Abstract
Biogenesis of cytochrome c oxidase (CcO), the terminal enzyme of the mitochondrial respiratory chain, is a complex process facilitated by several assembly factors. Pathogenic mutations were recently reported in one such assembly factor, COA6, and our previous work linked Coa6 function to mitochondrial copper metabolism and expression of Cox2, a copper-containing subunit of CcO. However, the precise role of Coa6 in Cox2 biogenesis remained unknown. Here we show that yeast Coa6 is an orthologue of human COA6, and like Cox2, is regulated by copper availability, further implicating it in copper delivery to Cox2. In order to place Coa6 in the Cox2 copper delivery pathway, we performed a comprehensive genetic epistasis analysis in the yeast Saccharomyces cerevisiae and found that simultaneous deletion of Coa6 and Sco2, a mitochondrial copper metallochaperone, or Coa6 and Cox12/COX6B, a structural subunit of CcO, completely abrogates Cox2 biogenesis. Unlike Coa6 deficient cells, copper supplementation fails to rescue Cox2 levels of these double mutants. Overexpression of Cox12 or Sco proteins partially rescues the coa6Δ phenotype, suggesting their overlapping but non-redundant roles in copper delivery to Cox2. These genetic data are strongly corroborated by biochemical studies demonstrating physical interactions between Coa6, Cox2, Cox12 and Sco proteins. Furthermore, we show that patient mutations in Coa6 disrupt Coa6–Cox2 interaction, providing the biochemical basis for disease pathogenesis. Taken together, these results place COA6 in the copper delivery pathway to CcO and, surprisingly, link it to a previously unidentified function of CcO subunit Cox12 in Cox2 biogenesis.
Introduction
Defects in the function and formation of the mitochondrial respiratory chain (MRC) manifest clinically in mitochondrial diseases, one of the most common classes of inborn errors of metabolism (1). A subset of MRC disorders can be attributed to the deficiency of MRC complex IV, commonly known as cytochrome c oxidase (CcO). CcO is the terminal enzyme of the MRC that catalyzes the reduction of molecular oxygen to water and generates an electrochemical gradient that drives mitochondrial adenosine triphosphate (ATP) synthesis. CcO is an evolutionarily conserved multi-subunit enzyme complex whose catalytic core is composed of three subunits: Cox1, Cox2 and Cox3, which are encoded by mitochondrial DNA in both yeast and humans (2). The other structural subunits, which are encoded by nuclear DNA, surround the catalytic core to form the CcO holoenzyme. In addition to the protein subunits, CcO contains several cofactors including two copper centers (CuA and CuB), two heme groups (heme a and a3), a magnesium ion and a zinc ion (3). The assembly of a fully mature, catalytically active CcO is an extremely complex process that requires a number of assembly factors to bring together the mitochondrial and nuclear DNA-encoded subunits with their metal cofactors.
CcO biogenesis is a modular process that begins with the independent maturation of the core subunits Cox1, Cox2 and Cox3, followed by the addition of other nuclear-encoded subunits (4,5). There are ∼40 assembly factors discovered to date that facilitate different steps of CcO assembly (2). For example, 22 assembly factors are required for the expression and membrane insertion of the catalytic core subunits, 9 assembly factors are required for copper delivery to copper A (CuA) and copper B (CuB) site in Cox2 and Cox1 subunits, respectively, and 4 factors are required for heme biosynthesis and insertion into the catalytic core (2). Unlike assembly factors required for the expression and insertion of mitochondrial DNA-encoded catalytic subunits, all the factors required for copper delivery and heme insertion to the Cox1 and Cox2 subunits are conserved in yeast and humans (2).
Patients suffering from CcO deficiency exhibit multi-systemic and tissue-specific disorders, primarily affecting organs with higher energy demands including the brain, skeletal muscle and heart (6,7). CcO deficiency leads to early onset, autosomal recessive disorders with fatal clinical outcomes (6,7). The combination of human genetics and knowledge of CcO assembly factors from Saccharomyces cerevisiae has led to the discovery of multiple CcO disease genes. While a few mutations can be attributed to the structural subunits of CcO, including COX1, COX2, COX3, COX4 and COX6B (7–10), the majority of patient mutations are found in genes encoding assembly factors including LRPPRC, TACO1, FASTKD2, PET100, COX10, COX14, COX15, COX20, SURF1, SCO1, SCO2, COA3, COA5 and COA6 (11–28). Although CcO deficient patients display heterogeneous clinical presentations, it has been noted that mutations in the assembly factors involved in the same pathway exhibit similar clinical phenotypes. For example, patients with pathogenic mutations in the copper metallochaperones SCO1 and SCO2, which are involved in copper delivery to the CuA site of CcO subunit COX2, typically develop neonatal encephalopathy and hypertrophic cardiomyopathy (18–21). Similarly, patient mutations in COA6 also result in neonatal hypertrophic cardiomyopathy (23,24); however, the precise role of COA6 in CcO assembly has remained unknown.
We first reported that COA6 is essential for CcO assembly in yeast, zebrafish and human cells possibly by delivering copper to COX2 subunit (29). While up to nine factors have been implicated in copper delivery to CcO subunits COX1 and COX2, the precise role of many of them remains obscure (2). The components of the copper delivery pathway that have been reconstituted in vitro suggest that COX17 receives copper from a mitochondrial matrix pool (30) and donates it to copper metallochaperones COX11 and SCO1/SCO2 (31), which ultimately transfer copper to the CuB site in COX1 and CuA site in COX2, respectively (32,33). Multiple other proteins, including COA6, have been implicated in this copper delivery pathway, but it is not clear where they act in the pathway.
Our initial findings linking COA6 to COX2 biogenesis and mitochondrial copper metabolism have been corroborated by two recent studies that show reduced stability of nascent COX2 in COA6 deficient cells and that COA6 binds to copper in vitro (34,35). While these studies have also identified physical interactions between COA6 and copper metallochaperones SCO1 and SCO2, the functional significance of the interaction is not known. Here, we have used genetic epistasis analysis in yeast to demonstrate that COA6 is essential for Cox2 expression in the absence of SCO2 and COX12, a structural subunit of CcO. This synthetic interaction between Coa6, Sco2 and Cox12 proteins suggests their overlapping functions in Cox2 biogenesis. Consistent with this observation, we find that overexpression of Cox12 and Sco proteins partially rescues the respiratory deficient growth phenotype of coa6Δ cells. We further substantiate these genetic data by demonstrating a physical interaction between Coa6, Cox12, Cox2 and Sco proteins. Taken together, our study not only places Coa6 in the copper delivery pathway to Cox2, but also implicates COX12, a yeast orthologue of the human mitochondrial disease gene COX6B, in copper metabolism and Cox2 biogenesis.
