Abstract
FIG4 is a phosphoinositide phosphatase that is mutated in several diseases including Charcot-Marie-Tooth Disease 4J (CMT4J) and Yunis-Varon syndrome (YVS). To investigate the mechanism of disease pathogenesis, we generated Drosophila models of FIG4-related diseases. Fig4 null mutant animals are viable but exhibit marked enlargement of the lysosomal compartment in muscle cells and neurons, accompanied by an age-related decline in flight ability. Transgenic animals expressing Drosophila Fig4 missense mutations corresponding to human pathogenic mutations can partially rescue lysosomal expansion phenotypes, consistent with these mutations causing decreased FIG4 function. Interestingly, Fig4 mutations predicted to inactivate FIG4 phosphatase activity rescue lysosome expansion phenotypes, and mutations in the phosphoinositide (3) phosphate kinase Fab1 that performs the reverse enzymatic reaction also causes a lysosome expansion phenotype. Since FIG4 and FAB1 are present together in the same biochemical complex, these data are consistent with a model in which FIG4 serves a phosphatase-independent biosynthetic function that is essential for lysosomal membrane homeostasis. Lysosomal phenotypes are suppressed by genetic inhibition of Rab7 or the HOPS complex, demonstrating that FIG4 functions after endosome-to-lysosome fusion. Furthermore, disruption of the retromer complex, implicated in recycling from the lysosome to Golgi, does not lead to similar phenotypes as Fig4, suggesting that the lysosomal defects are not due to compromised retromer-mediated recycling of endolysosomal membranes. These data show that FIG4 plays a critical noncatalytic function in maintaining lysosomal membrane homeostasis, and that this function is disrupted by mutations that cause CMT4J and YVS.
Introduction
Phosphoinositides play critical roles in the orchestration of vesicle trafficking events, often by defining the identity of specific vesicular compartments (1). Their significance is highlighted by the expanding list of diseases resulting from the disruption of phosphoinsotide-dependent events. Mutations in the phosphoinositide (3,5) bisphosphatase FIG4 can cause three different autosomal recessive diseases: Charcot-Marie-Tooth disease type 4J (OMIM #611228), a primarily demyelinating sensorimotor polyneuropathy (2,3), Yunis-Varon syndrome (YVS) (OMIM #216340), a multisystem congenital disorder affecting the nervous and musculoskeletal systems (4,5); and a familial epilepsy syndrome with cerebral polymicrogyria (OMIM #612691) (6). In addition, dominant mutations have been implicated in familial amyotrophic lateral sclerosis (ALS) type 11 (OMIM #612577) (7).
YVS is caused by homozygous or compound heterozygous null alleles of FIG4, whereas Charcot-Marie-Tooth Disease 4J (CMT4J) results when one allele of FIG4 is null and the other allele carries a hypomorphic missense mutation (e.g. I41T or L17P) (2,3,7). The functional consequence of the I41T mutation has been analyzed in mice and is believed to cause neurodegeneration by destabilizing the FIG4 protein (8,9), whereas the L17P mutation has not yet been characterized in vivo. Studies of mouse Fig4 suggest that it is required in both neurons and glia for proper neuronal survival and function (10,11). In addition to neuronal defects, YVS patients also exhibit vacuoles in muscles and cartilage, which clinically manifest as dilated cardiomyopathy and skeletal abnormalities (4,12). Degenerative phenotypes are also observed in Fig4 null mice that die ∼5 weeks of age due to neurodegeneration associated with dramatic vacuolation in the brain and dorsal root ganglia (2). It is unclear which aspects of FIG4 function are selectively altered by different disease-causing mutations and how these mutations result in disparate clinical syndromes.
