding motifs are important for HLCS binding [21]. The predicted motif in N-CoR includes Lys2323 and Lys2325 in full-length N-CoR. When HEK-293 cells were co-transfected with the plasmid pCMV-HA-HLCS and plasmids coding for the wild-type N-CoR C-terminus (pCMV-Myc-N-CoR-CT), the Lys2323 mutant (pCMV-Myc-N-CoR-CTK2323R), the Lys2325 mutant (pCMV-Myc-N-CoR-CTK2325R) or the deletion construct (pCMV-Myc-N-CoRCTdel), meaningful signals were detected only for the wild-type N-CoR and the Lys2323 mutant, suggesting that Lys2325 and the HLCS-binding motif are essential for mediating interactions with HLCS (Figure 3C, upper panel). The same pattern was seen when cell lysates were precipitated with an anti-HA antibody and probed with an anti-Myc antibody (Figure 3C, lower panel).
Limited proteolysis assays were conducted using recombinant proteins. When recombinant HLCS and the C-terminal fragment of N-CoR were incubated with trypsin in the absence of putative binding partners, both proteins degraded completely within 10 min of incubation (Figure 3D, top and middle panel). In contrast, when recombinant HLCS was pre-incubated with the N-CoR C-terminal fragment before trypsin treatment, the proteolytic degradation of both HLCS and N-CoR was substantially delayed compared with the individual proteins, and strong protein signals were detectable after 30 min of incubation with trypsin (Figure 3D, bottom panel). When recombinant HLCS was pre-incubated with the N-CoR N-terminus and central domain, tryptic digestion of the proteins was not delayed (results not shown). Note that the molecular masses of recombinant HLCS (~81 kDa) and the N-CoR C-terminus (~82 kDa) are similar, making it difficult to distinguish the two proteins at early time points of incubation (≤20 min) when protein concentrations are high.
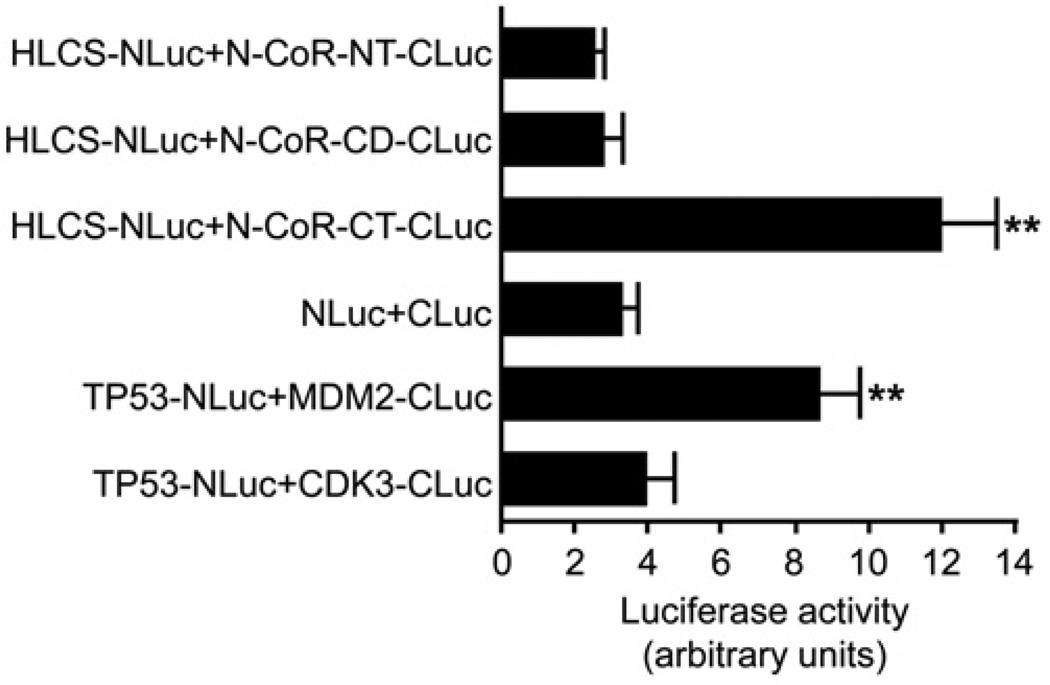
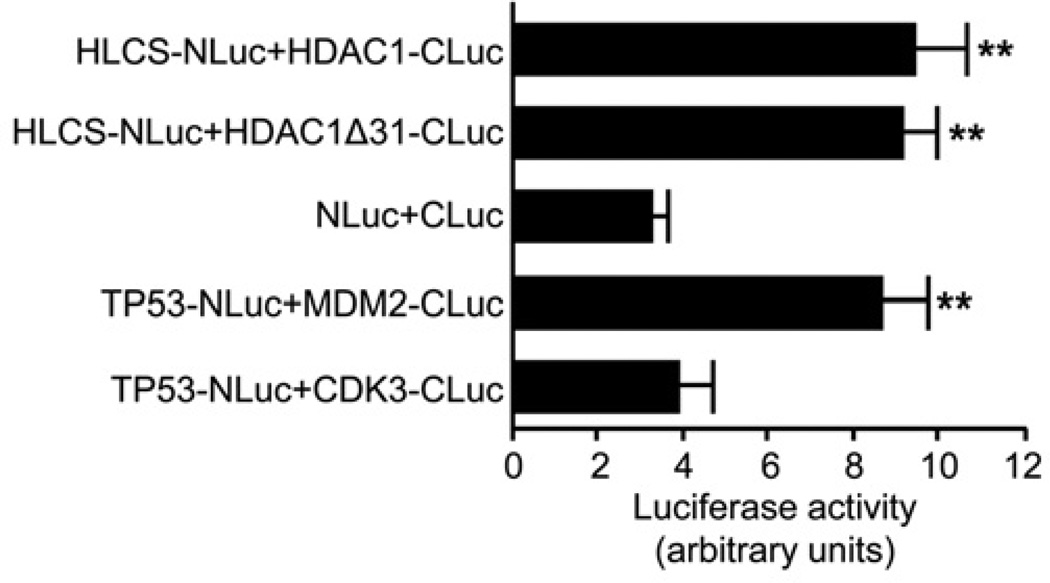
Split luciferase complementation assays were conducted to confirm the interactions between HLCS and N-CoR fragments. When co-expressed in HEK-293 cells, the constructs HLCS-NLuc and N-CoR-CT-CLuc produced a strong chemiluminescence signal, whereas HLCS-NLuc with N-CoR-NT-CLuc or N-CoR-CD-CLuc produced signals that were equivalent to the background signal produced by co-expressing the N- and C-terminal fragments of luciferase not fused to protein (Figure 4). The positive (TP53-NLuc plus MDM2-CLuc) and negative (TP53-NLuc plus CDK3-CLuc) controls produced the expected results. Specifically, the signal produced by co-expression of the constructs HLCS-NLuc and N-CoR-CT-CLuc was approximately 3.7-fold the signal produced by cells that were co-transfected with unfused NLuc and CLuc fragments (background control for self-association) and approximately 3.1-fold the signal produced by cells transfected with TP53-NLuc and CDK3-CLuc.