Results
Yeast and human COA6 are orthologues
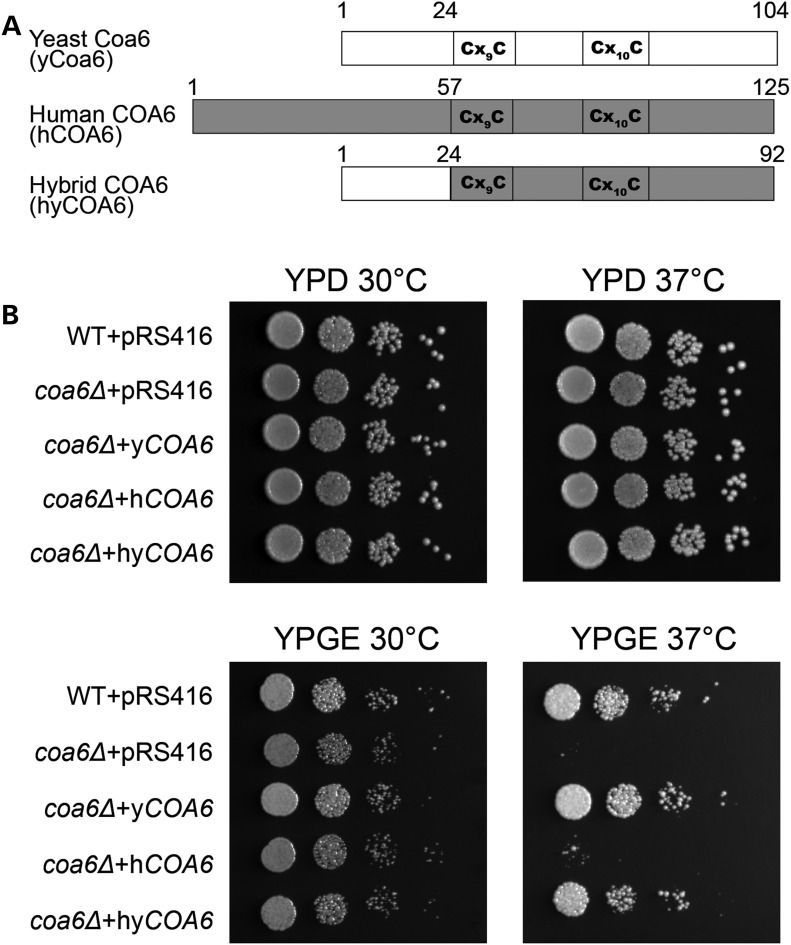
Previously, we showed that COA6 is an evolutionarily conserved protein that is required for the expression of CcO subunits including COX2 in yeast, zebrafish and human cells (29). Sequence alignment of yeast and human COA6 showed that these two proteins are highly conserved except in the N-terminal region (29), possibly because of differences in N-terminal mitochondrial targeting sequences in these two species. The sequence conservation and their common role in CcO expression suggested that yeast and human COA6 are orthologues. To experimentally test the functional similarity of these two proteins, we performed a complementation experiment by heterologous expression of human COA6 in yeast coa6Δ cells. We episomally expressed yeast COA6 (yCOA6), human COA6 (hCOA6) or yeast–human hybrid COA6 (hyCOA6) in coa6Δ cells and tested for their ability to rescue the respiratory growth deficiency of coa6Δ cells. The hyCOA6 was constructed by fusing the gene segments corresponding to the N-terminus of yeast Coa6 (amino acid residues 1–24) and the C-terminus of human COA6 (amino acids residues 57–125) containing the evolutionarily conserved Cx9CxnCx10C motif (Fig. 1A). As expected, yCOA6 completely rescued the respiratory growth defect of coa6Δ cells, but hCOA6 was not able to rescue this growth defect (Fig. 1B). However, hyCOA6 was able to completely restore respiratory growth (Fig. 1B) suggesting that yeast and human COA6 are orthologues.
Figure 1.
Heterologous expression of yeast–human hybrid COA6 rescues respiratory growth defect of yeast coa6Δ cells. (A) Schematic representation of yeast Coa6, human COA6 and hybrid COA6 proteins. The sequence from yeast Coa6 is shown in white, and the sequence from human COA6 is shown in gray. (B) Ten-fold serial dilutions of WT and coa6Δ cells transformed with either empty vector (pRS416) or with pRS416 expressing yeast COA6 (yCOA6), human COA6 (hCOA6) or hybrid COA6 (hyCOA6) were spotted on fermentable (YPD) and non-fermentable (YPGE) growth media. Plates were incubated at 30°C and 37°C and images were taken after 2–4 days of growth. These data are representative of three independent experiments.
Coa6 is regulated by mitochondrial biogenesis factors
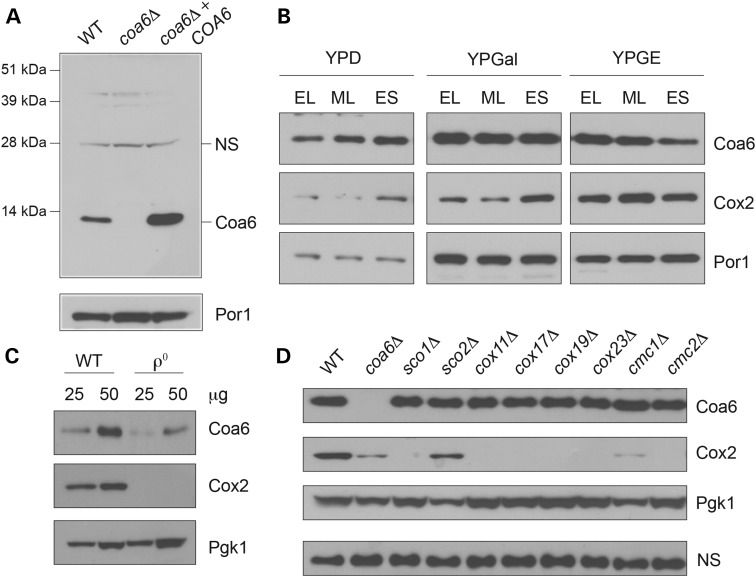
Having established the orthologous relationship between the human and yeast Coa6 protein, we decided to use yeast as a model system to study the function and regulation of Coa6. To study the regulation of the endogenous yeast Coa6 protein, we generated polyclonal antibodies against native Coa6 protein. We confirmed antibody specificity by detecting a Coa6 specific band of ∼12 kDa in cellular extracts from wild type (WT) and Coa6 overexpressing cells (Fig. 2A). We hypothesized that Coa6, as a mitochondrial protein, would be regulated by mitochondrial biogenesis factors including carbon source, growth phase and the presence of mitochondrial DNA. Growth of yeast cells in respiro-fermentable (YPGal) or non-fermentable (YPGE) media is known to stimulate mitochondrial biogenesis, and accordingly, Coa6 levels were higher in cells grown in YPGal and YPGE medium compared with fermentable medium (YPD) (Fig. 2B). In glucose containing YPD medium, Coa6 expression increased late in the growth phase, likely because of derepression of glucose-mediated inhibition of mitochondrial biogenesis (Fig. 2B). Interestingly, we found that an increase in Coa6 levels precedes an increase in Cox2 levels (Fig. 2B), implying that the presence of Coa6 is essential for Cox2 expression. Consistent with a previous report, which showed a suppression of mitochondrial biogenesis in mitochondrial DNA deficient ρ0 yeast cells (36), we observed a dramatic decrease in Coa6 levels in ρ0 cells (Fig. 2C), further confirming that Coa6 levels are regulated by mitochondrial biogenesis.
Figure 2.
Coa6 expression is regulated by mitochondrial biogenesis factors and is independent of CcO assembly factors. (A) Total cellular protein was extracted from BY4741 WT, coa6Δ and coa6Δ transformed with pRS426-COA6 and subjected to SDS PAGE/western blot. Yeast Coa6 protein was detected using purified polyclonal Coa6 antibody; NS designates a non-specific protein band detected by polyclonal Coa6 antibody. Porin (Por1) was used as a loading control. (B) Western blot analysis of Coa6 and Cox2 protein levels in total cellular extracts from BY4741 WT yeast cells grown in YPD, YPGal or YPGE to early logarithmic, mid-logarithmic (ML) and early stationary (ES) growth phase. Por1 was used as a loading control. (C) Western blot analysis of Coa6 and Cox2 levels in BY4741 WT and ρ0 cells grown in YPGal medium to ML growth phase. Pgk1 was used as a loading control. As indicated, 25 or 50 μg of protein was loaded in each lane. (D) Western blot analysis of Coa6, Cox2 and Pgk1 protein levels in the indicated knockout strains grown to ML growth phase in YPGal medium. Since Pgk1 levels increased in cells completely lacking Cox2, a non-specific (NS) band was used as a loading control. The data in panels B-D are representative of three independent experiments.
It has been shown that steady-state levels of some of the CcO assembly factors are reciprocally regulated. For example, levels of Cmc1, a copper-binding mitochondrial intermembrane space (IMS) protein, increase in response to deletion of Cmc2, another CcO assembly factor (37). To tie Coa6 to CcO assembly factors involved in copper metabolism, we explored the possibility that Coa6 levels are regulated by other CcO assembly factors. Therefore, we measured Coa6 levels in various yeast strains lacking known copper metallochaperones including Cox11, Cox17, Sco1 and Sco2 and twin Cx9C motif-containing proteins implicated in CcO assembly, including Cox19, Cox23, Cmc1 and Cmc2. We found that the levels of Coa6 did not change in the knockout strains tested (Fig. 2D). Interestingly, we noticed that the levels of Pgk1, a glycolytic enzyme used as a loading control, increased in all CcO assembly factor mutants that are completely devoid of Cox2 (Fig. 2D). Therefore, we used a non-specific band to confirm equal loading (Fig. 2D). The increase in Pgk1 could be a homeostatic mechanism to maintain cellular ATP production in Cox2 deficient cells that are unable to produce mitochondrial ATP. In a reciprocal experiment, we measured the levels of copper metallochaperones and CcO subunits, Cox17, Sco1, Sco2, Cmc1, Cox12 and Cox2 in coa6Δ cells and, except for a decrease in the Cox2 and Cox12 levels, we did not find any significant change in the levels of the other proteins measured (Supplementary Material, Fig. S1). This result suggests that Coa6 abundance is independent of the presence of these CcO assembly factors and vice-versa.
