Abstract
Individuals carrying the same pathogenic mutation can present with a broad range of disease outcomes. While some of this variation arises from environmental factors, it is increasingly recognized that the background genetic variation of each individual can have a profound effect on the expressivity of a pathogenic mutation. In order to understand this background effect on disease-causing mutations, studies need to be performed across a wide range of backgrounds. Recent advancements in model organism biology allow us to test mutations across genetically diverse backgrounds and identify the genes that influence the expressivity of a mutation. In this study, we used the Drosophila Genetic Reference Panel, a collection of ∼200 wild-derived strains, to test the variability of the retinal phenotype of the Rh1G69D Drosophila model of retinitis pigmentosa (RP). We found that the Rh1G69D retinal phenotype is quite a variable quantitative phenotype. To identify the genes driving this extensive phenotypic variation, we performed a genome-wide association study. We identified 106 candidate genes, including 14 high-priority candidates. Functional testing by RNAi indicates that 10/13 top candidates tested influence the expressivity of Rh1G69D. The human orthologs of the candidate genes have not previously been implicated as RP modifiers and their functions are diverse, including roles in endoplasmic reticulum stress, apoptosis and retinal degeneration and development. This study demonstrates the utility of studying a pathogenic mutation across a wide range of genetic backgrounds. These candidate modifiers provide new avenues of inquiry that may reveal new RP disease mechanisms and therapies.
Introduction
Mendelian diseases are commonly understood to be caused by mutations in a single causative gene. However, differences in the genetic background can also severely alter the phenotype of any particular mutation (1–3), as has been demonstrated in a variety of model organisms, including mouse (1,2,4), Drosophila (5,6) and yeast (7). It is increasingly apparent that human patients carrying the same causative mutation, even when environmental factors are considered, can present with drastically different clinical outcomes (3,8). These effects of genetic background can sometimes make it difficult to arrive at strong genotype–phenotype correlations.
In most laboratory studies investigating the effect of genetic background, only a limited number of backgrounds are examined (1,2). However, even these studies demonstrate that closely related genetic backgrounds may still have profound effects on the penetrance of a causative mutation. Some studies have begun to unravel general mechanisms behind the effects of genetic background (7). Apart from the principle that genetic background effects are virtually ubiquitous, it is likely that each primary disease-causing mutation will be differentially impacted by a unique set of variants in the background. Variation in the expression of phenylketonuria led Scriver and Waters to generalize that ‘monogenic traits are not simple.’ (8). Recent whole exome studies in patients with peripheral neuropathy show that patients carrying a primary causative mutation also carry a larger burden of rare alleles that may contribute to phenotypic variability (9). To fully understand which genes modify disease and how they interact in the background, we need to study individual pathogenic mutations in a large number of backgrounds.
Retinitis pigmentosa (RP) is the most common form of retinal degeneration in people, affecting ∼1/4000 individuals (10). Most commonly, RP symptoms begin with night blindness and tunnel vision. They eventually lead to complete loss of vision, due to degeneration of rods and cones in the retina (10). Autosomal dominant retinitis pigmentosa (ADRP) makes up 30–40% of RP cases (10). Dominant mutations in the Rhodopsin gene (RHO) comprise 25% of all ADRP cases and represent the most common cause of RP (10). These dominant mutations result in retinal degeneration through diverse mechanisms, including altered signaling, gain of function, protein mislocalization and protein misfolding. Understanding the effect of each of these mutations on disease pathogenesis is under intense investigation.
The most prevalent class of RP-causing RHO mutations results in misfolded rhodopsin proteins (11). Misfolded rhodopsin accumulates in the endoplasmic reticulum (ER) of photoreceptor neurons (11). Animal studies in mouse (12) and Drosophila (Rh1 is the ortholog of human RHO) (13–21), have established that this abnormal accumulation and retention of misfolded rhodopsin protein results in ER stress, leading to degeneration and cell death of photoreceptor neurons. One such Drosophila model expresses the Rh1G69D mutant protein in the eye imaginal disc, which include the retinal progenitor cells (16,17). The Rh1G69D mutation results in a misfolded protein that is retained in the ER, resulting in ER stress and apoptosis (13–17). Experimental manipulation of the ER stress and ER associated degradation pathways drastically alter the severity of retinal degeneration phenotype in the Rh1G69D model (14–16). Furthermore, elimination of pro-apoptotic signals can also reduce the amount of retinal degeneration (13). Together, this suggests that modifier genes and therapies might target the ER stress and apoptosis pathways in this and other misfolded RHO mutations.
Typical laboratory studies of gene function in model organisms employ a single highly inbred or isogenic genetic background, and hence they cannot test the impact of genetic variation present in a population. To model natural genetic variation, the impact of the defective alleles needs to be assessed on a variety of genetic backgrounds (4–6). Development of tools that accelerate model organism research now allow us to do exactly that. One such tool, the Drosophila Genetics Reference Panel (DGRP), was developed to study the genetic architecture underlying a variety of complex traits (22,23). The DGRP is comprised of ∼200 inbred strains, such that each strain represents a single wild-derived genome. Thus, the DGRP strains capture genetic variation that is present in a natural, wild population, which is likely to better model the variation in the human population.
To assess background effects on RP, we crossed Rh1G69D onto 173 naturally derived DGRP strains. We found that the Rh1G69D retinal phenotype is a quantitative phenotype that shows extensive variation across these different backgrounds. We used an unbiased association study to identify 106 candidate genetic modifiers, including 14 high-priority candidates that drive this extensive phenotypic variation. We found candidate genes with roles in retinal development and other retinal diseases in both human and Drosophila. We also found modifier genes in both the ER stress and apoptotic pathways, previously identified as modifier pathways, suggesting that this approach uncovers bona fide modifier genes for this form of RP. Functional testing indicates that 10/13 of the tested top candidates have the potential to modify the primary Rh1G69D retinal phenotype. Mendelian diseases need to be viewed more like quantitative traits and the use of natural genetic variation can expose novel modifiers that serve as candidate loci for targeted study in human patients. Identifying these modifier genes also nominates cellular pathways that may be potential targets for therapy.