Figure 4. Split luciferase complementation assays for detection of HLCS and N-CoR interaction in HEK-293 cells.
Fusion constructs of TP53 and MDM2 were used as positive controls and TP53 and CDK3 were used as negative controls. Values are means ± S.D. of independent experiments (n = 3). **P < 0.01 compared with the background control for self-association and negative control.
Interactions between HLCS and HDAC1
HLCS interacts physically with full-length HDAC1 and the novel splicing variant HDAC1Δ31, based on the following lines of evidence. HEK-293 cells were co-transfected with pCMV-Myc-HLCS and either pCMV-HA-HDAC1 or pCMV-HA-HDAC1Δ31. When cell lysates were precipitated with an anti-Myc antibody and probed with an anti-HDAC1 antibody (Abcam), distinct signals were obtained for both HDAC1 and HDAC1Δ31 (Figure 5A, upper panel). Likewise, when cell lysates were precipitated with an anti-HA antibody and probed with an anti-HLCS antibody, distinct signals were obtained for HLCS (Figure 5A, lower panel). Empty vectors in various permutations and non-transfected cells produced no detectable signal (negative controls). Next, tags were swapped and HEK-293 cells were co-transfected with pCMV-HA-HLCS and either pCMV-Myc-HDAC1 or pCMV-Myc-HDAC1Δ31. Again, only cells co-transfected with HLCS and HDAC1 or HDAC1Δ31 produced distinct signals, whereas no signal was produced by negative controls (Figure 5B).
Figure 5. HLCS interacts physically with HDAC1.
(A) In co-immunoprecipitation assays, Myc–HLCS and HA–HDAC1 were overexpressed in HEK-293 cells and cell lysates were precipitated with anti-Myc or anti-HA antibodies. Proteins were resolved by electrophoresis and probed with anti-HDAC1 or anti-HLCS antibodies. Empty vectors in various permutations and non-transfected cells were used as negative controls. (B) As for (A), but tags were swapped. (C) Limited proteolysis assays. Recombinant (r) HLCS and HDAC1 were pre-incubated before trypsin digestion (middle and right-hand panels). Negative controls were generated by omission of HLCS or HDAC1 (left-hand panel). Gels depict representative examples from three biological repeats.
Limited proteolysis assays were conducted with recombinant HLCS and HDAC1. Recombinant HLCS was degraded within 10 min of incubation with trypsin if no HDAC1 was added to the sample. Similarly, HDAC1 and HDAC1Δ31 were completely degraded within 20 min of trypsin treatment if no HLCS was present (Figure 5C, left-hand panel). When recombinant HLCS was pre-incubated with either HDAC1 or HDAC1Δ31 before trypsin treatment, the proteolytic degradation of both HDAC1 and HDAC1Δ31 was substantially delayed compared with the individual treatments, and strong protein signals were detectable even after 30 min of incubation with trypsin (Figure 5C, middle and right-hand panels). The proteolytic degradation of HLCS was not delayed in samples containing HDAC1 (see the Discussion).