Coa6 levels change in response to extracellular copper abundance
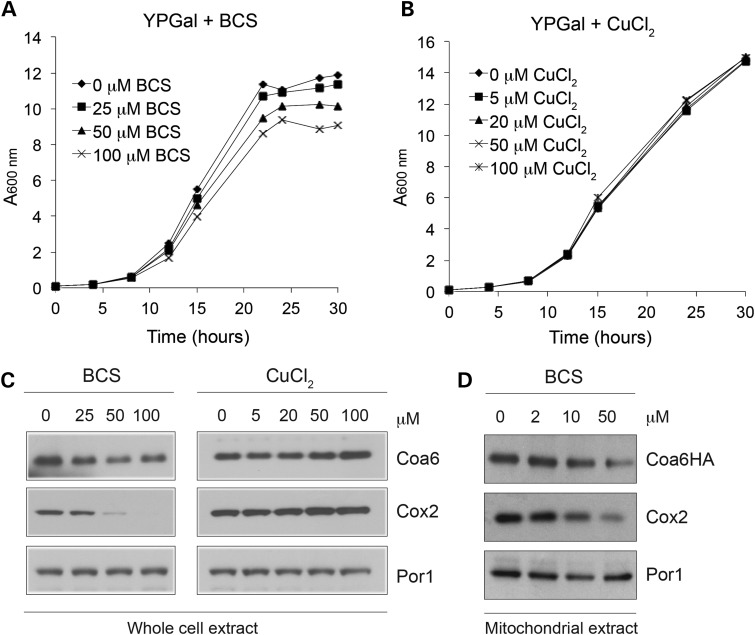
A recent study has shown that levels of iron-containing proteins in the MRC decrease in response to increasing amounts of the iron chelator deferoxamine in mouse muscle cells (38). Since Coa6 has been implicated in the copper delivery pathway to CcO (29) and has recently been shown to bind copper (34,35), we hypothesized that Coa6 levels would alter in response to copper chelation and supplementation. In order to identify the optimal concentration of copper-specific chelator bathocuproinedisulfonic acid (BCS) and copper, WT yeast were grown in respiro-fermentable YPGal medium supplemented with increasing amounts of BCS or copper chloride (CuCl2). We found that 25 μm or more of BCS reduced yeast growth in YPGal medium by limiting bioavailable copper required for respiratory growth, whereas up to 100 μm copper supplementation did not alter yeast growth (Fig. 3A and B). We observed that Coa6 levels decreased with increasing concentrations of BCS, whereas copper supplementation modestly increased Coa6 levels (Fig. 3C). Similarly, Cox2 levels decreased drastically under copper limiting conditions, while copper supplementation slightly increased Cox2 (Fig. 3C). The decrease in Coa6 with BCS supplementation is more pronounced in mitochondrial samples isolated from chromosomal hemagglutinin (HA) tagged-Coa6 cells (Fig. 3D). These results suggest that, like the copper-containing protein Cox2, Coa6 expression is dependent on bioavailable copper, further implicating it in mitochondrial copper metabolism.
Figure 3.
Coa6 levels are regulated by copper availability. WT yeast cells were grown in YPGal medium with increasing concentrations of (A) the copper chelator, BCS or (B) CuCl2. Growth was analyzed by measuring absorbance at 600 nm. (C) Western blot analysis of Coa6 and Cox2 levels in whole cell protein lysate prepared from WT yeast cells grown to ML growth phase in YPGal medium with increasing amounts of BCS or CuCl2. (D) Western blot analysis of Coa6HA and Cox2 levels in mitochondria isolated from chromosomally HA-tagged COA6 cells grown in YPD to ES growth phase with increasing amounts of BCS. Por1 was used as a loading control. The blots are representative of at least two independent experiments.
Coa6 acts in parallel with Cox12 and Sco2 to maintain Cox2 levels
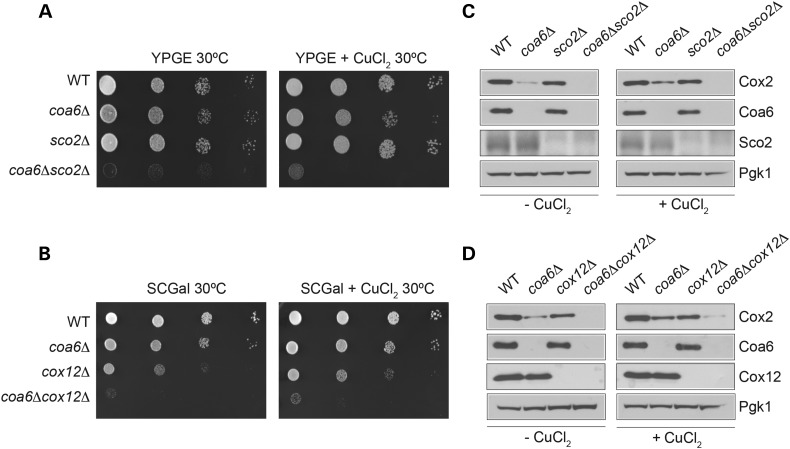
Our previous results showed that copper supplementation rescues the coa6Δ respiratory-deficient phenotype (29) and our current study shows that Coa6 levels are regulated by copper availability. Together, these studies strongly suggest that Coa6 plays a role in copper delivery to CcO. In order to place Coa6 in the CcO copper delivery pathway, a genetic epistasis analysis was performed where coa6Δ cells were crossed with the deletion strains of the known copper metallochaperones and Cx9C proteins implicated in CcO assembly. The resulting double knockouts were extensively phenotyped in different growth conditions (Supplementary Material, Table S1). Analysis of the growth phenotypes of both the parental single knockouts and the double knockouts identified strong synthetic lethal interactions of COA6 with SCO2 and COX12 (Fig. 4A and B). To probe for the mechanism that leads to synthetic lethality of double knockouts in respiratory media, we measured levels of the copper-containing CcO subunit Cox2. Cox2 levels are decreased in coa6Δ, sco2Δ and cox12Δ to different degrees, but completely absent in both coa6Δsco2Δ and coa6Δcox12Δ cells (Fig. 4C and D). Exogenous supplementation of copper fails to rescue the growth defect and Cox2 deficiency of either double knockout (Fig. 4A–D). These data suggest that Coa6, Sco2 and Cox12 play overlapping roles in the copper delivery to Cox2.
Figure 4.
Coa6, Sco2 and Cox12 have an overlapping but non-redundant role in Cox2 expression. (A) Ten-fold serially diluted WT, coa6Δ, sco2Δ and coa6Δsco2Δ cells were spotted on YPGE plates with and without 5 μm CuCl2. (B) Ten-fold serially diluted WT, coa6Δ, cox12Δ and coa6Δcox12Δ cells were spotted on synthetic complete (SC) galactose medium with and without 5 μm CuCl2. Plates were incubated at 30°C and images were taken after 3 days of growth. (C and D) WT, single knockouts of Coa6, Sco2, Cox12 and double knockouts coa6Δsco2Δ and coa6Δcox12Δ were grown to ML phase in YPGal liquid medium with and without 5 μm CuCl2. Cox2 levels in total cellular protein extracts of WT and mutant cells were analyzed by western blot. Pgk1 was used as a loading control.