Results
Rh1G69D retinal degeneration shows strong genetic background-dependence
We crossed the Rh1G69D mutation onto the DGRP strains to assess the effect of natural variation on the Rh1G69D retinal phenotype. We utilized the GAL4;UAS system (24) to specifically target expression of the Rh1G69D mutant protein to the larval eye imaginal disc, which includes the retinal precursors. We utilized the GMR-GAL4 driver (25) to drive expression of UAS-Rh1G69D. We generated, by genetic crosses, a single ‘donor’ strain (GMR-GAL4, UAS-Rh1G69D/CyO) (Supplementary Material, Fig. S1A) that we crossed to each DGRP strain. Females from the ‘donor’ strain were crossed with males of each of 173 DGRP strains to generate F1 progeny that expressed Rh1G69D mutant protein in the eye disc (Supplementary Material, Fig. S1B). The progeny from these crosses have 50% of their genome from the maternal ‘donor’ strain and 50% from the paternal DGRP strain. Thus, we are measuring the dominant effect of the DGRP background on the Rh1G69D retinal phenotype.
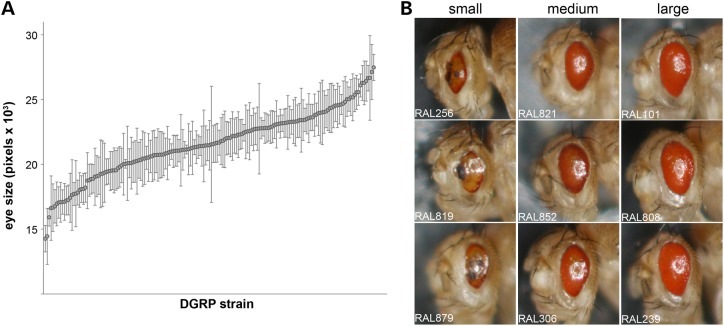
To quantify the mutant Rh1G69Dretinal phenotype, we measured the left eye of a minimum of 10 female F1 progeny from each DGRP/donor cross. Eye size was determined by digital imaging on a light microscope and measuring the area of each eye (in pixels). We observed a very strong effect of DGRP background on eye size (P < 2.2 × 10−16; Supplementary Material, Table S1; Fig. 1A). Median eye size ranged from 1.42 × 104 pixels (RAL256) to 2.75 × 104 pixels (RAL239). There is a clear relationship between the measured eye size and the qualitative appearance of the eye—strains with the smaller eyes also displayed qualitatively more degenerate eyes (Fig. 1B). Strains with smaller eyes often had misshapen eyes and evidence of apparent necrotic tissue, as indicated by the presence of large areas of black tissue on the eye. Strains with the largest eyes had eyes closer in shape to the wild-type eye and showed no evidence of necrosis (Fig. 1B). In no strain was the eye completely rescued to wild-type size. The variation in Rh1G69D eye size is not simply a function of variation in body size due to genetic background, as there was no correlation between mutant eye size and body weight in a subset of DGRP F1 females tested (r2 = 0.09; P = 0.65; Supplementary Material, Table S2 and Fig. S2). We chose to measure females because we observed extensive male larval lethality in many of the DGRP crosses, likely due to apparent leakiness of GMR-GAL4 expression (26). In a subset of strains where male F1 progeny were measurable, we found a strong correlation of their phenotype with that of the female progeny (r2 = 0.73; P < 0.0001; Supplementary Material, Table S3 and Fig. S3).
Figure 1.
Severity of the Rh1G69D retinal phenotype depends on genetic background. (A) The eye sizes for 173 DGRP/Donor F1 crosses. There is a very strong strain effect on the severity of the retinal degeneration as measured by eye size (P < 2.2 × 10−16). Median ± SD. (B) Examples of the qualitative differences in eye phenotype across the size spectrum shown in (A). Small = three strains with the smallest eyes. Medium = strains with eye size from the middle of the distribution. Large = three strains with the largest eyes. RAL# = the strain number of the DGRP strain.
Genome-wide association identifies novel modifiers of Rh1G69D retinal phenotype
We hypothesized that the genomes of the DGRP strains must harbor natural genetic variation underlying the phenotypic differences we observed. In order to generate an unbiased list of candidate genes harboring natural variation that influence the Rh1G69D retinal phenotype, we performed a genome-wide association study. The median eye size from all 173 DGRP strains was used as the quantitative phenotype to which we tested for association with single nucleotide polymorphism (SNPs) genome-wide. We only considered SNPs with a minor allele frequency (MAF) >0.05, yielding a total of 1 891 334 SNPs (Supplementary Material, Table S4). We used a linear mixed model (LMM) to test whether a particular SNP was associated with the Rh1G69D retinal phenotype. Despite using nearly the full set of DGRP lines, this study suffers from a multiple testing problem. In particular, with so many SNPs to test association across so few lines, there is linkage disequilibrium across distant SNPs that arise from sampling (27). Thus, the statistical significance of association of specific SNPs with our phenotype is challenging to interpret. Rather than considering the initial association tests as definitive, we use these SNPs to nominate candidate genes, which we subject to further functional testing. This approach provides us with a prioritized, unbiased list of candidate genes that might influence the expressivity of the Rh1G69D mutation.
Because we are focused on identifying candidate genes, analysis and discussion from here on will focus mainly on the tagged genes, rather than associated SNPs. At a nominal P-value of P < 10−5, 106 candidate genes harbor SNPs associated with the Rh1G69D retinal phenotype (271 total associated SNPs; Supplementary Material, Table S5). These 106 genes will herein be referred to as ‘candidate genes’. With a more stringent cutoff of P < 10−6, there are 14 top candidate genes (36 total associated SNPs; Supplementary Material, Table S5; Table 1). These 14 genes will be herein referred to as ‘top candidate genes’. Because the GMR-GAL4 construct used in this study contains a series of glass-binding elements taken from the Rh1 locus, there was the possibility that SNPs in the glass gene itself could cause some of the variability. However, none of our associated SNPs were found in or around the glass gene. This observation suggests that differences in GAL4 expression do not contribute to the phenotypic variation we observed. This is in agreement with previous studies that show that GMR-GAL4 does not show strain-dependent differences in GAL4 protein expression (5,6).
Table 1.