Split luciferase complementation assays confirmed that HLCS interacts with HDAC1. HEK-293 cells were co-transfected with the constructs HLCS-NLuc and either HDAC1-CLuc or HDAC1Δ31-CLuc. Co-expression resulted in a strong chemiluminescence signal for both HDAC constructs, which was significantly higher than that of controls (Figure 6). The signal was approximately 2.9-fold the signal produced by cells that were co-transfected with unfused NLuc and CLuc fragments (background control for self-association) and approximately 2.4-fold the signal produced by cells transfected with TP53-NLuc and CDK3-CLuc (negative controls).
Figure 6. Split luciferase complementation assays for detection of HLCS and HDAC1 interaction in HEK-293 cells.
Fusion constructs of TP53 and MDM2 were used as positive controls and TP53 and CDK3 were used as negative controls. Values are means ± S.D. of independent experiments (n = 3). **P < 0.01 compared with the background control for self-association and negative control.
The roles of HLCS domains in mediating interactions with N-CoR and HDAC1
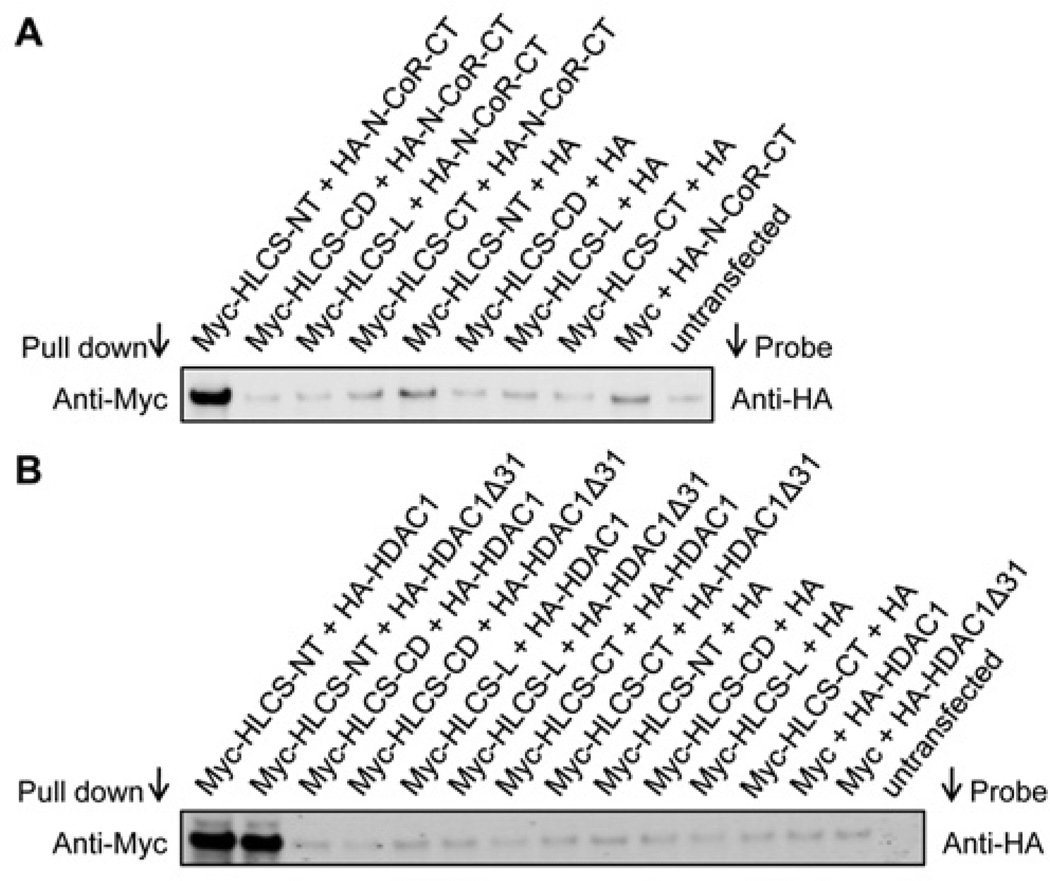
The N-terminus in HLCS is essential for mediating physical interactions with N-CoR and HDAC1. HEK-293 cells were co-transfected with pCMV-Myc constructs coding for the four known HLCS domains [27] and either pCMV-HA-N-CoR-CT, pCMV-HA-HDAC1 or pCMV-HA-HDAC1Δ31. When cell lysates were precipitated with an anti-Myc antibody and probed with an anti-HA antibody, distinct signals were obtained only for cells co-transfected with pCMV-Myc-HLCS-NT and the N-CoR C-terminus or HDAC1 constructs, but not for cells in which HLCS domains other than the N-terminus were overexpressed and also not for the various negative controls (Figure 7).
Figure 7. The roles of HLCS domains in mediating interactions with N-CoR and HDAC1.
Myc-tagged HLCS and HA-tagged N-CoR C-terminus (A) or HDAC1 (B) were overexpressed in HEK-293 cells. Cell lysates were precipitated with an anti-Myc antibody and probed with an anti-HA antibody. Non-transfected cells were used as negative controls. Gels depict representative examples from three biological repeats.