Coa6 physically interacts with Cox2, Cox12 and Sco proteins
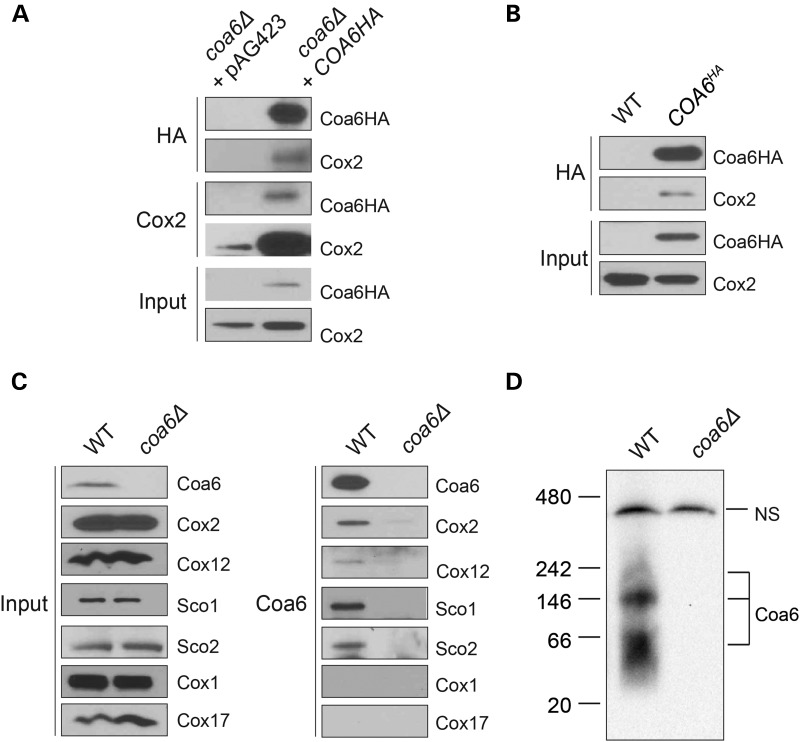
Genetic interactors tend to participate in a common pathway and are more likely to physically interact (39). To test whether Coa6 physically interacts with Cox2 and copper metallochaperones, we performed co-immunoprecipitation experiments using anti-HA and anti-Cox2 antibodies. Reciprocal co-immunoprecipitation experiments showed that Coa6 specifically interacts with Cox2 protein (Fig. 5A) and not with Cox1 or other proteins of the MRC (data not shown). In order to rule out the possibility that the interaction was merely due to Coa6 overexpression, we constructed chromosomal HA-tagged Coa6 cells, where Coa6HA is expressed from its endogenous promoter. In this case, we found a similar interaction between Coa6HA and Cox2 (Fig. 5B). Furthermore, to confirm that the interaction we observed was not because of an interaction between Cox2 and the HA tag itself, we performed an immunoprecipitation experiment using polyclonal anti-Coa6 antibodies on WT and coa6Δ mitochondria. We again found Coa6–Cox2 interaction and additionally, upon probing with other antibodies, we identified physical interactions between Coa6 and Sco1, Sco2 and Cox12, but not between Coa6 and CuB center-containing Cox1 or copper metallochaperone Cox17 (Fig. 5C). These results suggest that Coa6 exists as a part of a multimeric protein complex in mitochondria. To detect Coa6-containing complex(es), we performed a blue native polyacrylamide gel electrophoresis (BN PAGE)/western analysis on mitochondrial extracts and found that Coa6 is part of three high molecular weight complexes of ∼60, 140 and 200 kDa (Fig. 5D). We thus conclude that Coa6 and its interacting partners form complexes that may participate in Cox2 metallation.
Figure 5.
Coa6 physically interacts with Cox2, Cox12 and Sco proteins. (A) Western blot detection of Coa6-HA and Cox2 proteins in mitochondrial extracts from coa6Δ cells transformed with either empty vector (pAG423) or pAG423-COA6HA before (input) or after immunoprecipitation with anti-HA and anti-Cox2 antibodies. (B) Western blot detection of Coa6-HA and Cox2 proteins in mitochondrial extracts from WT and chromosomally HA-tagged COA6 cells before (input) or after immunoprecipitation with anti-HA antibody. (C) Western blot detection of native Coa6, Cox1, Cox2, Cox12, Sco1 and Sco2 proteins in mitochondrial extracts from WT and coa6Δ cells before (input) or after immunoprecipitation with anti-Coa6 antibody. (D) BN PAGE/western blot detection of Coa6-containing complexes from 1% digitonin solubilized mitochondria isolated from WT and coa6Δ cells grown in YPGE with 5 μm CuCl2. NS designates a non-specific protein band.
Coa6, Cox12 and Sco proteins have overlapping functions
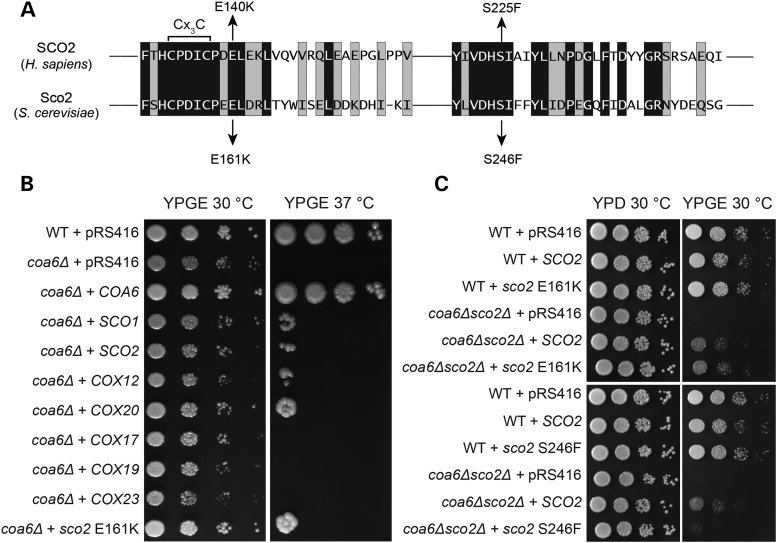
Our genetic interaction study suggested that Coa6 acts in parallel with Cox12 and Sco2 in Cox2 biogenesis; these proteins are thus likely to have overlapping functions. To test this hypothesis, we overexpressed COX12, SCO2 and SCO1, in coa6Δ cells and scored for rescue of respiratory growth on YPGE plates at 37°C. As a control, we expressed other proteins involved in Cox2 biogenesis including COX17, COX19, COX20 and COX23 in coa6Δ cells. We also expressed the yeast mimic (Sco2 E161K) of the most common human SCO2 patient mutation (E140K) (20) in coa6Δ cells to test for its ability to suppress coa6Δ growth defect (Fig. 6A). As seen in Figure 6B, the coa6Δ respiratory growth defect was partially suppressed by overexpression of SCO1, SCO2, COX12 and COX20 but not by COX17, COX19 and COX23. Interestingly, the Sco2 E161K mutant also partially rescued coa6Δ growth phenotype suggesting that E161 is not essential for yeast Sco2 function (Fig. 6B). To further interrogate yeast Sco2 function, we expressed yeast mimics (E161K and S246F) of two human SCO2 patient mutations (E140K and S225F) (20) in coa6Δsco2Δ cells. We found that while E161 is not essential, S246 is critical for coa6Δsco2Δ growth and yeast Sco2 function (Fig. 6C). While the rescue of the respiratory growth defect of coa6Δ cells by overexpression of Sco proteins and Cox20 is not surprising since these proteins are known to cooperate in the late stages of Cox2 biogenesis (40), the ability of COX12 to suppress the coa6Δ growth defect is surprising and links Cox12 to the copper delivery pathway to Cox2.
Figure 6.
Overexpression of Cox12, Cox20 and Sco proteins partially rescues coa6Δ respiratory defect. (A) Sequence alignment of conserved regions of human SCO2 and its yeast homolog. Arrows indicate amino acid residues (glutamic acid E140 and serine S225) shown to be mutated in human mitochondrial disease patients. Two mutations in yeast Sco2, E161K and S246F, that mimic the patient mutations are also indicated with arrows. (B) Ten-fold serially diluted WT cells transformed with pRS416 empty vector and coa6Δ cells transformed with pRS416 empty vector or pRS416 vector expressing either COA6, SCO1, SCO2, COX12, COX20, COX17, COX19, COX23 and sco2 E161K were spotted on YPGE plates at 30°C and 37°C. Images were taken after 3 days for cells grown at 30°C and 6 days for cells grown at 37°C. (C) Ten-fold serially diluted WT and coa6Δsco2Δ cells transformed with pRS416 empty vector or pRS416 vector expressing either SCO2, or sco2 with patient mutation E161K or S225F were spotted on YPD or YPGE plates and incubated at 30°C. Images were taken after 2 days for cells grown on YPD and 4 days for cells grown on YPGE.
Sequence alignment and structural analysis of both yeast and mammalian Coa6 and Cox12 proteins shows presence of a conserved Cx9CxnCx10C motif (Supplementary Material, Fig. S2A) with a similar predicted tertiary structure (Supplementary Material, Fig. S2B), strongly supporting their overlapping function. To further dissect the interaction between Coa6 and Cox12, we performed a reciprocal experiment, whereby we tried to rescue the respiratory growth deficient phenotype of cox12Δ with COA6 overexpression. Interestingly, we found that instead of rescuing, COA6 overexpression enhanced the cox12Δ growth defect (Supplementary Material, Fig. S2C). Taken together, our results suggest overlapping but non-redundant roles of Coa6, Sco1 Sco2, and Cox12 in Cox2 maturation.