Top candidate genes
| Rank order | Candidate gene | SNPa | P-valuea | Human orthologs | OMIM | Retinal diseaseb |
|---|---|---|---|---|---|---|
| 1 | CG2004 | X:8608255 | 2.08E−08 | Many | — | — |
| 2 | Cdk5 | 2R:11457488 | 3.97E−07 | CDK5 | 616 342 | Yes |
| 3 | CG15666 | 2R:17230301 | 1.47E−06 | BBS9 | 615 986 | Yes |
| 4 | CG31468 | 3R:19525550 | 2.10E−06 | ODF3/3B | No | No |
| 5 | CG1785 | X:8609333 | 2.86E−06 | GLTSCR2 | No | No |
| 6 | Adgf-D | 3R:9530149 | 3.79E−06 | CECR1 | 615 688 | Yes |
| 7 | fred | 2L:3942577 | 5.05E−06 | KIRREL/2/3 | 612 581 | No |
| 8 | prosap | 2R:10004495 | 5.14E−06 | SHANK1/2/3 | 613 436 | Yes |
| 9 | CG16885 | 2L:13933849 | 6.28E−06 | ZNF512,ZNF512B,TPRX1 | No | No |
| 10 | Hexo2 | X:8605385 | 6.75E−06 | HEXA/B | 272 800 | Yes |
| 11 | hppy | 2R:15095769 | 8.12E−06 | MAP4K1/2/3/5 | No | Yes |
| 12 | lola | 2R:6387329 | 8.47E−06 | None | — | — |
| 13 | Pde1c | 2L:11820427 | 8.96E−06 | PDE1A/B/C | No | No |
| 14 | CG43795 | 2R:19369219 | 9.73E−06 | GPR179, GPR158 | 614 565 | Yes |
aMost significant associated SNP in the candidate gene and the associated P-value.
bBased on PubMed search.
The candidate gene CG2004 harbors the most significant SNP with a nominal P-value of P = 2.08 × 10−8 which is also significant after multiple testing correction (Bonferroni; P = 0.039). It is worth noting that 13 of the 36 SNPs at P < 10−6 fall in CG2004 (Supplementary Material, Table S5). In fact, the top six SNPs are all in CG2004. This strong linkage disequilibrium, which is quite unusual for Drosophila and especially for the DGRP (23), extends to several genes surrounding CG2004, including Hexo2, which also harbors two SNPs at P < 10−6 (Supplementary Material, Table S5 and Fig. S4). Because of this strong linkage, it is impossible to know which gene is the more likely candidate. However, functional data suggest that both are very good candidate genes (see below and discussion).
To determine if the candidate genes belong to a common pathway, we performed Gene Ontology (GO) analysis. With only 14 top candidate genes, it is not surprising that there is no functional enrichment. However, when we considered all 106 candidate genes, we find significant enrichment in genes involved in the categories of eye development (GO:0001745, q = 0.048), planar cell polarity (GO:0001736, q = 0.034) and neural development (GO:0030182, q = 0.023) (Supplementary Material, Table S6). This observation suggests that there might be some common developmental mechanisms influencing the Rh1G69D retinal phenotype and future work might target developmental pathways as avenues for therapy. We did find candidates in the ER stress and apoptosis pathways, which is important because both pathways have previously been shown to play a role in Rh1G69D pathogenesis (13–16). However, ER stress and apoptosis pathways were not among the GO categories enriched in our candidates overall, suggesting that genes and pathways contributing to variation in this phenotype might involve a broad range of functions (see Discussion).
Functional testing
To validate the potential role of the top candidate genes in modifying the Rh1G69D retinal phenotype, we used RNAi to ask whether reduction in candidate gene activity altered the mutant Rh1G69D phenotype. We focused our validation on the 14 top candidate genes (Table 1). These candidates fall into numerous functional categories and thus might modify the original Rh1G69D retinal phenotype through a variety of mechanisms. Because our ultimate goal was to nominate candidate genes that can be eventually screened in human populations, we did not aim to specifically test a particular SNP, but instead we sought to demonstrate the potential for a particular gene to modify the original pathogenic mutation. Our functional analysis uses RNAi to examine loss-of-function effects. This approach underestimates the extent of the contribution of these genes to modify the original phenotype, but is a very simple, straightforward, direct test of how these candidate genes might modify the Rh1G69D retinal phenotype.
To test candidate gene function, we again used the GAL4;UAS system (24). We crossed each respective RNAi strain to the donor strain (Supplementary Material, Fig. S1C). This results in a fly that expresses both Rh1G69D mutant protein and the candidate RNAi in the same cells. As a control, we crossed the donor strain to the RNAi background strain (strain AttP; no RNAi insert). We considered this to be the simplest test of function, although we recognize that a particular gene's effect may originate from cells or tissues that do not overlap with the GMR-GAL4 directed expression of Rh1G69D.