HLCS-dependent biotinylation of N-CoR and HDAC1
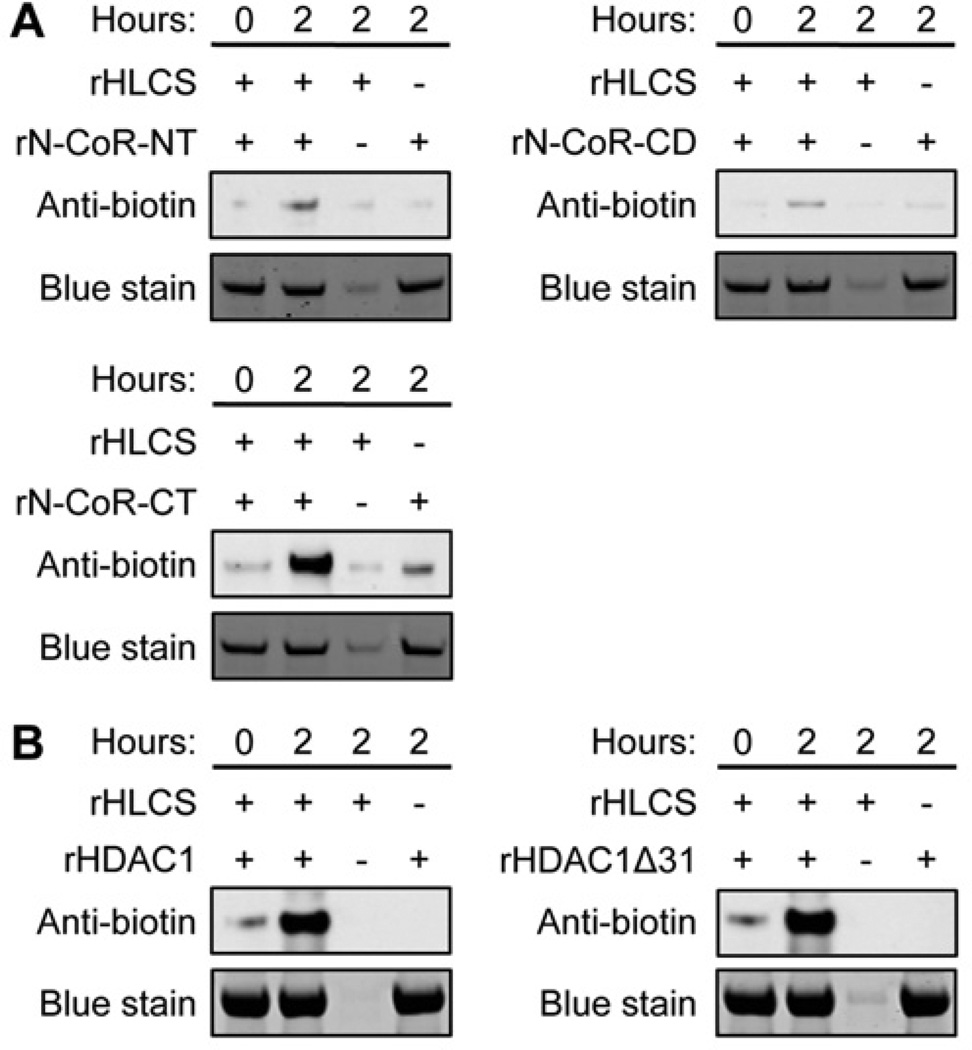
Recombinant HLCS biotinylates recombinant N-CoR and HDACs in vitro, consistent with the promiscuous nature of HLCS and its microbial orthologue BirA [6,19,21] (see the Discussion). When recombinant HLCS was incubated with recombinant N-CoR N-terminus, central domain and C-terminus for 2 h, protein-bound biotin was detected in the N-CoR C-terminus (Figure 8A, lower panel) and, to a much lesser extent, in the other domains of N-CoR (upper panel) using an anti-biotin antibody as a probe. When recombinant HLCS was incubated with HDAC1 or HDAC1Δ31 for 2 h, protein-bound biotin was detected in both HDAC1 and HDAC1Δ31 (Figure 8B). Negative controls produced biotin signals that were negligible or not detectable.
Figure 8. N-CoR and HDAC1 are targets for biotinylation of HLCS.
(A) Recombinant N-terminus, central domain and C-terminus in N-CoR were incubated with recombinant (r) HLCS, biotin and cofactors for biotinylation for up to 2 h. N-CoR-bound biotin was detected with an anti-biotin antibody. Negative controls were created by omitting N-CoR fragments or HLCS. Equal loading was confirmed by Coomassie Blue stain. (B) Similarly, biotinylation of recombinant HDAC1 by HLCS was conducted. Gels depict representative examples from three biological repeats.