Pathogenic mutations within the conserved Cx9CxnCx10C motif of Coa6 disrupt its interaction with Cox2
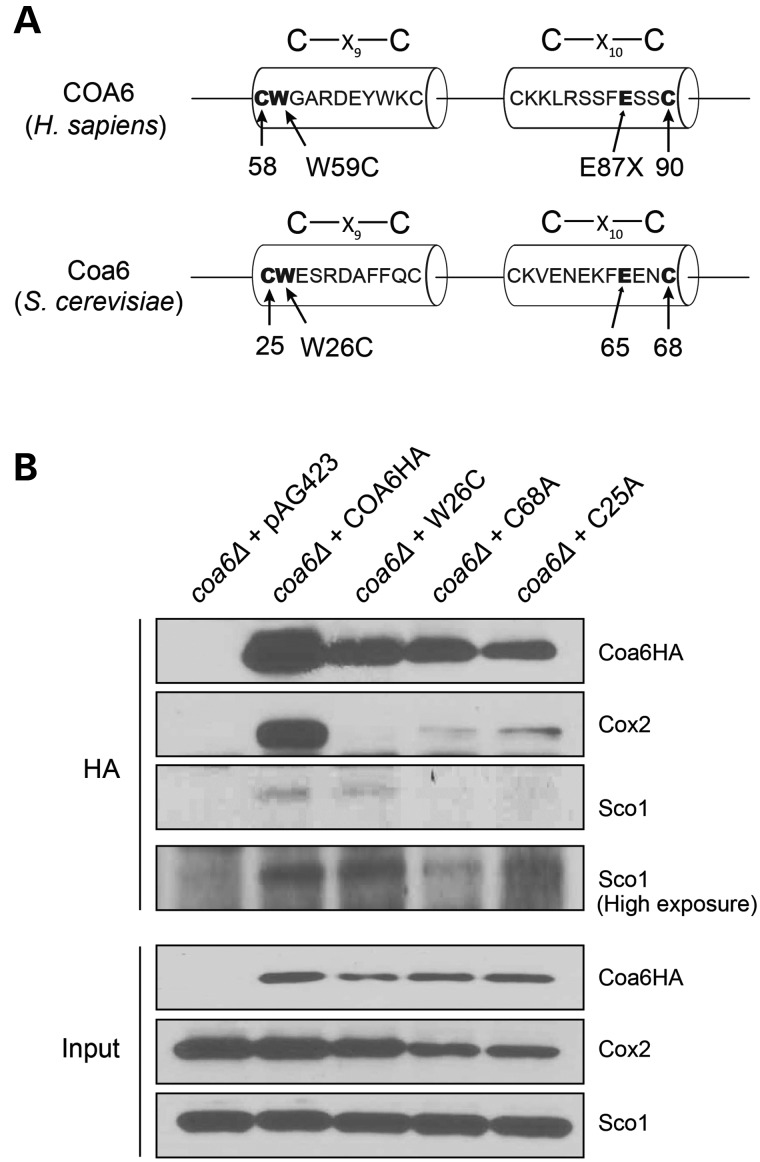
Previously, we showed that patient mutations in the Cx9CxnCx10C motif of Coa6 are pathogenic and that the conserved cysteine residues of the motif are essential for Coa6 function (29). Patient mutations reduced the stability of Coa6, partly explaining the mechanism of pathogenesis (29). However, it was not clear if reduced levels of Coa6 were sufficient to cause pathogenesis. In addition to Coa6 stability, it is possible that patient mutations may disrupt Coa6 function by preventing its interactions with its binding partners. Therefore, we tested the effect of patient mutations (p.W26C and p.C68A) as well as a mutation in a conserved cysteine residue (p.C25A) on Coa6 interactions (Fig. 7A). Co-immunoprecipitation with anti-HA antibodies confirmed that the patient mutations severely disrupted Coa6–Cox2 interaction while only mildly affecting the Coa6–Sco1 interaction (Fig. 7B). These results demonstrate that the Coa6 residues mutated in the mitochondrial disease patient are essential for its interaction with Cox2, providing the biochemical basis for the disease pathogenicity.
Figure 7.
Patient mutations disrupt Coa6 interaction with Cox2. (A) Schematic representation of human and yeast Coa6 proteins highlighting the conserved Cx9CxnCx10C motif with conserved residues that are mutated in human mitochondrial disease patient. (B) Western blot detection of Coa6-HA, Cox2 and Sco1 proteins from mitochondrial extracts of coa6Δ cells transformed with either empty vector pAG423 or pAG423 expressing COA6HA, or COA6 mutants before (input) or after immunoprecipitation with anti-HA antibodies.
Discussion
Recent advances in genomic technologies have identified pathogenic mutations in a plethora of uncharacterized genes; however, understanding the function of these disease genes has remained a major bottleneck in elucidating disease pathogenesis. Recently, we uncovered an evolutionarily conserved role of a previously uncharacterized mitochondrial disease gene, COA6, in CcO assembly and mitochondrial copper metabolism (29); however, its precise role in these processes remained unknown. In this study, we place Coa6 in the mitochondrial copper delivery pathway to CcO subunit Cox2. We also show that Coa6 has an overlapping function with a mitochondrial copper metallochaperone, Sco2 and a CcO subunit, Cox12, in Cox2 biogenesis. Like Coa6, mutations in the human orthologues of Sco2 and Cox12 have been shown to result in hypertrophic cardiomyopathy (10,20,21,23,24). Thus, our study not only links three different mitochondrial disease genes known to cause hypertrophic cardiomyopathy to the mitochondrial copper delivery pathway, but also provides mechanistic insights into Cox2 biogenesis and uncovers genetic redundancies in the copper delivery pathway.
We present several lines of evidence demonstrating that Coa6 is a novel member of the mitochondrial copper delivery pathway to Cox2. First, simultaneous deletion of Coa6 and Sco2, a well-known mitochondrial copper metallochaperone, leads to a synthetic growth defect and complete absence of Cox2 in yeast cells (Fig. 4A and C). Second, overexpression of copper metallochaperones Sco1 and Sco2 partially rescues the coa6Δ respiratory deficient phenotype (Fig. 6). Third, Coa6 physically interacts with Sco1, Sco2 and Cox2, providing biochemical evidence suggesting these proteins operate in the same pathway (Fig. 5C). These results obtained using the yeast model system are consistent with two recent studies in human cell lines showing physical interactions of COA6 with SCO2 and COX2 (34), and COA6 with SCO1 and COX2 (35). Together, these findings establish an evolutionarily conserved role of Coa6 in copper delivery to Cox2. Furthermore, overexpression experiments with known Cox2 biogenesis factors helped place Coa6 in the copper delivery pathway to Cox2. Inability of Cox17, Cox19 and Cox23, well-known IMS proteins involved in copper delivery to mitochondria, to rescue the coa6Δ growth defect suggests that these proteins act upstream of Coa6. Conversely, rescue of the coa6Δ phenotype by Sco1, Sco2, Cox12 and Cox20 suggests that these proteins either act downstream of Coa6 or in a parallel pathway of copper delivery and Cox2 maturation.
Previously, we showed that copper supplementation was able to rescue coa6Δ cells, but how copper supplementation is able to bypass the complete absence of Coa6 remained an open question (29). In light of our current findings that Coa6, Sco2 and Cox12 have an overlapping function, it is conceivable that either Sco2 or Cox12 can substitute for Coa6 when excess copper is available. We suggest Sco2 as the more likely candidate to perform this function, because we are not able to detect any Cox2 in coa6Δsco2Δ cells even after copper supplementation (Fig. 4C). We do detect a faint band corresponding to Cox2 in copper supplemented coa6Δcox12Δ cells (Fig. 4D), presumably because Sco2 is able to function in these cells to form Cox2, albeit at a much lower level. This model can also explain the mild respiratory deficient phenotypes of individual deletions of COA6 and SCO2 cells, since the absence of one is partially compensated by the other and vice-versa. The function of Sco2 in yeast cells has remained enigmatic for many years because of the absence of an overt respiratory deficient phenotype of sco2Δ cells (41), but our results now firmly tie Sco2 function to Cox2 biogenesis and provide a system to uncover the role of Sco2 in vivo (Fig. 6C).