FIG4 is a member of the SAC phosphatase family and removes the 5-phosphate from PI(3,5)P2 to form PI(3)P (13–15). FIG4 forms a ternary complex with two other proteins: VAC14, a scaffolding protein, and FAB1, a kinase that generates PI(3,5)P2 from PI(3)P (16,17). Surprisingly, Fig4 null mice have a reduction in PI(3,5)P2 levels (2), and increasing evidence suggests that a primary function of FIG4 in the FIG4-VAC14-FAB1 complex is to generate PI(3,5)P2 (16,18). Thus, the functional role of the phosphatase activity of FIG4 remains unclear. In yeast, deletion of any member of this ternary complex results in severe vacuolar enlargement (16). However, functional studies on these proteins in higher organisms have yielded disparate results. Depending on the experimental system, depletion of Fab1 has been shown to result in the enlargement of early endosomes, late endosomes, or lysosomes (19,20). Moreover, excessive accumulation of p62 has been observed in astrocytes in the brains of Fig4 null mice, suggesting that autophagy may be defective in the absence of FIG4 and its partners (15). In contrast, other studies have not observed autophagy disruption in Fig4 mutants (8,21,22). Furthermore, ultrastructural analyses have revealed that cytoplasmic accumulations resulting from Fig4 dysfunction are morphologically different in different cell types; whereas sensory neurons in Fig4 null mice accumulate large vacuoles, motor neurons accumulate electron-dense structures in their soma (21,23). Therefore, while FIG4 clearly plays an important role in endolysosomal trafficking in multiple systems, the step at which it functions and the role of its phosphatase activity remain unclear.
Given the important role of FIG4 in synthesis of PI(3,5)P2, FIG4 function has been studied by analyzing downstream targets of PI(3,5)P2. The lysosomal cation channel TRPML has been shown to interact with PI(3,5)P2 and has been proposed to regulate calcium efflux from lysosomes, an important step in regulating lysosomal homeostasis (24), and recently, it has been proposed that TRPML function is disrupted in cells with Fig4 deficiency(23,25). The transport of cargoes to lysosomes primarily occurs through the fusion of late endosomes with lysosomes, a process dependent on the GTPase Rab7 and its effector HOPS (homotypic fusion and protein sorting) complex. The recycling of membranes from lysosomes is less understood, but the retromer complex has been implicated in this process (26). The clinical significance of these lysosomal trafficking events is underscored by the finding that mutations in genes functioning in transport, fusion and recycling cause several neurodegenerative diseases, including lysosomal storage disease mucolipidosis IV due to TRPML mutations, a familial form of Parkinson's disease due to mutations in VPS35 (a retromer component), and CMT type 2B due to RAB7 mutations (27–30).
To better understand the pathogenesis of FIG4-related disease, we generated mutations in the Drosophila Fig4 gene. We assessed the functional significance of different FIG4 CMT4J-causing missense mutations and found that they partially rescue the lysosomal defects observed in Fig4 mutants. Surprisingly, phosphatase activity is not required for this function. Through a candidate screening approach, we have identified Rab7 and members of the HOPS complex, required for endosome-to-lysosome fusion, as potent suppressors of Fig4. Together, our data demonstrate that FIG4 plays a phosphatase-independent function in maintenance of lysosome size downstream of endosome-to-lysosome fusion, and that this function is disrupted by mutations that cause CMT4J and YVS.