Transcriptional regulation of repeats by HLCS
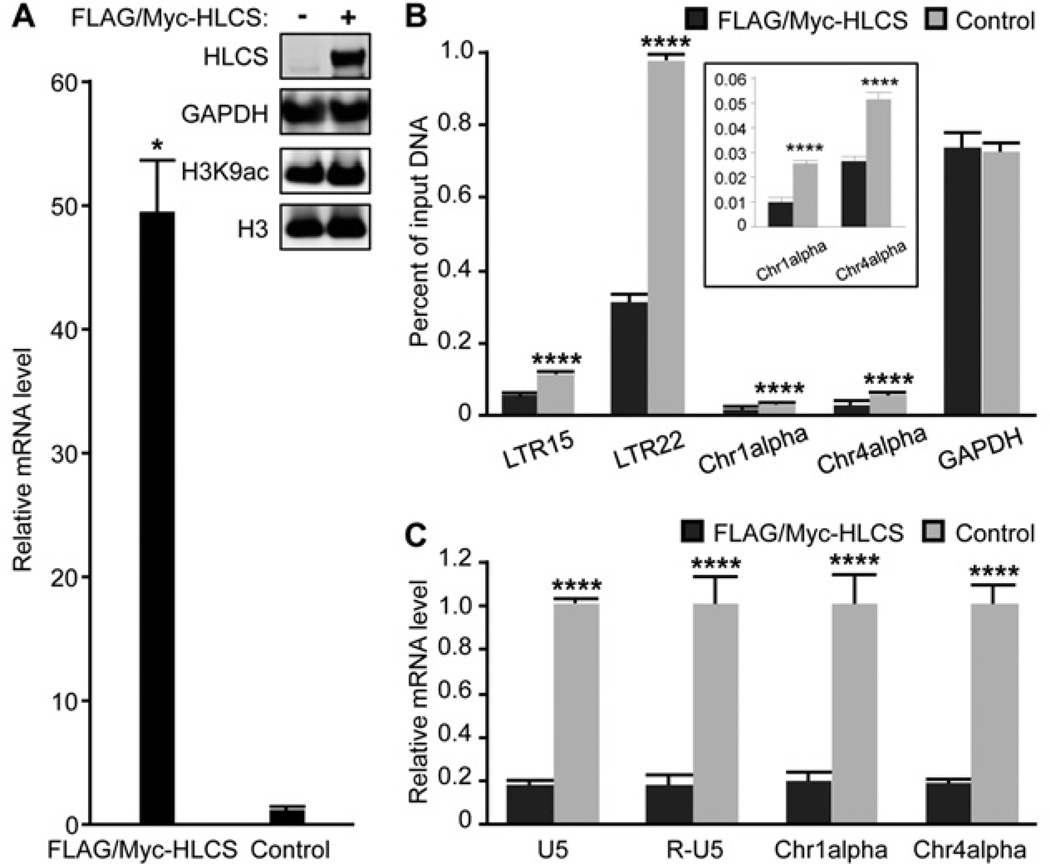
The abundance of H3K9ac marks in repeats depended on HLCS in HEK-293 cells. HLCS was overexpressed using plasmid p3XFLAG-Myc-CMV-26-HLCS [20] and the efficacy of HLCS overexpression was assessed at the mRNA and protein levels. The abundance of mRNA coding for HLCS was approximately 49-fold greater in overexpression cells compared with the non-transfected controls and the increase in protein abundance was equally compelling (Figure 9A and insert). Although HLCS overexpression caused no meaningful change in the global abundance of H3K9ac marks in whole-cell extracts (Figure 9A, insert), there was a considerable loss of H3K9ac marks in the repeats.
Figure 9. Transcriptional repression of repeats by HLCS overexpression.
(A) Transfection with the plasmid p3XFLAG-Myc-CMV-26-HLCS produced a stable overexpression of HLCS in HEK-293 cells, judged by qPCR (n = 3; *P < 0.05 compared with the untransfected controls) and Western blot analysis. GAPDH and histone H3 were used as loading controls. (B) The enrichment of H3K9ac marks in loci coding for LTR15, LTR22, Chr1alpha, Chr4alpha and GAPDH in HLCS-overexpressing HEK-293 cells and controls (n = 3). ****P < 0.0001 compared with the FLAG/Myc–HLCS controls. The insert depicts a magnified version of the Chr1alpha and Chr4alpha data. (C) The abundance of transcripts coding for LTRs, Chr1alpha and Chr4alpha in HLCS-overexpressing HEK-293 cells and controls. Values are means ± S.D. of independent experiments (n = 3). ****P < 0.0001 compared with the FLAG/Myc–HLCS controls.
HLCS overexpression caused a decrease in H3K9ac marks in LTR15, LTR22, Chr1alpha and Chr4alpha compared with the HLCS-normal controls (Figure 9B, including a magnified view of the Chr1alpha and Chr4alpha data). The decreases amounted to the loss of 50%, 68%, 62% and 50% of H3K9ac marks in LTR15, LTR22, Chr1alpha and Chr4alpha respectively. The abundance of H3K9ac marks did not change in the GAPDH promoter in euchromatin, consistent with a locus-specific effect of HLCS.
The HLCS-dependent depletion of H3K9ac marks in LTRs caused an 83% decrease in the global abundance of mRNA coding for LTRs in HEK-293 cells (Figure 9C), using primers that do not distinguish among the transcriptionally active LTR in the human genome [14]. Again, the effects of HLCS overexpression on transcriptional activity were not specific for LTRs, but were also seen in the Chr1alpha and Chr4alpha repeats for which the abundance of transcripts decreased by 81% and 82% respectively, compared with the HLCS-normal controls.