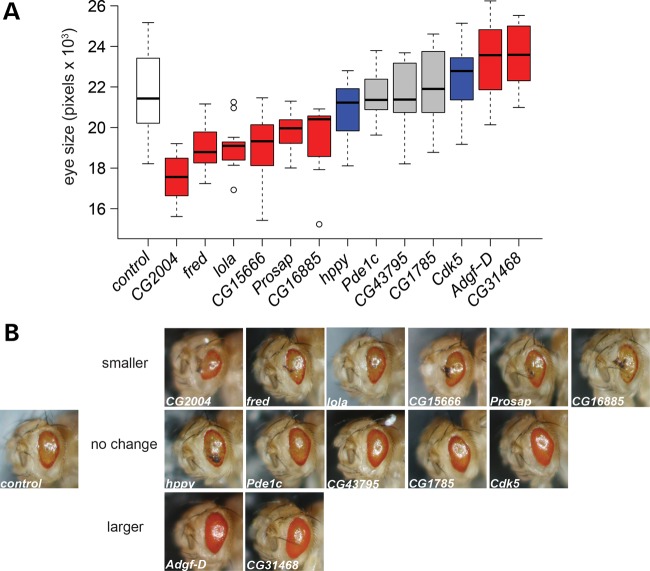
RNAi constructs were available for 13 of the 14 top candidate genes (all except for Hexo2) (Fig. 2A and B). RNAi knockdown (KD) of 6 candidate genes enhanced the original Rh1G69D retinal phenotype, showing a significantly smaller and more degenerated eye (CG2004, CG15666, fred, prosap, CG16885 and lola). It is notable that CG2004, the top candidate based on SNP association, also showed the strongest effect by RNAi KD (Fig. 2A and B). RNAi KD of two candidate genes suppressed the original Rh1G69D retinal phenotype, showing a significantly larger, less degenerated eye (CG31468 and Adgf-D). RNAi KD of five candidate genes showed no quantitative change in the Rh1G69D eye size when compared with control (Cdk5, CG1785, hppy, Pde1c and CG43795). The Rh1G69D eyes from CG1785, Pde1c and CG43795 KDs showed no quantitative change and were qualitatively indistinguishable from the control Rh1G69D eyes (Fig. 2A and B). On the other hand, while Cdk5 and hppy KD Rh1G69D eyes show no quantitative change (Fig. 2A) from the control Rh1G69D, their eye phenotype appeared qualitatively different from that of the control (Fig. 2B). KD of Cdk5 caused an apparent qualitative improvement in the Rh1G69D eye phenotype (Fig. 2B). Cdk5 KD Rh1G69D eyes were less misshapen, had more pigment, and showed no signs of necrosis, all of which were indicative of partial rescue of the eye phenotype (13,14). In line with this qualitative appearance of rescue, Cdk5 KD Rh1G69D eyes were qualitatively similar to those of KD and DGRP strains that have quantitatively larger eyes. This qualitative rescue by Cdk5 KD was quite reproducible and was apparent in every individual we measured (N = 12) (Supplementary Material, Fig. S5). Our observation that Cdk5 KD qualitatively rescues the Rh1G69D phenotype is in agreement with a previous report that rescue of Rh1G69D was also seen with loss of Cdk5 expression (13) (see Discussion). hppy KD in the Rh1G69D background also caused no quantitative change in eye size (Fig. 2A), but showed an apparent qualitative enhancement of the Rh1G69D eye abnormality (Fig. 2B). Similar to KD Rh1G69D and DGRP Rh1G69D strains with small eyes, all hppy KD Rh1G69D eyes showed loss of pigment and had areas of black necrotic tissue (N = 10) (Supplementary Material, Fig. S5). It is important to note that GMR-GAL4 directed KD of these candidate genes in the absence of Rh1G69D does not result in a retinal phenotype (Supplementary Material, Fig. S6), indicating a specific interaction of these genes with the RH1G69D mutation.
Figure 2.
Functional analysis of candidate genes. (A) The eye sizes for RNAi KD of candidate genes on the Rh1G69D background. White = AttP control; red = significantly different in size from AttP control (P); blue = not significantly different in size but shows clear qualitative change in eye phenotype (see B); gray = not significantly different from AttP control. In the box plots, the boxes represent the interquartile range, the whiskers represent 1.5 × interquartile range and open circles are outliers. See Supplementary Material, Table S7 for values and significance. (B) Representative examples of eyes from functional tests. All eyes are from flies carrying Rh1G69D. Control is on the left. RNAi KD eyes are shown in the order that they appear in (A). Note that hppy and Cdk5 KD eyes appear qualitatively different from the control, but do not show a difference in measured eye size. The control strain shown in (A and B) is the 60 100 AttP strain. This strain has the same genetic background for all the RNAi strains except CG2004, and thus serves as their control. CG2004 is on a different genetic background; its appropriate control strain is 60 000 AttP. Since we found that the two AttP control strains are indistinguishable in eye phenotype in the Rh1G69D background (Supplementary Material, Fig. S9), for simplicity, only the results for 60 100 AttP are shown here.
To demonstrate that our approach identifies an enriched pool of candidate genes for modifiers of retinal degeneration, we compared our gene list to that a previous targeted screen. Kang et al. (13) used an RNAi screen to identify genes encoding phosphatases and kinases that might modify the Rh1G69D phenotype. They found that only 3 of 158 genes screened showed a modifying effect on the original phenotype (13). In our study, we found that 10 of the 13 candidate genes we tested affected the original Rh1G69D retinal phenotype. This proportion is significantly higher than that observed by Kang et al. (χ2; P = 1.05 × 10−33), indicating that the use of natural variation is an efficient way to identify promising candidate modifier genes for RP.
Discussion
At the core of personalized medicine is the fact that individuals carrying the same pathogenic mutation can have very different disease manifestations (28). Understanding what contributes to these inter-individual differences can be critical to developing appropriate targeted therapies. Among the factors that influence variable disease outcomes are an individual's genetic background and his/her collection of environmental exposures. Both factors are complex and they can also interact to modify phenotypes, sometimes in unexpected ways. The study of background effects has been recently made much easier by groups that have organized superb model organism resources of strains and whole-genome sequence data and annotation (23,29–31). The DGRP has been used repeatedly to document variation in adaptive (32–34) and medically relevant (35–38) traits. In every case, the DGRP has been instrumental in understanding how genetic variation influences phenotypic variability. In this study, we took advantage of natural genetic variation in the DGRP to demonstrate that a simple autosomal dominant mutation has the potential to produce a highly variable quantitative phenotype. We show that the Drosophila Rh1G69D RP model has a wide ranging mutant retinal phenotype that depends on genetic background. This unbiased approach allowed us to identify candidate modifier genes of RP. Through direct functional tests, we demonstrate that most of the top candidate genes do in fact modify the RP phenotype.
The extensive variation in the Rh1G69D quantitative retinal phenotype displayed across the DGRP demonstrates the potential for clinical variation in patients carrying similar RHO mutations. However, simply knowing that the phenotype might vary is not sufficient. We identified a list of prioritized candidate genes that might play a role in modifying the phenotypic effect of the original pathogenic mutation. With the exception of one gene (lola), all the top candidate genes have human orthologs (13/14) (Table 1). Overall, 76% of all candidate genes (82/106) have clear human orthologs, indicating that modifier genes are in conserved pathways. This high level of conservation suggests that the findings in this study could be highly informative as human candidates in translational studies. Indeed, orthologs of 7 of the top 14 candidates have been implicated in a variety of human genetic diseases (Table 1). There may be common pathways or mechanisms shared between these other human diseases and RP.