Results
Fig4 null flies exhibit markedly enlarged Lysotracker-positive structures
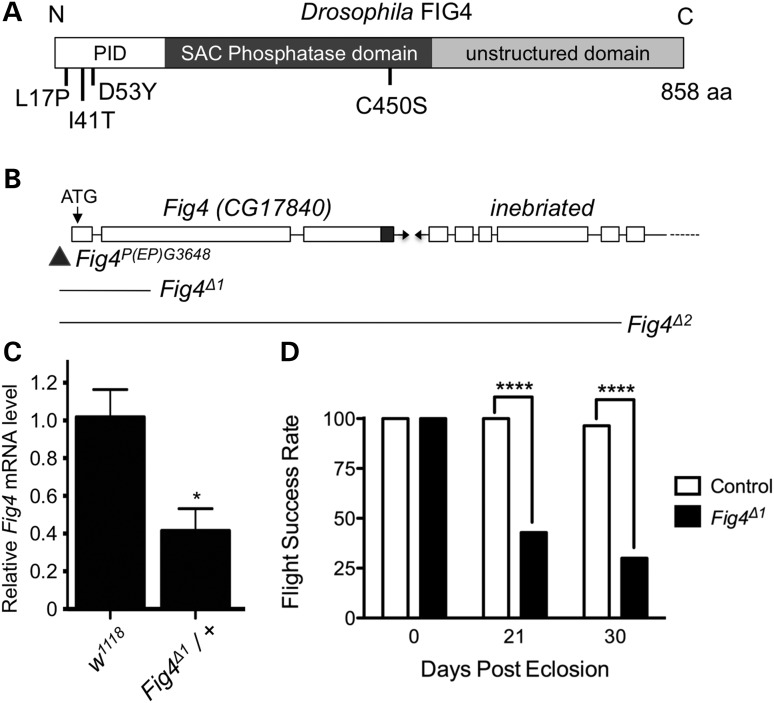
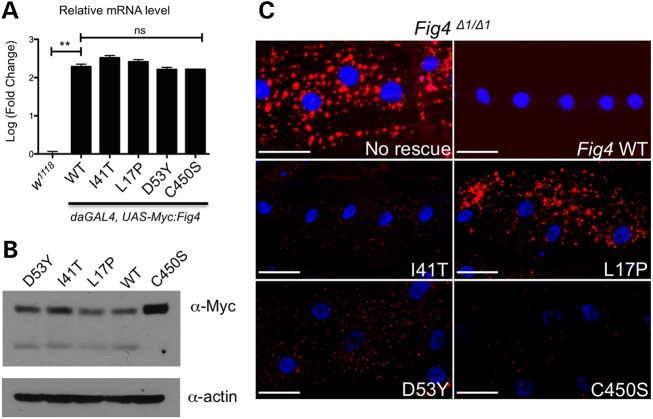
The Drosophila genome contains a single Fig4 orthologue (CG17840) that contains 59% similarity and 42% identity with human FIG4 over the entire length of the protein (Supplementary Material, Fig. S1A). Like vertebrate FIG4, Drosophila FIG4 protein carries an amino-terminal protein interaction domain (PID), a SAC phosphatase domain and an unstructured carboxy-terminus (9) (Fig. 1A). To assess the function of Fig4 in Drosophila, we generated several deletion mutations in the Fig4 gene through imprecise excisions of a P-element P(EP)G3648 (Fig. 1B). Deletion Fig4Δ1 removes 322 nucleotides including the entire first and part of the 2nd exon, whereas Fig4Δ2removes 3194 nucleotides including the entire Fig4 gene and part of the adjacent gene inebriated. Fig4Δ1 is most likely a null allele because it deletes the translation initiation site in the first exon and introduces a frame shift mutation in the 2nd exon. Furthermore, the mRNA level is reduced by ∼50% in the Fig4Δ1/+ heterozygous animals (Fig. 1C), and the phenotypic severity of Fig4Δ1 homozygous mutants is similar to the Fig4Δ1 allele in trans with Fig4Δ2 or a deficiency spanning the entire Fig4 locus (see below). Therefore, we have used the Fig4Δ1 allele for most of our analyses. Interestingly, both Fig4Δ1 and Fig4Δ2 mutants are homozygous viable, surviving to adulthood with the expected Mendelian ratio. Newborn Fig4Δ1 adults are able to fly; however, they lose this ability with age (Fig. 1D). Forty-five percent of 21 days old and 30% of 30 days old Fig4Δ1 mutant flies can fly, in contrast to 100% of 21 days old and 96% of 30 days old WT flies. Thus, Fig4 appears to be required for maintenance of flight with aging, but is not an essential gene in Drosophila.
Figure 1.