DISCUSSION
This is the first report suggesting that HLCS interacts physically with N-CoR and HDAC1, thereby promoting the repression of repeats through removing H3K9ac marks. The present study supports a model in which HLCS and biotin participate in the repression of genes at the epigenetic level through orchestrating the assembly of a multi-protein gene repression complex that integrates DNA methylation, histone H3 methylation and histone deacetylation events [20,21]. There is high confidence that the reported interactions are real, based on the following lines of reasoning. First, the interactions reported in the present study are consistent predictions by a protocol for identifying HLCS-binding proteins in silico [21]. Secondly, the in silico predictions were validated experimentally using three independent procedures, i.e. co-immunoprecipitation assays, limited proteolysis assays and split luciferase complementation assays. Thirdly, the interaction between HLCS and the C-terminus in N-CoR was abrogated when the HLCS-binding motif was altered by point mutation or deletion. Fourthly, the N-CoR C-terminus, HDAC1 and HDAC1Δ31 are targets for biotinylation by HLCS in vitro. Fifthly, the interactions between HLCS and other chromatin proteins is mediated by the HLCS N-terminus, which is also responsible for mediating interactions between HLCS and its classical carboxylase substrate [27,40]. Sixthly, overexpression of HLCS causes a substantial locus-specific loss of H3K9ac marks in LTRs and pericentromeric alpha satellite repeats. In addition, this report is consistent with previous studies in healthy adults, D. melanogaster and mammalian cell cultures suggesting that the repression of LTRs and select other loci depend on HLCS and biotin [10,11,13,14,41].
On the basis of the present study, it is a plausible assumption that loss of HLCS or deficiency of biotin may cause a derepression of repeats, which may impair genome stability. For example, LTRs make up approximately 8% of the human genome and at least 51 LTRs are transcriptionally competent [38]. Repetitive elements, such as LTRs, pose a burden to genome stability, as their mobilization facilitates recombination between non-homologous loci leading to chromosomal deletions and translocations [42,43]. Mobilization of LTR transposons is associated with 10% of all spontaneous mutations in mice [44]. The transcriptional activity of LTRs is controlled by histone acetylation and other epigenetic marks; inhibition of HDACs leads to an increase in LTR transcription [45]. Derepression of LTRs may impair genome stability through insertional mutagenesis, recombination events that cause translocations and other rearrangements, deregulation of genes in the host genome mediated by LTR promoter activity, and antisense effects if transcription extends into the exon sequence downstream of the transposon [46].
Our discovery that N-CoR and HDAC1 are targets for biotinylation of HLCS is also of great interest in the field of protein biotinylation. Previous studies suggest that biotinylation sites in the histone methyltransferase EHMT-1 are important for strengthening HLCS–EHMT-1 interaction [21]. The same appears to be the case for Lys2325 in N-CoR which was important for mediating HLCS–N-CoR interactions in limited proteolysis assays. We are in no position to formally conclude that biotinylation of Lys2325 in N-CoR is important for mediating these interactions, because we do not know the exact biotinylation site(s) in N-CoR. However, it is becoming increasingly clear that HLCS and its microbial orthologue BirA are fairly promiscuous enzymes that catalyse the biotinylation of proteins other than their classical substrates, i.e. the five mammalian biotin-dependent carboxylases [6,19,21]. We have discovered 108 novel biotinylated proteins in human cells (Y. Li, unpublished work), but think it is noteworthy that not all lysine-containing peptides and proteins are substrates for biotinylation by HLCS [35], suggesting that some of these biotinylation events have biological importance. For example, we have reported preliminary findings that HLCS biotinylates Lys10 and Lys12 in MBP-1 (c-Myc promoter-binding protein-1), and that biotinylation of MBP-1 decreases the expression of the c-Myc and cyclo-oxygenase 2 oncogenes [47].
Some uncertainties remain. First, we do not know for certain that biotinylation of N-CoR and HDAC1 contributes towards strengthening the binding between HLCS and other proteins. Theoretically, it is possible that point mutations and deletion can have the same effect without depending on changes in protein biotinylation. Secondly, the HLCS–HDAC interaction caused a considerable decrease in the proteolytic degradation of HDAC1 in limited proteolysis assays, but did not slow the degradation of HLCS. We speculate that target sites for proteolytic degradation are exposed on the surface of HLCS upon interaction with HDAC1. Unfortunately, no 3D structures are available for human HLCS, making it difficult to test the validity of our speculation. Thirdly, we do not know yet whether HLCS also interacts with HDAC2, which has a high sequence similarity compared with HDAC1 [26].
We conclude that HLCS exerts some of its roles in gene regulation through the formation of a multi-protein gene repression complex in human chromatin. Possible members of this complex include proteins involved in DNA methylation, EHMT-1, N-CoR and HDAC1. This model also suggests that the roles of HLCS in metabolism go far beyond that of a ligase that attaches biotin to carboxylases. We are currently in the process of creating an HLCS-knockout mouse model that will allow us to shed some additional light on the roles of HLCS in cell biology and signalling.
Supplementary Material
Acknowledgments
FUNDING
This work was supported, in part, by the University of Nebraska Agricultural Research Division with funds provided through the Hatch Act. Additional support was provided by the NIH (National Institutes of Health) [grant numbers DK063945 and DK077816].