The interaction between Coa6 and Cox12 is more intriguing, considering that Cox12, a structural subunit of CcO, has never been linked to the copper delivery pathway (42). Interestingly, COX6B, the mammalian homolog of Cox12, and Coa6 have highly homologous sequences, including a conserved Cx9CxnCx10C motif, and are predicted to have similar structures (Supplementary Material, Fig. S2A and B), which combined with their synthetic lethal interaction (Fig. 4B) strongly supports their overlapping function in Cox2 maturation. Consistent with this possibility, we observed that overexpression of Cox12 is able to partially rescue respiratory growth deficiency of coa6Δ cells. Additional evidence in support of overlapping functions of Coa6 and Cox12 comes from a human genetics study showing that a mutation in COX12/COX6B results in hypertrophic cardiomyopathy (10), a signature clinical presentation in patients with mutations in COA6 and SCO2 (20,21,23,24). Interestingly, Coa6 overexpression in cox12Δ fails to rescue the growth defect (Supplementary Material, Fig. S2C), which suggests that Cox12 has an additional function that is not compensated by Coa6.
A recent paper showed that Coa6 is part of a single oligomeric complex in human cells (34). Similarly, we identified Coa6 in multimeric complexes in yeast mitochondria (Fig. 5D). We predict that these complexes are likely to be Cox2 assembly intermediates composed of assembly factors required for copper delivery to Cox2. Our prediction is supported by co-immunoprecipitation experiments showing physical interactions of Coa6 with Cox2, Cox12, Sco1 and Sco2 proteins (Fig. 5A–C). The differences in the size and number of Coa6-containing complexes in yeast and humans could be due to differences in the Cox2 assembly processes in these two organisms. In yeast, modular assembly of mitochondrial encoded Cox1 and Cox3 subunits have been delineated (5,43), however Cox2 assembly intermediates have not yet been characterized. Our findings together with the reagents developed in this study provide the necessary tools to delineate the Cox2 assembly process in detail.
Towards understanding the molecular basis of pathogenesis, it was recently shown that the human COA6 (p.W59C) mutant is mislocalized to the mitochondrial matrix and thus would not retain its function (34). In contrast, a subsequent study suggested that the COA6 (p.W59C) mutant retains some functionality and that pathogenesis results from an increased aggregation state of the mutant protein rather than mislocalization (35). Here, we find that yeast Coa6 with the patient mutation (p.W26C) no longer physically interacts with Cox2 (Fig. 7B). However, since we are able to still detect interaction of the mutant Coa6 with Sco1, an integral membrane protein that has most of its soluble part facing the IMS, it is unlikely that the mutant Coa6 is mislocalized to mitochondrial matrix. Therefore, we argue that the cause of pathogenesis in the patient with these Coa6 mutations is loss of interaction between mutant Coa6 and Cox2.
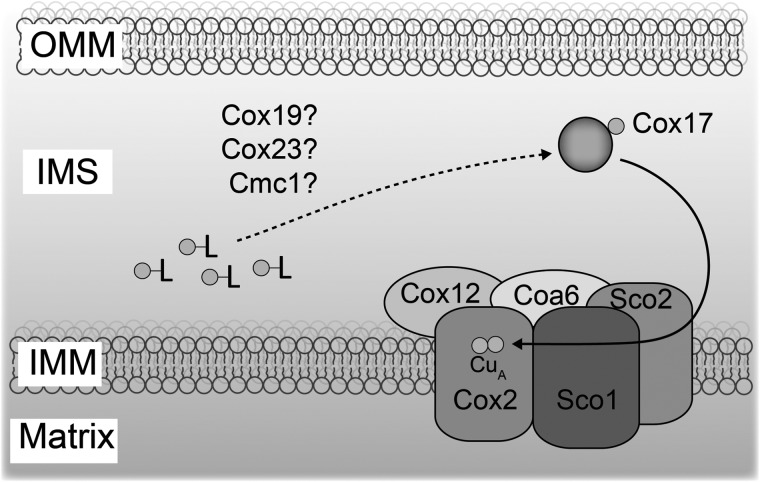
Based on our genetic and biochemical interaction studies, we propose a model placing the Coa6 and Cox12 in the copper delivery to the CuA site for Cox2 maturation (Fig. 8). Since human COA6 has been shown to bind copper with high affinity, it is possible that Coa6 acts as a metallochaperone. However, an in vitro demonstration of CuA site formation by Coa6 and its interacting partners will be required to dissect its precise molecular function in copper delivery.
Figure 8.
A proposed role of Coa6 in the mitochondrial copper delivery pathway to Cox2. Coa6 associates with Cox2, Cox12 and Sco proteins to form a copper delivery complex for CuA center biogenesis. Previously, it has been shown that Cox17 transfers copper to Sco proteins, but the mechanism by which Cox17 receives copper from a ligand bound (-L) copper pool is not known. Although Cmc1, Cox19 and Cox23 are implicated in the copper delivery pathway, we did not detect Coa6 interaction with any of these proteins. OMM, outer mitochondrial membrane; IMM, inner mitochondrial membrane; IMS, intermembrane space.
Materials and Methods
Yeast strains and culture conditions
All strains used in this study are listed in Supplementary Material, Table S2 and were confirmed by polymerase chain reaction as well as by replica plating on dropout plates. The WT yeast BY4741 cells expressing the COA6 gene with a 3′ chromosomal HA tag was constructed as previously described (44) using primers listed in Supplementary Material, Table S3. Yeast cells were grown in YP (1% yeast extract, 2% bactopeptone) medium with 2% dextrose (YPD), 2% galactose (YPGal) or 3% glycerol + 1% ethanol (YPGE) as a carbon source. Synthetic media was prepared with 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 0.2% dropout amino acid mix and contained either 2% dextrose, 2% galactose or 3% glycerol as a carbon source. Solid media additionally contained 2% agar. Growth medium was supplemented with BCS or CuCl2 wherever indicated. Growth was measured spectrophotometrically at 600 nm in liquid medium or by spotting on solid plates. Transformed yeast cells were grown in selection media to prevent loss of the plasmid. Double knockout yeast strains (Supplementary Material, Table S2) were constructed by sporulation of the diploids followed by tetrad dissection on YPD medium. The identities of all double mutant strains were confirmed by their genotypes. BY4741 ρ0 cells were obtained by culturing WT cells in the presence of ethidium bromide (25 μg/ml) for 2 days. BY4741 cox17Δ cells were constructed by one-step gene disruption using a hygromycin cassette (44).
Plasmids
Yeast COA6 gene was cloned into three different plasmids: (1) a single copy plasmid (pRS416) under control of the native promoter; (2) a multi-copy plasmid (pRS426) under control of the native promoter and (3) a Gateway cloning plasmid (pAG423-GPD-ccdB-HA) as described previously (29). The hCOA6 (PubMed Gene ID: 388753) and the hyCOA6 gene constructs were codon optimized for yeast, synthesized using GeneArt® Gene Synthesis (Life Technologies) and cloned into pRS416 plasmid under the control of the yeast Coa6 promoter. Yeast SCO1, SCO2, COX12, COX17, COX19, COX20 and COX23 were cloned into pRS416 vector under native yeast promoter. Point mutations in SCO2 were introduced by site-directed-mutagenesis using QuickChange Lightning kit from the Agilent Technologies. All the primers used in this study are listed in Supplementary Material, Table S3. All constructs were confirmed by DNA sequencing.
Coa6 purification and antibody generation
Yeast COA6 was first cloned into a pET28a-His6-GFP-TEV plasmid using EcoRI and XhoI restriction sites. This construct was then transformed into Rosetta DE3 Escherichia coli cells to express recombinant Coa6 protein. Protein purification was performed using a HisTrap™ HP column (GE Healthcare Life Sciences) and a gel filtration Superdex 200 column (GE Healthcare Life Sciences). Purified Coa6 (1.5 mg) was used to generate rabbit polyclonal antisera (Rockland Immunochemicals, Inc.). In order to obtain purified Coa6 antibody, rabbit antisera was incubated with Coa6 protein coupled to Affi-Gel 10 beads (Biorad) for 2–4 h at room temperature in a 5 ml Qiagen column. The column was then washed with 50–100 ml phosphate buffered saline (PBS) and antibodies were eluted using 0.2 m glycine, 500 mm NaCl, (pH 2.0) buffer into a tube containing 1.5 m Tris (pH 8.8) to neutralize the pH. Aliquots of purified antibody were stored at −20°C.
Whole cell yeast protein extraction and isolation of mitochondrial fractions
Yeast cells (60–80 mg wet weight) were suspended in 350 μl SUMEB buffer (1.0% sodium dodecyl sulfate (SDS), 8 m urea, 10 mm MOPS, pH 6.8, 10 mm EDTA) containing 1 mm PMSF and protease inhibitor cocktail (Roche Diagnostics). Cells were transferred to a fresh tube containing 350 mg of acid-washed glass beads (Sigma-Aldrich) and were vortexed three times for 1 min each, with 30 s incubation on ice between every vortex step. Lysed cells were kept on ice for 10 min to reduce bubbles and then heated at 70°C for 10 min. Cell debris and glass beads were spun down at 14 000 g for 10 min at 4°C. The supernatant was transferred to a separate tube and protein was quantified by the Pierce BCA protein assay kit (Thermo Scientific). Mitochondria were isolated from yeast cells as described previously (45) and mitochondrial protein concentration was quantified using a BCA assay.