Generation and characterization of Drosophila Fig4 mutants. (A) Domain organization of FIG4 protein and mutations analyzed in this study. PID is the protein interaction domain. Drosophila FIG4 has 59% similarity with human FIG4 over the entire length of the protein (Supplementary Material, Fig. S1), including conserved amino acids mutated in disease: L17P and I41T missense mutations are found in patients with CMT4J, and D53Y has been reported in a family with ALS. The C450S mutation in the conserved CX5RT SAC phosphatase domain inactivates the catalytic cysteine residue. (B) Schematic representation of Fig4 (CG17840) genomic locus harboring P-element P(EP)G3648 (triangle) and the extent of genomic deletion in various Fig4 mutants. (C) Fig4 mRNA is reduced by ∼50% in Fig4Δ1 heterozygous animals, consistent with genetic and molecular evidence that demonstrate that it is a null allele (D) Fig4 mutants lose flight ability with age. n = 30 animals for each genotype and time-point. *P < 0.05 using Student's t-test, ****P < 0.0001 using Fischer's exact test.
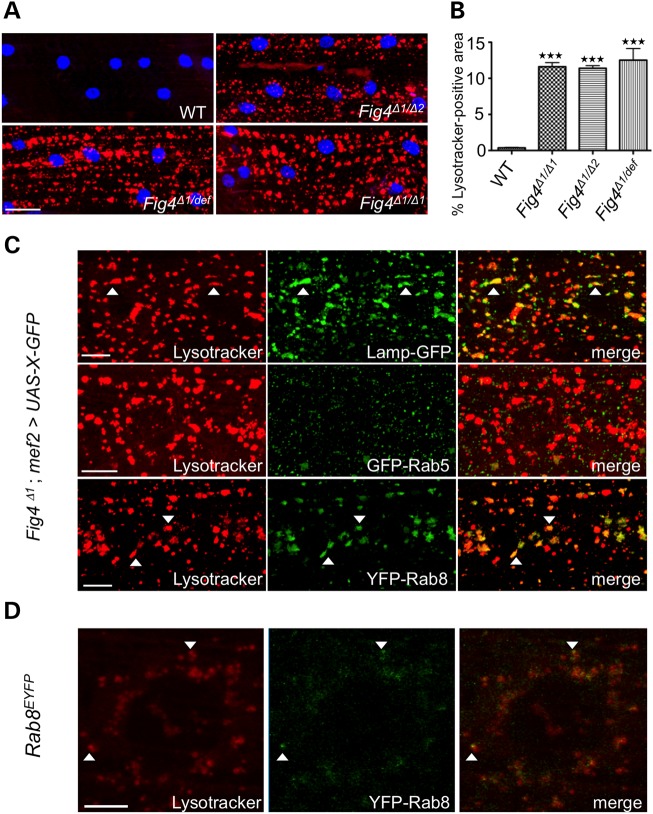
Since FIG4 and interacting proteins have been implicated in lysosomal biology, we stained wild-type and Fig4Δ1 third-instar larvae with Lysotracker, a marker of acidic organelles. In all animals tested, we find a dramatic accumulation of Lysotracker-positive structures in the muscles, but not the neuromuscular junctions, of Fig4 mutant animals (Fig. 2A and Supplementary Material, Fig. S2). Compared with wild type, Fig4Δ1 mutants show a marked increase in both the size and number of Lysotracker-positive structures. This phenotype is indistinguishable in Fig4Δ1/Δ1, Fig4Δ1/Δ2 and Fig4Δ1/def animals (Fig. 2A and B), consistent with the notion that Fig4Δ1 is a null allele (Fig. 1B and C). These results demonstrate that acidic organelles become enlarged in Fig4 mutant animals.
Figure 2.
Fig4Δ1 mutants show accumulation of Lysotracker-positive structures. (A) WT and Fig4 mutant larvae stained with Lysotracker (red) and TO-PRO-3 (blue). (B) Quantification of percentage area occupied by Lysotracker-positive structures. Muscles 6/7 in 30 hemisegments were examined for each experiment. Data are mean ± s.e.m. ***P < 0.005. (C) Larval muscles of Fig4Δ1/Δ1 mutants with mef2-Gal4-driven expression of GFP-tagged vesicle markers were stained with Lysotracker. Colocalization between Lysotracker and GFP-positive structures was examined (n = 30 hemisegments). (D) Endogenous YFP-tagged Rab8 partially colocalizes with Lysotracker. Scale bars: 30 μm in (A), 10 μm in (C), 5 μm in (D). Arrowheads highlight colocalization.