Perhaps even more striking, 7 of the 14 top candidate genes or their mouse/human orthologs have been implicated in retinal disease and retinal function, further supporting the promise of studying these candidate genes and how they relate to RP. BBS9 is the human ortholog of CG15666, the third highest top candidate (based on SNP association). BBS9 is a member of the BBSome involved in trafficking cargo to the cilium (39,40). Mutations in BBS9 cause a form of Bardet–Biedl Syndrome, which consists of a series of conditions including RP (41,42). While the Drosophila retina does not have cilia, our functional follow up clearly demonstrates that loss of CG15666 activity exacerbates the Drosophila Rh1G69D retinal phenotype. Thus, the role of BBS9/CG15666 in RP pathology may go beyond ciliary biology and might play a more basic role in cellular trafficking. CECR1 is the human ortholog of Adgf-D. CECR1 has been implicated in a variety of eye diseases including Cat eye syndrome (43), which involves abnormal development of the retina in addition to other ocular tissues. More recently, CECR1 has been shown to be upregulated in retinas experiencing diabetic retinopathy (44). It is thought that CECR1 contributes to inflammatory problems associated with this type of retinopathy (44). Inflammation is an important aspect of apoptosis (45) and may influence the outcome of this form of RP. GPR179 is the human ortholog of CG43795. Mutations in GPR179 cause complete congenital stationary night blindness (46,47). GPR179 protein plays an important role in retinal bipolar cells (RPCs) (46,48). While RPCs are not directly affected by RP, these and other neurons in the retina undergo significant remodeling when photoreceptors undergo cell death (49).
Two candidate genes, prosap and hppy, have orthologs that have been implicated in neuronal (50) and vascular (51) development in the retina, respectively. Neither has been implicated in any human disease, but their clear roles in retinal development make them good candidates for modifying RP-associated degeneration. Two candidates have been implicated in rhodopsin biology and retinal degeneration in Drosophila. Loss of Cdk5 has been demonstrated to modify the Rh1G69D phenotype (13) (see below). Hexo2, which we could not functionally test, is a hexosaminidase which likely plays a role in modifying wild-type RH1 glycosylation. fused lobes (fdl) and Hexo1, homologs of Hexo2, modify the glycosylation state of wild-type RH1 (52). Strikingly, fdl is also a candidate gene (though not a top candidate, but 1 of the 106 candidates). Wild-type RH1 glycosylation state is important for proper trafficking of wild-type RH1 from the ER to the cell membrane. Inappropriate glycosylation results in retention in the ER and Golgi (52). Glycosylation may affect how the misfolded RH1G69D protein is retained or degraded. Together, these candidate genes play diverse roles that influence retinal function and may prove to be important candidates that can be targeted for therapy.
We choose the Rh1G69D model of RP because it is well established that alterations in the response to ER stress and apoptosis signaling can independently influence the outcome of the severity of the retinal degeneration (13–16). We hypothesized that at least some of the genes driving variation in the RH1G69D phenotype would be due to differences in the ER stress and apoptosis pathways. In fact, the two top candidates, CG2004 and Cdk5, are involved in ER stress response and apoptosis, respectively. CG2004 is the top candidate gene and in our functional follow up, KD of CG2004 had the strongest effect on modifying the original RH1G69D phenotype. CG2004 is a member of a large group of genes that contain a choline kinase (CHK)-kinase domain. While this particular gene has no known function and remains unstudied, we previously demonstrated that CHK-kinase genes are enriched in genes that contribute to genetic variation in the ER stress response in Drosophila (35). Protein interaction data indicate that CG2004 putatively interacts with a variety of ER resident proteins and proteins involved in protein transport (Supplementary Material, Fig. S7). Given these observations, CG2004 likely modifies the RH1G69D retinal phenotype through altering the ER stress response.
The Cdk5 protein product has pro-apoptotic activity through the c-Jun N-terminal kinase pathway (13,53). Strikingly, KD of Cdk5 was previously shown to modify the RH1G69D phenotype (13). Kang et al. demonstrated that Cdk5 protein has pro-apoptotic activity and that loss of Cdk5 rescues the retinal degeneration and results in a larger eye (13,53). In our functional follow up, we also found that KD of Cdk5 also rescues the RH1G69D retinal phenotype, though to a lesser degree, likely due to experimental and genetic background differences. Previous functional work confirms and validates the utility of this approach. Furthermore, combined with previous studies, our finding indicates that the apoptotic pathway may be a very promising target for designing therapies for RP.
We also hypothesized that most of the genetic variation would arise from genes that were previously not known to this disease network. In fact, 12/14 prioritized candidates are in pathways not previously implicated in ER stress response or apoptosis. Previous studies used loss of function or KD mutations to establish the role of ER stress and apoptosis in Rh1G69D pathogenesis. However, loss-of-function mutations are not common in the human population, and we expect that a substantial portion of the variance in RP pathogenesis will be mediated by more common variation of small allelic effects (54). Indeed, the associated SNPs we identified are all in positions (untranslated regions, introns, etc.) that are likely to alter expression, rather than eliminate the function of the encoded protein.
Our results have important implications for our understanding of the role of genetic variation in altering disease outcomes. This study and others (4–6) now clearly demonstrate that Mendelian diseases are best considered to be quantitative traits whose penetrance is modulated by the genetic background. The human population is genetically diverse, and pathogenic mutations generally present a phenotypic spectrum when those mutations appear in the genetic backgrounds of a diverse patient population. The observations from this study, and from human disease etiologies, clearly demonstrate the presence and effect of cryptic genetic variation. Cryptic variants on their own might not have a fitness or phenotypic effect on an organism; their effects are only seen if another mutation, or an environmental insult, is present (55–57). The type of study described here is well suited for exploiting a model system to uncover disease-relevant cryptic genetic variation. Beyond establishing a phenotypic spectrum, we show that most of the candidate genes that contribute to this variation have never been considered to contribute to the pathogenesis of this particular form of RP in humans. Further detailed analysis is needed to fit each candidate gene into the RP disease network. We can now model the impact of naturally occurring variation on important human genetic disorders in certain model systems like the DGRP (23) and the Mouse Collaborative Cross (29), and here we show how this nominates novel candidate modifiers genes for those disease pathways. By applying classical genetics to these quantitative tools, we gain a more comprehensive view of the potential for any single disease mutation to cause disease and we can identify novel pathways that contribute to its pathogenesis. Studies such as this provide new targets for investigation and will likely prove to be informative for developing personalized therapies.