SDS-polyacrylamide gel electrophoresis, BN PAGE and western blotting
SDS-polyacrylamide gel electrophoresis (SDS PAGE) and BN PAGE were performed to separate denatured and native protein complexes. For SDS PAGE, whole cell yeast protein lysate (50 μg) or mitochondria (20 μg) were separated and blotted onto a polyvinylidene difluoride membrane. For BN PAGE sample preparation, yeast mitochondria (40 μg) were solubilized in 1% digitonin and soluble lysate was resolved on a 3–12% gradient native PAGE Bis-Tris gel (Life Technologies). Membranes were blocked for 1 h in 5% nonfat milk dissolved in Tris-buffered saline with 0.1% Tween 20 (TBST-milk), followed by overnight incubation with primary antibody in TBST-milk at 4°C. Primary antibodies were used at the following dilutions: coa6, 1:1,000; Cox1, 1:1000 (Abcam 110270), Cox2, 1:10 000 (Abcam 110271); HA, 1:10 000 (Sigma H9658); Sco1, 1:500; Sco2, 1:600 and Cox17, 1:250 (from Dr Alexander Tzagoloff); Cox12, 1:1000 (from Dr Chris Meisinger); Cmc1, 1:500 (from Dr Antoni Barrientos); Porin, 1:50 000 (Abcam 110326); Pgk1, 1:50 000 (Life Technologies 459250).
Immunoprecipitation
Immunoprecipitation with anti-HA was performed using mitochondrial extracts of coa6Δ cells transformed with empty vector (pAG423), pAG423 expressing COA6HA or COA6 mutants (W26C, C25A, C68A). Mitochondrial protein (3 mg) were solubilized with RIPA buffer (Thermo Scientific) with protease inhibitor cocktail (Roche Diagnostics) for 1 h at 4°C and then centrifuged at 14 000 g for 10 min. After removal of insoluble materials, mitochondrial lysates (input) were incubated overnight with anti-HA antibody at 4°C. Protein A-agarose beads were then mixed for 2 h at 4°C to bind antibodies. After washing the beads four times with buffer containing 50 mm Tris, pH 7.4, 0.25% deoxycholate, 1% NP-40, 150 mm NaCl, 1 mm EDTA and once with PBS, beads were suspended in NuPAGE LDS sample buffer and boiled for 5 min prior to performing SDS PAGE. For immunoprecipitation with anti-Coa6 antibodies, mitochondria were isolated from WT and coa6Δ cells grown in YPGE with 5 μm CuCl2 and then solubilized with buffer containing 20 mm 4-(2-hyrdoxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 100 mm NaCl, 1 mm CaCl2, 1.5% digitonin and 10% glycerol with protease inhibitor cocktail (Roche Diagnostics) for 30 min in a rotator at 4°C. Insoluble mitochondrial fraction was pelleted at 20 000 g and the remaining soluble supernatant was used for immunoprecipitation. Immunoprecipitation was performed using Dynabeads® Protein G Immunoprecipitation Kit (Life Technologies) as per manufacturer's protocol. Briefly, anti-Coa6 was coupled to Dynabeads® Protein G and incubated with mitochondrial extract in a rotator for 2 h at room temperature. After three washes, proteins were eluted and boiled with NuPAGE LDS sample buffer prior to SDS PAGE and western blotting.
Supplementary Material
Funding
This work was supported by the National Institutes of Health award (R01GM111672) and the Welch Foundation grant (A-1810) to V.M.G.
Supplementary Material
Acknowledgements
We thank Dr Miriam L. Greenberg for the BY4741 and BY4742 wild type yeast cells; Dr Alexander Tzagoloff, Dr Chris Meisinger and Dr Antoni Barrientos for their generous gift of antibodies and Dr David Barondeau and Dr Jared Rutter for pET28aGFP-TEV and pRS plasmids, respectively. We thank Charli Baker and Shrishiv Timbalia for their technical support.
Conflict of Interest statement. None declared.
References
- 1.Vafai S.B., Mootha V.K. (2012) Mitochondrial disorders as windows into an ancient organelle. Nature, 491, 374–383. [DOI] [PubMed] [Google Scholar]
- 2.Soto I.C., Fontanesi F., Liu J., Barrientos A. (2012) Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core. Biochim. Biophys. Acta, 1817, 883–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. (1996) The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science, 272, 1136–1144. [DOI] [PubMed] [Google Scholar]
- 4.Fornuskova D., Stiburek L., Wenchich L., Vinsova K., Hansikova H., Zeman J. (2010) Novel insights into the assembly and function of human nuclear-encoded cytochrome c oxidase subunits 4, 5a, 6a, 7a and 7b. Biochem. J., 428, 363–374. [DOI] [PubMed] [Google Scholar]
- 5.McStay G.P., Su C.H., Tzagoloff A. (2013) Modular assembly of yeast cytochrome oxidase. Mol. Biol. Cell, 24, 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghezzi D., Zeviani M. (2012) Assembly factors of human mitochondrial respiratory chain complexes: physiology and pathophysiology. Adv. Exp. Med. Biol., 748, 65–106. [DOI] [PubMed] [Google Scholar]
- 7.Shoubridge E.A. (2001) Cytochrome c oxidase deficiency. Am. J. Med. Genet., 106, 46–52. [DOI] [PubMed] [Google Scholar]
- 8.Massa V., Fernandez-Vizarra E., Alshahwan S., Bakhsh E., Goffrini P., Ferrero I., Mereghetti P., D'Adamo P., Gasparini P., Zeviani M. (2008) Severe infantile encephalomyopathy caused by a mutation in COX6B1, a nucleus-encoded subunit of cytochrome c oxidase. Am. J. Hum. Genet., 82, 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shteyer E., Saada A., Shaag A., Al-Hijawi F.A., Kidess R., Revel-Vilk S., Elpeleg O. (2009) Exocrine pancreatic insufficiency, dyserythropoeitic anemia, and calvarial hyperostosis are caused by a mutation in the COX4I2 gene. Am. J. Hum. Genet., 84, 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdulhag U.N., Soiferman D., Schueler-Furman O., Miller C., Shaag A., Elpeleg O., Edvardson S., Saada A. (2015) Mitochondrial complex IV deficiency, caused by mutated COX6B1, is associated with encephalomyopathy, hydrocephalus and cardiomyopathy. Eur. J. Hum. Genet., 23, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mootha V.K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F. et al. (2003) Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl Acad. Sci., 100, 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gohil V.M., Nilsson R., Belcher-Timme C.A., Luo B., Root D.E., Mootha V.K. (2010) Mitochondrial and nuclear genomic responses to loss of LRPPRC expression. J. Biol. Chem., 285, 13742–13747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valnot I., von Kleist-Retzow J.C., Barrientos A., Gorbatyuk M., Taanman J.W., Mehaye B., Rustin P., Tzagoloff A., Munnich A., Rotig A. (2000) A mutation in the human heme A: farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum. Mol. Genet., 9, 1245–1249. [DOI] [PubMed] [Google Scholar]
- 14.Weraarpachai W., Sasarman F., Nishimura T., Antonicka H., Aure K., Rotig A., Lombes A., Shoubridge E.A. (2012) Mutations in C12orf62, a factor that couples COX I synthesis with cytochrome c oxidase assembly, cause fatal neonatal lactic acidosis. Am. J. Hum. Genet., 90, 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antonicka H., Mattman A., Carlson C.G., Glerum D.M., Hoffbuhr K.C., Leary S.C., Kennaway N.G., Shoubridge E.A. (2003) Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. Am. J. Hum. Genet., 72, 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk R., Wanschers B.F., Nijtmans L.G., Rodenburg R.J., Zschocke J., Dikow N., van den Brand M.A., Hendriks-Franssen M.G., Gilissen C., Veltman J.A. et al. (2013) A mutation in the FAM36A gene, the human ortholog of COX20, impairs cytochrome c oxidase assembly and is associated with ataxia and muscle hypotonia. Hum. Mol. Genet., 22, 656–667. [DOI] [PubMed] [Google Scholar]