Fig4 Δ1 mutants show enlargement of the lysosomal compartment
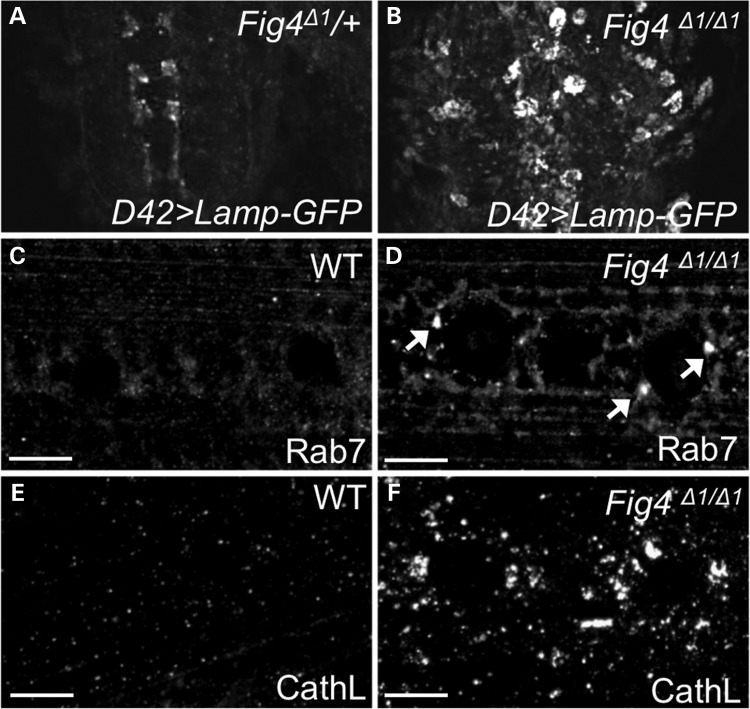
Lysotracker stains acidic organelles, and thus represents late endosomes, lysosomes or autophagolysosomes. FIG4 has been proposed to regulate trafficking of each of these organelles in addition to other endosomal compartments (15,16,21,31). To determine the identity of the Lysotracker-positive structures, we expressed fluorescently tagged markers of specific endolysosomal compartments in Fig4Δ1 larvae. We find that Lystotracker-positive structures largely colocalize with GFP-tagged lysosome-associated membrane protein (Lamp-GFP), but do not colocalize with early endosomal markers such as GFP-Rab5 or FYVE-GFP (Fig. 2C and data not shown). Similar to what was observed in muscle cells, large Lamp-GFP puncta are also observed in motor neurons of the CNS in Fig4Δ1 larvae (Fig. 3A and B). Interestingly, we observe colocalization of Lysotracker with overexpressed YFP-Rab8 in Fig4Δ1 animals (Fig. 2C and Supplementary Material, Fig. S3), suggesting that Rab8 may play a role in vesicle trafficking to acidic compartments as has been proposed (32). To confirm that this colocalization was not an artifact of overexpression, we analyzed expression of YFP-Rab8 expressed under control of the endogenous promoter (33). In both wild-type and Fig4Δ1 animals, there is significant co-localization of YFP-Rab8 with Lysotracker staining in muscles (Fig. 2D and Supplementary Material, Fig. S3), consistent with a role for Rab8 in trafficking to or from lysosomes (32). These data suggest that lysosomes, but not early endosomes, are enlarged in Fig4Δ1 animals.
Figure 3.
Fig4 mutants exhibit expansion of lysosomal compartment in brain and muscles. (A and B) Confocal images of Fig4Δ1/WT and Fig4Δ1/Δ1 larval brain, expressing Lamp-GFP driven by D42 Gal4 driver. In heterozygous animals, Lamp-GFP is difficult to detect in motor neuron cell bodies, whereas it is robustly expressed in motor neuron cytoplasm in Fig4 null animals. The Lamp-GFP staining present in control animals highlights axon terminals of rare sensory neurons labeled by D42-GAL4. (C–F) WT and Fig4Δ1/Δ1 larval muscles stained with anti-Rab7 (C and D) to label late endosomes and anti-CathL (E and F) to label lysosomes. Arrows highlight occasional expanded Rab7-positive late endosomes. Scale bars: 10 μm.