Materials and Methods
Drosophila melanogaster fly cultures
One hundred seventy-three lines from the DGRP were used in this study (Supplementary Material, Table S1). The strain carrying UAS-Rh1G69D on chromosome 2 has been previously described (13,14,16). The GMR-GAL4 driver on chromosome 2 was obtained from the Bloomington Drosophila Stock Center (strain 1104). The donor strain was constructed by crossing the strain carrying UAS-Rh1G69D on chromosome 2 with the strain carrying the GMR-GAL4 driver on chromosome 2, to generate recombinants in which the two constructs had been recombined onto a single chromosome 2 (for further details on strain construction, see crossing scheme in Supplementary Material, Fig. S1A). To maintain a standard background, the donor strain was derived from a single male recombinant fly. Once the donor strain carrying the recombinant chromosome 2 was established, it was backcrossed to the w1118 laboratory strain for six generations. For the functional tests, the strains carrying RNAi constructs were ordered from the Vienna Drosophila RNAi Center (VDRC; see Supplementary Material, Table S7). All flies were collected as virgins under CO2 anesthesia and aged 3 days. All flies were maintained at room temperature on standard agar-dextrose yeast media on a 12 h light/dark cycle.
DGRP phenotypic analysis
To produce the experimental DGRP F1 lines, crosses were performed between virgin females of the donor strain and males of each individual DGRP strain (Supplementary Material, Fig. S1B). F1 progeny were collected upon emergence, aged 3 days, flash frozen and stored at −80 until they were visualized. The left eye of each fly was digitally imaged using a light microscope. Eye size was determined using the NIH ImageJ software package and was defined as the area (in pixels) of the eye. At least 10 female F1 progeny from each DGRP cross were measured for our study. In some cases, male progeny were also measured. In all cases, the investigator was not blinded to genotype when capturing the image, but was blinded to genotype when measuring or assessing phenotypic changes.
To identify the line effect, analysis of variance (ANOVA) was used to apply a simple linear model to the eye size. The eye size from the jth replicate of the ith DGRP line was
where µ is the grand mean, DGRPi is the deviation of the mean of the ith DGRP line from the grand mean and εij is the error term. The index i for DGRP strains runs from 1 to 173, and the index j for replicates varies among lines (≥10).
To determine the weight of female F1 flies, 10 female flies were weighed before imaging for eye size. Correlation analysis was performed on the mean weight measurements with the median eye size of the same 10 female flies.
Genome-wide association
DGRP genotypes were downloaded from the website, http://dgrp.gnets.ncsu.edu/ 17 December 2015, date last accessed. Variants were filtered for MAF (≥0.05), missingness (≥0.2) and non-biallelic sites were removed. A total of 1 891 334 SNPs were included in the analysis (Supplementary Material, Table S4). Median eye size for 173 DGRP lines was regressed on each SNP. To account for cryptic relatedness (5,22), GEMMA (v. 0.94) (58) was used to both estimate a centered genetic relatedness matrix and perform association tests using the following LMM:
where as described and adapted from Zhou and Stephens (58), is the n-vector of median eye size for the n lines, is the intercept, is the -vector of marker genotypes, is the effect size of the marker. is an n × n matrix of random effects with a multivariate normal distribution () that depends on , the ratio between the two variance components, , the variance of residuals errors, and where the covariance matrix is informed by , the calculated n × n marker-based relatedness matrix. accounts for all pairwise non-random sharing of genetic material among lines. , is an n-vector of residual errors, with a multivariate normal distribution that depends on and , the identity matrix. Quantile–quantile (Q–Q) plot analysis indicated an acceptable fit to the LMM (Supplementary Material, Fig. S8). Bonferroni correction was applied to account for multiple testing, appreciating that it is quite conservative.
RNAi functional testing
To test the effect of RNAi KD of each candidate gene, each RNAi strain was crossed to the donor strain (Supplementary Material, Table S7 and Fig. S1C). As a control, we crossed the donor strain to the AttP strain which has the same genetic background as the RNAi strains, but does not contain the RNAi-generating transgene. Measurement of eye size was performed as described above. At least 10 eyes were measured for the progeny of each cross. Eyes from the RNAi cross were compared with the AttP control cross using a similar ANOVA model as described above. Note, the CG2004 RNAi line is derived from a different AttP control strain (VDRC transformant ID: 60000) than the rest of the RNAi lines (VDRC transformant ID: 60100). There was no significant difference in progeny eye size from crosses of the donor strain to either AttP strain. CG2004 RNAi showed a similar significant reduction in eye size when compared with either control AttP strains. The comparison between both AttP strains and CG2004 RNAi is shown in Supplementary Material, Figure S9. However, only the result from one AttP (VDRC transformant ID: 60100) strain is shown in Figure 2.
Bioinformatics analysis
Human orthologs of candidate genes and their roles in disease were identified by searching Online Mendelian Inheritance of Man (OMIM). GO analysis was performed using DAVID (david.ncifcrf.gov) and FlyMine (www.flymine.org). Gene interaction network for CG2004 was created using GeneMANIA (http://www.genemania.org/, 17 December 2015, date last accessed) (59).
Supplementary Material
Funding
This study was supported by a Meinig Family Investigator Award to A.G.C. and by NIH R01 HD059060 to A.G.C. and M.F.W.
Supplementary Material
Acknowledgements
We would like to thank Dr Hyung Don Ryoo (NYU) for generously providing the UAS-Rh1G69D stock.
Conflict of Interest statement. None declared.