- 17.Zhu Z., Yao J., Johns T., Fu K., De Bie I., Macmillan C., Cuthbert A.P., Newbold R.F., Wang J., Chevrette M. et al. (1998) SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat. Genet., 20, 337–343. [DOI] [PubMed] [Google Scholar]
- 18.Valnot I., Osmond S., Gigarel N., Mehaye B., Amiel J., Cormier-Daire V., Munnich A., Bonnefont J.P., Rustin P., Rotig A. (2000) Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal-onset hepatic failure and encephalopathy. Am. J. Hum. Genet., 67, 1104–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stiburek L., Vesela K., Hansikova H., Hulkova H., Zeman J. (2009) Loss of function of Sco1 and its interaction with cytochrome c oxidase. Am. J. Physiol. Cell Physiol., 296, C1218–C1226. [DOI] [PubMed] [Google Scholar]
- 20.Papadopoulou L.C., Sue C.M., Davidson M.M., Tanji K., Nishino I., Sadlock J.E., Krishna S., Walker W., Selby J., Glerum D.M. et al. (1999) Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet., 23, 333–337. [DOI] [PubMed] [Google Scholar]
- 21.Jaksch M., Ogilvie I., Yao J., Kortenhaus G., Bresser H.G., Gerbitz K.D., Shoubridge E.A. (2000) Mutations in SCO2 are associated with a distinct form of hypertrophic cardiomyopathy and cytochrome c oxidase deficiency. Hum. Mol. Genet., 9, 795–801. [DOI] [PubMed] [Google Scholar]
- 22.Huigsloot M., Nijtmans L.G., Szklarczyk R., Baars M.J., van den Brand M.A., Hendriksfranssen M.G., van den Heuvel L.P., Smeitink J.A., Huynen M.A., Rodenburg R.J. (2011) A mutation in C2orf64 causes impaired cytochrome c oxidase assembly and mitochondrial cardiomyopathy. Am. J. Hum. Genet., 88, 488–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calvo S.E., Compton A.G., Hershman S.G., Lim S.C., Lieber D.S., Tucker E.J., Laskowski A., Garone C., Liu S., Jaffe D.B. et al. (2012) Molecular diagnosis of infantile mitochondrial disease with targeted next-generation sequencing. Sci. Transl. Med., 4, 118ra110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baertling F., A M van den Brand M., Hertecant J.L, Al-Shamsi A., P van den Heuvel L., Distelmaier F., Mayatepek E., Smeitink J.A., Nijtmans L.G., Rodenburg R.J. (2015) Mutations in COA6 cause cytochrome c oxidase deficiency and neonatal hypertrophic cardiomyopathy. Hum. Mutat., 36, 34–38. [DOI] [PubMed] [Google Scholar]
- 25.Weraarpachai W., Antonicka H., Sasarman F., Seeger J., Schrank B., Kolesar J.E., Lochmuller H., Chevrette M., Kaufman B.A., Horvath R. et al. (2009) Mutation in TACO1, encoding a translational inactivator of COX I, results in cytochrome c oxidase deficiency and late-onset Leigh syndrome. Nat. Genet., 41, 833–837. [DOI] [PubMed] [Google Scholar]
- 26.Ghezzi D., Saada A., D'Adamo P., Fernandez-Vizarra E., Gasparini P., Tiranti V., Elpeleg O., Zeviani M. (2008) FASTKD2 nonsense mutation in an infantile mitochondrial encephalomyopathy associated with cytochrome C oxidase deficiency. Am. J. Hum. Genet., 83, 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostergaard E., Weraarpachai W., Ravn K., Born A.P., Jonson L., Duno M., Wibrand F., Shoubridge E.A., Vissing J. (2015) Mutations in COA3 cause isolated complex IV deficiency associated with neuropathy, exercise intolerance, obesity, and short stature. J. Med. Genet., 52, 203–207. [DOI] [PubMed] [Google Scholar]
- 28.Lim S.C., Smith K.R., Stroud D.A., Compton A.G., Tucker E.J., Dasvarma A., Gandolfo L.C., Marum J.E., McKenzie M., Peters H.L. et al. (2014) A founder mutation in PET100 causes isolated complex IV deficiency in Lebanese individuals with Leigh syndrome. Am. J. Hum. Genet., 94, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh A., Trivedi P.P., Timbalia S.A., Griffin A.T., Rahn J.J., Chan S.S., Gohil V.M. (2014) Copper supplementation restores cytochrome c oxidase assembly defect in a mitochondrial disease model of COA6 deficiency. Hum. Mol. Genet., 23, 3596–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cobine P.A., Ojeda L.D., Rigby K.M., Winge D.R. (2004) Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. J. Biol. Chem., 279, 14447–14455. [DOI] [PubMed] [Google Scholar]
- 31.Horng Y.C., Cobine P.A., Maxfield A.B., Carr H.S., Winge D.R. (2004) Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J. Biol. Chem., 279, 35334–35340. [DOI] [PubMed] [Google Scholar]
- 32.Hiser L., Di Valentin M., Hamer A.G., Hosler J.P. (2000) Cox11p is required for stable formation of the Cu(B) and magnesium centers of cytochrome c oxidase. J. Biol. Chem., 275, 619–623. [DOI] [PubMed] [Google Scholar]
- 33.Leary S.C., Kaufman B.A., Pellecchia G., Guercin G.H., Mattman A., Jaksch M., Shoubridge E.A. (2004) Human SCO1 and SCO2 have independent, cooperative functions in copper delivery to cytochrome c oxidase. Hum. Mol. Genet., 13, 1839–1848. [DOI] [PubMed] [Google Scholar]
- 34.Pacheu-Grau D., Bareth B., Dudek J., Juris L., Vogtle F.N., Wissel M., Leary S.C., Dennerlein S., Rehling P., Deckers M. (2015) Cooperation between COA6 and SCO2 in COX2 Maturation during cytochrome c oxidase assembly links two mitochondrial cardiomyopathies. Cell Metab., 21, 823–833. [DOI] [PubMed] [Google Scholar]
- 35.Stroud D.A., Maher M.J., Lindau C., Vogtle F.N., Frazier A.E., Surgenor E., Mountford H., Singh A.P., Bonas M., Oeljeklaus S. et al. (2015) COA6 is a mitochondrial complex IV assembly factor critical for biogenesis of mtDNA-encoded COX2. Hum. Mol. Genet., 24, 5404–5415. [DOI] [PubMed] [Google Scholar]
- 36.Epstein C.B., Waddle J.A., Hale W., Davé V., Thornton J., Macatee T.L., Garner H.R., Butow R.A. (2001) Genome-wide responses to mitochondrial dysfunction. Mol. Biol. Cell., 12, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horn D., Zhou W., Trevisson E., Al-Ali H., Harris T.K., Salviati L., Barrientos A. (2010) The conserved mitochondrial twin Cx9C protein Cmc2 Is a Cmc1 homologue essential for cytochrome c oxidase biogenesis. J. Biol. Chem., 285, 15088–15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rensvold J.W., Ong S.E., Jeevananthan A., Carr S.A., Mootha V.K., Pagliarini D.J. (2013) Complementary RNA and protein profiling identifies iron as a key regulator of mitochondrial biogenesis. Cell Rep., 3, 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M. et al. (2004) Global mapping of the yeast genetic interaction network. Science, 303, 808–813. [DOI] [PubMed] [Google Scholar]
- 40.Bourens M., Boulet A., Leary S.C., Barrientos A. (2014) Human COX20 cooperates with SCO1 and SCO2 to mature COX2 and promote the assembly of cytochrome c oxidase. Hum. Mol. Genet., 23, 2901–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glerum D.M., Shtanko A., Tzagoloff A. (1996) SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J. Biol. Chem., 271, 20 531–20 535. [DOI] [PubMed] [Google Scholar]
- 42.LaMarche A.E., Abate M.I., Chan S.H., Trumpower B.L. (1992) Isolation and characterization of COX12, the nuclear gene for previously unrecognized subunit of Saccharomyces cerevisiae cytochrome c oxidase. J. Biol. Chem., 267, 22473–22480. [PubMed] [Google Scholar]
- 43.Su C.H., McStay G.P., Tzagoloff A. (2014) The Cox3p assembly module of yeast cytochrome oxidase. Mol. Biol. Cell., 25, 965–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janke C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E. et al. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast, 21, 947–962. [DOI] [PubMed] [Google Scholar]
- 45.Meisinger C., Pfanner N., Truscott K.N. (2006) Isolation of yeast mitochondria. Methods Mol. Biol, 313, 33–39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.