Abbreviations
- CDK3
cyclin-dependent kinase 3
- DNMT1
DNA methyltransferase 1
- EHMT-1
euchromatic histone-lysine N-methyltransferase-1
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GST
glutathione transferase
- HA
haemagglutinin
- HDAC
histone deacetylase
- HEK
human embryonic kidney
- H3K9ac
Lys9-acetylated histone H3
- H3K9me
Lys9-methylated histone H3
- HLCS
holocarboxylase synthetase
- LTR
long terminal repeat
- MBP-1
c-Myc promoter-binding protein-1
- MDM2
murine double minute 2
- MeCP2
methyl CpG-binding protein 2
- N-CoR
nuclear receptor co-repressor
- qPCR
quantitative real-time PCR
- TP53
tumour protein p53
Footnotes
AUTHOR CONTRIBUTION
Dandan Liu designed the research, conducted the experiments, analysed the data and wrote a draft version of the paper. Janos Zempleni designed the research, analysed the data and wrote the final paper. Both authors read and approved the final paper.
REFERENCES
- 1.Suzuki Y, Aoki Y, Ishida Y, Chiba Y, Iwamatsu A, Kishino T, Niikawa N, Matsubara Y, Narisawa K. Isolation and characterization of mutations in the human holocarboxylase synthetase cDNA. Nat. Genet. 1994;8:122–128. doi: 10.1038/ng1094-122. [DOI] [PubMed] [Google Scholar]
- 2.Zempleni J, Wijeratne SSK, Kuroishi T. Biotin. In: Erdman JW Jr, Macdonald I, Zeisel SH, editors. Present Knowledge in Nutrition. Washington: International Life Sciences Institute; 2012. pp. 587–609. [Google Scholar]
- 3.Camporeale G, Giordano E, Rendina R, Zempleni J, Eissenberg JC. Drosophila holocarboxylase synthetase is a chromosomal protein required for normal histone biotinylation, gene transcription patterns, lifespan and heat tolerance. J. Nutr. 2006;136:2735–2742. doi: 10.1093/jn/136.11.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh D, Pannier AK, Zempleni J. Identification of holocarboxylase synthetase chromatin binding sites using the DamID technology. Anal. Biochem. 2011;413:55–59. doi: 10.1016/j.ab.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao B, Rodriguez-Melendez R, Wijeratne SS, Zempleni J. Biotin regulates the expression of holocarboxylase synthetase in the miR-539 pathway in HEK-293 cells. J. Nutr. 2010;140:1546–1551. doi: 10.3945/jn.110.126359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobza K, Sarath G, Zempleni J. Prokaryotic BirA ligase biotinylates K4, K9, K18 and K23 in histone H3. BMB Reports. 2008;41:310–315. doi: 10.5483/bmbrep.2008.41.4.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N- and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J. Nutr. Biochem. 2006;17:225–233. doi: 10.1016/j.jnutbio.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, Zempleni J. K4, K9, and K18 in human histone H3 are targets for biotinylation by biotinidase. FEBS J. 2005;272:4249–4259. doi: 10.1111/j.1742-4658.2005.04839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur. J. Biochem. 2004;271:2257–2263. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 10.Rios-Avila L, Pestinger V, Zempleni J. K16-biotinylated histone H4 is overrepresented in repeat regions and participates in the repression of transcriptionally competent genes in human Jurkat lymphoid cells. J. Nutr. Biochem. 2012;23:1559–1564. doi: 10.1016/j.jnutbio.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pestinger V, Wijeratne SSK, Rodriguez-Melendez R, Zempleni J. Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes. J. Nutr. Biochem. 2011;22:328–333. doi: 10.1016/j.jnutbio.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wijeratne SS, Camporeale G, Zempleni J. K12-biotinylated histone H4 is enriched in telomeric repeats from human lung IMR-90 fibroblasts. J. Nutr. Biochem. 2010;21:310–316. doi: 10.1016/j.jnutbio.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gralla M, Camporeale G, Zempleni J. Holocarboxylase synthetase regulates expression of biotin transporters by chromatin remodeling events at the SMVT locus. J. Nutr. Biochem. 2008;19:400–408. doi: 10.1016/j.jnutbio.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chew YC, West JT, Kratzer SJ, Ilvarsonn AM, Eissenberg JC, Dave BJ, Klinkebiel D, Christman JK, Zempleni J. Biotinylation of histones represses transposable elements in human and mouse cells and cell lines, and in Drosophila melanogaster. J. Nutr. 2008;138:2316–2322. doi: 10.3945/jn.108.098673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camporeale G, Oommen AM, Griffin JB, Sarath G, Zempleni J. K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells. J. Nutr. Biochem. 2007;18:760–768. doi: 10.1016/j.jnutbio.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey LM, Ivanov RA, Wallace JC, Polyak SW. Artifactual detection of biotin on histones by streptavidin. Anal. Biochem. 2008;373:71–77. doi: 10.1016/j.ab.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Kuroishi T, Rios-Avila L, Pestinger V, Wijeratne SS, Zempleni J. Biotinylation is a natural, albeit rare, modification of human histones. Mol. Genet. Metab. 2011;104:537–545. doi: 10.1016/j.ymgme.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur. J. Biochem. 2001;268:5424–5429. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 19.Bao B, Pestinger VI, Hassan YI, Borgstahl GEO, Kolar C, Zempleni J. Holocarboxylase synthetase is a chromatin protein and interacts directly with histone H3 to mediate biotinylation of K9 and K18. J. Nutr. Biochem. 2011;22:470–475. doi: 10.1016/j.jnutbio.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue J, Wijeratne S, Zempleni J. Holocarboxylase synthetase synergizes with methyl CpG binding protein 2 and DNA methyltransferase 1 in the transcriptional repression of long-terminal repeats. Epigenetics. 2013;8:504–511. doi: 10.4161/epi.24449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Hassan YI, Moriyama H, Zempleni J. Holocarboxylase synthetase interacts physically with euchromatic histone-lysine N-methyltransferase, linking histone biotinylation with methylation events. J. Nutr. Biochem. 2013;24:1446–1452. doi: 10.1016/j.jnutbio.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 23.Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 24.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 25.Kouzarides T, Berger SL. Chromatin modifications and their mechanism of action. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor: Cold Spring Harbor Press; 2007. pp. 191–209. [Google Scholar]
- 26.Brunmeir R, Lagger S, Seiser C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int. J. Dev. Biol. 2009;53:275–289. doi: 10.1387/ijdb.082649rb. [DOI] [PubMed] [Google Scholar]
- 27.Hassan YI, Moriyama H, Olsen LJ, Bi X, Zempleni J. N- and C-terminal domains in human holocarboxylase synthetase participate in substrate recognition. Mol. Genet. Metab. 2009;96:183–188. doi: 10.1016/j.ymgme.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 29.Ishizuka T, Lazar MA. The nuclear receptor corepressor deacetylase activating domain is essential for repression by thyroid hormone receptor. Mol. Endocrinol. 2005;19:1443–1451. doi: 10.1210/me.2005-0009. [DOI] [PubMed] [Google Scholar]
- 30. Reference deleted. [Google Scholar]
- 31.Luker KE, Piwnica-Worms D. Optimizing luciferase protein fragment complementation for bioluminescent imaging of protein–protein interactions in live cells and animals. Methods Enzymol. 2004;385:349–360. doi: 10.1016/S0076-6879(04)85019-5. [DOI] [PubMed] [Google Scholar]
- 32.Griffin JB, Rodriguez-Melendez R, Zempleni J. The nuclear abundance of transcription factors Sp1 and Sp3 depends on biotin in Jurkat cells. J. Nutr. 2003;133:3409–3415. doi: 10.1093/jn/133.11.3409. [DOI] [PubMed] [Google Scholar]
- 33.ElSawy KM, Verma CS, Joseph TL, Lane DP, Twarock R, Caves LS. On the interaction mechanisms of a p53 peptide and nutlin with the MDM2 and MDMX proteins: a Brownian dynamics study. Cell Cycle. 2013;12:394–404. doi: 10.4161/cc.23511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsuji K, Mizumoto K, Yamochi T, Nishimoto I, Matsuoka M. Differential effect of ik3-1/cables on p53- and p73-induced cell death. J. Biol. Chem. 2002;277:2951–2957. doi: 10.1074/jbc.M108535200. [DOI] [PubMed] [Google Scholar]
- 35.Hassan YI, Moriyama H, Zempleni J. The polypeptide Syn67 interacts physically with human holocarboxylase synthetase, but is not a target for biotinylation. Arch. Biochem. Biophys. 2009;495:35–41. doi: 10.1016/j.abb.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dahl JA, Collas P. MicroChIP: a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res. 2008;36:e15. doi: 10.1093/nar/gkm1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovalskaya E, Buzdin A, Gogvadze E, Vinogradova T, Sverdlov E. Functional human endogenous retroviral LTR transcription start sites are located between the R and U5 regions. Virology. 2006;346:373–378. doi: 10.1016/j.virol.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E. GREM, a technique for genome-wide isolation and quantitative analysis of promoter active repeats. Nucleic Acids Res. 2006;34:e67. doi: 10.1093/nar/gkl335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Reference deleted. [Google Scholar]
- 40.Ingaramo M, Beckett D. Distinct amino termini of two human HCS isoforms influence biotin acceptor substrate recognition. J. Biol. Chem. 2009;284:30862–30870. doi: 10.1074/jbc.M109.046201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camporeale G, Zempleni J, Eissenberg JC. Susceptibility to heat stress and aberrant gene expression patterns in holocarboxylase synthetase-deficient Drosophila melanogaster are caused by decreased biotinylation of histones, not of carboxylases. J. Nutr. 2007;137:885–889. doi: 10.1093/jn/137.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 44.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 45.Brunmeir R, Lagger S, Simboeck E, Sawicka A, Egger G, Hagelkruys A, Zhang Y, Matthias P, Miller WJ, Seiser C. Epigenetic regulation of a murine retrotransposon by a dual histone modification mark. PLoS Genet. 2010;6:e1000927. doi: 10.1371/journal.pgen.1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 47.Zhou J, Wijeratne SSK, Zempleni J. Biotinylation of the c-myc promoter binding protein MBP-1 decreases c-myc expression in mammary carcinoma MCF-7 cells; Experimental Biology 2013 meeting; Boston, MA, U.S.A.. April 21.2013. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.