References
- 1.Hamilton B.A., Yu B.D. (2012) Modifier genes and the plasticity of genetic networks in mice. PLoS Genet., 8, e1002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nadeau J.H. (2003) Modifier genes and protective alleles in humans and mice. Curr. Opin. Genet. Dev., 13, 290–295. [DOI] [PubMed] [Google Scholar]
- 3.Gallati S. (2014) Disease-modifying genes and monogenic disorders: experience in cystic fibrosis. Appl. Clin. Genet., 7, 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferguson B., Ram R., Handoko H.Y., Mukhopadhyay P., Muller H.K., Soyer H.P., Morahan G., Walker G.J. (2015) Melanoma susceptibility as a complex trait: genetic variation controls all stages of tumor progression. Oncogene, 34, 2879–2886. [DOI] [PubMed] [Google Scholar]
- 5.He B.Z., Ludwig M.Z., Dickerson D.A., Barse L., Arun B., Vilhjalmsson B.J., Jiang P., Park S.Y., Tamarina N.A., Selleck S.B. et al. (2014) Effect of genetic variation in a Drosophila model of diabetes-associated misfolded human proinsulin. Genetics, 196, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park S.Y., Ludwig M.Z., Tamarina N.A., He B.Z., Carl S.H., Dickerson D.A., Barse L., Arun B., Williams C.L., Miles C.M. et al. (2014) Genetic complexity in a Drosophila model of diabetes-associated misfolded human proinsulin. Genetics, 196, 539–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vu V., Verster A.J., Schertzberg M., Chuluunbaatar T., Spensley M., Pajkic D., Hart G.T., Moffat J., Fraser A.G. (2015) Natural variation in gene expression modulates the severity of mutant phenotypes. Cell, 162, 391–402. [DOI] [PubMed] [Google Scholar]
- 8.Scriver C.R., Waters P.J. (1999) Monogenic traits are not simple: lessons from phenylketonuria. Trends Genet., 15, 267–272. [DOI] [PubMed] [Google Scholar]
- 9.Gonzaga-Jauregui C., Harel T., Gambin T., Kousi M., Griffin L.B., Francescatto L., Ozes B., Karaca E., Jhangiani S.N., Bainbridge M.N. et al. (2015) Exome Sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell. Rep., 12, 1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartong D.T., Berson E.L., Dryja T.P. (2006) Retinitis pigmentosa. Lancet, 368, 1795–1809. [DOI] [PubMed] [Google Scholar]
- 11.Sung C.H., Davenport C.M., Nathans J. (1993) Rhodopsin mutations responsible for autosomal dominant retinitis pigmentosa. Clustering of functional classes along the polypeptide chain. J. Biol. Chem., 268, 26645–26649. [PubMed] [Google Scholar]
- 12.Kroeger H., LaVail M.M., Lin J.H. (2014) Endoplasmic reticulum stress in vertebrate mutant rhodopsin models of retinal degeneration. Adv. Exp. Med. Biol., 801, 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang M.J., Chung J., Ryoo H.D. (2012) CDK5 and MEKK1 mediate pro-apoptotic signalling following endoplasmic reticulum stress in an autosomal dominant retinitis pigmentosa model. Nat. Cell Biol., 14, 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang M.J., Ryoo H.D. (2009) Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation. Proc. Natl Acad. Sci. USA, 106, 17043–17048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendes C.S., Levet C., Chatelain G., Dourlen P., Fouillet A., Dichtel-Danjoy M.L., Gambis A., Ryoo H.D., Steller H., Mollereau B. (2009) ER stress protects from retinal degeneration. EMBO J., 28, 1296–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryoo H.D., Domingos P.M., Kang M.J., Steller H. (2007) Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J., 26, 242–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colley N.J., Cassill J.A., Baker E.K., Zuker C.S. (1995) Defective intracellular transport is the molecular basis of rhodopsin-dependent dominant retinal degeneration. Proc. Natl Acad. Sci. USA, 92, 3070–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonard D.S., Bowman V.D., Ready D.F., Pak W.L. (1992) Degeneration of photoreceptors in rhodopsin mutants of Drosophila. J. Neurobiol., 23, 605–626. [DOI] [PubMed] [Google Scholar]
- 19.Kurada P., O'Tousa J.E. (1995) Retinal degeneration caused by dominant rhodopsin mutations in Drosophila. Neuron, 14, 571–579. [DOI] [PubMed] [Google Scholar]
- 20.Galy A., Roux M.J., Sahel J.A., Leveillard T., Giangrande A. (2005) Rhodopsin maturation defects induce photoreceptor death by apoptosis: a fly model for RhodopsinPro23His human retinitis pigmentosa. Hum. Mol. Genet., 14, 2547–2557. [DOI] [PubMed] [Google Scholar]
- 21.Davidson F.F., Steller H. (1998) Blocking apoptosis prevents blindness in Drosophila retinal degeneration mutants. Nature, 391, 587–591. [DOI] [PubMed] [Google Scholar]
- 22.Huang W., Massouras A., Inoue Y., Peiffer J., Ramia M., Tarone A.M., Turlapati L., Zichner T., Zhu D., Lyman R.F. et al. (2014) Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res., 24, 1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackay T.F., Richards S., Stone E.A., Barbadilla A., Ayroles J.F., Zhu D., Casillas S., Han Y., Magwire M.M., Cridland J.M. et al. (2012) The Drosophila melanogaster Genetic Reference Panel. Nature, 482, 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brand A.H., Perrimon N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 25.Hay B.A., Wolff T., Rubin G.M. (1994) Expression of baculovirus P35 prevents cell death in Drosophila. Development, 120, 2121–2129. [DOI] [PubMed] [Google Scholar]
- 26.Li W.Z., Li S.L., Zheng H.Y., Zhang S.P., Xue L. (2012) A broad expression profile of the GMR-GAL4 driver in Drosophila melanogaster. Genet. Mol. Res., 11, 1997–2002. [DOI] [PubMed] [Google Scholar]
- 27.Houle D., Marquez E.J. (2015) Linkage disequilibrium and inversion-typing of the Drosophila melanogaster Genome Reference Panel. G3 (Bethesda), 5, 1695–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins F.S., Varmus H. (2015) A new initiative on precision medicine. N. Engl. J. Med., 372, 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Threadgill D.W., Miller D.R., Churchill G.A., de Villena F.P. (2011) The collaborative cross: a recombinant inbred mouse population for the systems genetic era. ILAR J., 52, 24–31. [DOI] [PubMed] [Google Scholar]
- 30.Churchill G.A., Gatti D.M., Munger S.C., Svenson K.L. (2012) The Diversity Outbred mouse population. Mamm. Genome, 23, 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grenier J.K., Arguello J.R., Moreira M.C., Gottipati S., Mohammed J., Hackett S.R., Boughton R., Greenberg A.J., Clark A.G. (2015) Global diversity lines—a five-continent reference panel of sequenced Drosophila melanogaster strains. G3 (Bethesda), 5, 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unckless R.L., Rottschaefer S.M., Lazzaro B.P. (2015) A genome-wide association study for nutritional indices in Drosophila. G3 (Bethesda), 5, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayroles J.F., Buchanan S.M., O'Leary C., Skutt-Kakaria K., Grenier J.K., Clark A.G., Hartl D.L., de Bivort B.L. (2015) Behavioral idiosyncrasy reveals genetic control of phenotypic variability. Proc. Natl Acad. Sci. USA, 112, 6706–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow C.Y., Wolfner M.F., Clark A.G. (2013) Large neurological component to genetic differences underlying biased sperm use in Drosophila. Genetics, 193, 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chow C.Y., Wolfner M.F., Clark A.G. (2013) Using natural variation in Drosophila to discover previously unknown endoplasmic reticulum stress genes. Proc. Natl Acad. Sci. USA, 110, 9013–9018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber A.L., Khan G.F., Magwire M.M., Tabor C.L., Mackay T.F., Anholt R.R. (2012) Genome-wide association analysis of oxidative stress resistance in Drosophila melanogaster. PLoS One, 7, e34745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jordan K.W., Craver K.L., Magwire M.M., Cubilla C.E., Mackay T.F., Anholt R.R. (2012) Genome-wide association for sensitivity to chronic oxidative stress in Drosophila melanogaster. PLoS One, 7, e38722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carbone M.A., Ayroles J.F., Yamamoto A., Morozova T.V., West S.A., Magwire M.M., Mackay T.F., Anholt R.R. (2009) Overexpression of myocilin in the Drosophila eye activates the unfolded protein response: implications for glaucoma. PLoS One, 4, e4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C. et al. (2007) A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell, 129, 1201–1213. [DOI] [PubMed] [Google Scholar]
- 40.Veleri S., Bishop K., Dalle Nogare D.E., English M.A., Foskett T.J., Chitnis A., Sood R., Liu P., Swaroop A. (2012) Knockdown of Bardet–Biedl syndrome gene BBS9/PTHB1 leads to cilia defects. PLoS One, 7, e34389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura D.Y., Swiderski R.E., Searby C.C., Berg E.M., Ferguson A.L., Hennekam R., Merin S., Weleber R.G., Biesecker L.G., Stone E.M. et al. (2005) Comparative genomics and gene expression analysis identifies BBS9, a new Bardet–Biedl syndrome gene. Am. J. Hum. Genet., 77, 1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaghloul N.A., Katsanis N. (2009) Mechanistic insights into Bardet–Biedl syndrome, a model ciliopathy. J. Clin. Invest., 119, 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riazi M.A., Brinkman-Mills P., Nguyen T., Pan H., Phan S., Ying F., Roe B.A., Tochigi J., Shimizu Y., Minoshima S. et al. (2000) The human homolog of insect-derived growth factor, CECR1, is a candidate gene for features of cat eye syndrome. Genomics, 64, 277–285. [DOI] [PubMed] [Google Scholar]
- 44.Elsherbiny N.M., Naime M., Ahmad S., Elsherbini A.M., Mohammad S., Fulzele S., El-Remessy A.B., Al-Gayyar M.M., Eissa L.A., El-Shishtawy M.M. et al. (2013) Potential roles of adenosine deaminase-2 in diabetic retinopathy. Biochem. Biophys. Res. Commun., 436, 355–361. [DOI] [PubMed] [Google Scholar]
- 45.Rock K.L., Kono H. (2008) The inflammatory response to cell death. Annu. Rev. Pathol., 3, 99–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peachey N.S., Ray T.A., Florijn R., Rowe L.B., Sjoerdsma T., Contreras-Alcantara S., Baba K., Tosini G., Pozdeyev N., Iuvone P.M. et al. (2012) GPR179 is required for depolarizing bipolar cell function and is mutated in autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet., 90, 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Audo I., Bujakowska K., Orhan E., Poloschek C.M., Defoort-Dhellemmes S., Drumare I., Kohl S., Luu T.D., Lecompte O., Zrenner E. et al. (2012) Whole-exome sequencing identifies mutations in GPR179 leading to autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet., 90, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ray T.A., Heath K.M., Hasan N., Noel J.M., Samuels I.S., Martemyanov K.A., Peachey N.S., McCall M.A., Gregg R.G. (2014) GPR179 is required for high sensitivity of the mGluR6 signaling cascade in depolarizing bipolar cells. J. Neurosci., 34, 6334–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strettoi E., Pignatelli V. (2000) Modifications of retinal neurons in a mouse model of retinitis pigmentosa. Proc. Natl Acad. Sci. USA, 97, 11020–11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stella S.L. Jr., Vila A., Hung A.Y., Rome M.E., Huynh U., Sheng M., Kreienkamp H.J., Brecha N.C. (2012) Association of shank 1A scaffolding protein with cone photoreceptor terminals in the mammalian retina. PLoS One, 7, e43463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vitorino P., Yeung S., Crow A., Bakke J., Smyczek T., West K., McNamara E., Eastham-Anderson J., Gould S., Harris S.F. et al. (2015) MAP4K4 regulates integrin-FERM binding to control endothelial cell motility. Nature, 519, 425–430. [DOI] [PubMed] [Google Scholar]
- 52.Rosenbaum E.E., Vasiljevic E., Brehm K.S., Colley N.J. (2014) Mutations in four glycosyl hydrolases reveal a highly coordinated pathway for rhodopsin biosynthesis and N-glycan trimming in Drosophila melanogaster. PLoS Genet., 10, e1004349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inoue H., Tateno M., Fujimura-Kamada K., Takaesu G., Adachi-Yamada T., Ninomiya-Tsuji J., Irie K., Nishida Y., Matsumoto K. (2001) A Drosophila MAPKKK, D-MEKK1, mediates stress responses through activation of p38 MAPK. EMBO J., 20, 5421–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu W., O'Connor T.D., Akey J.M. (2013) Genetic architecture of quantitative traits and complex diseases. Curr. Opin. Genet. Dev., 23, 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paaby A.B., Rockman M.V. (2014) Cryptic genetic variation: evolution's hidden substrate. Nat. Rev. Genet., 15, 247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gibson G., Dworkin I. (2004) Uncovering cryptic genetic variation. Nat. Rev. Genet., 5, 681–690. [DOI] [PubMed] [Google Scholar]
- 57.Gibson G. (2009) Decanalization and the origin of complex disease. Nat. Rev. Genet., 10, 134–140. [DOI] [PubMed] [Google Scholar]
- 58.Zhou X., Stephens M. (2012) Genome-wide efficient mixed-model analysis for association studies. Nat. Genet., 44, 821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., Franz M., Grouios C., Kazi F., Lopes C.T. et al. (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res., 38, W214–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.