To investigate late endosomes, we performed immunostaining on WT and Fig4Δ1 mutant larvae with an antibody against the late endosomal marker Rab7. Though Fig4Δ1 mutants do not show a marked increase in the number or size of Rab7-positive structures as is observed for lysosomes, we did observe occasional abnormally large late endosomes in Fig4Δ1 mutants (Fig. 3D, arrows). To confirm that lysosomes are expanded in Fig4Δ1 animals, we next stained them with an antibody against the lysosomal protease Cathepsin L. As shown in Figure 3E and F, Cathepsin L staining is markedly increased in Fig4Δ1 muscles, suggesting that this enzyme is trafficked to expanded lysosomes in Fig4 mutants. Taken together, these data demonstrate that Fig4 is required for the proper maintenance of lysosomal size in Drosophila.
The ubiquitin-interacting protein p62 accumulates in neurons and glia of Fig4 mutant mice, suggesting that Fig4 may regulate autophagy (15). However, in the muscles of Fig4Δ1 mutants, we do not observe any colocalization between the enlarged Lysotracker-positive structures and the autophagosomal marker Atg8-GFP (data not shown). To test for a role of Fig4 in autophagy, we starved third-instar larvae and analyzed autophagolysosomes in fat bodies with Lysotracker staining (34) (Supplementary Material, Fig. S4). These data suggest that Drosophila Fig4 is not required for starvation-induced autophagy, in agreement with other studies of Fig4 in mice (22).
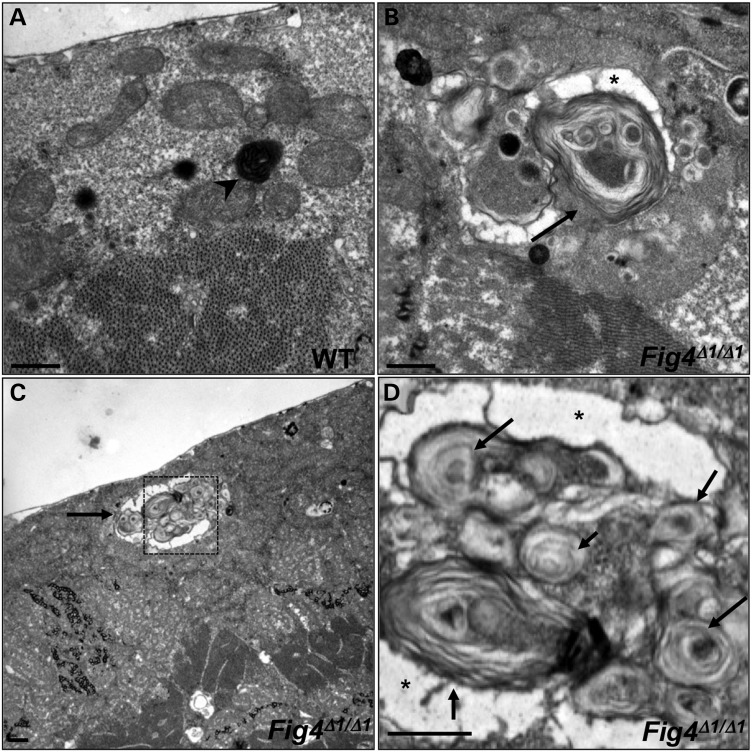
To further characterize the expanded lysosomes in Fig4 mutants, we performed ultrastructural analysis of muscles in WT and Fig4 Δ1 mutant larvae (Fig. 4A–D). In Fig4Δ1 mutants, but not in WT animals, we observe markedly enlarged structures that contain complex clusters of electron-dense organelles filled with whorled membranes, characteristic of lysosomes (Fig. 4B–D). Interestingly, these structures are often surrounded by electron-lucent, membrane-bound compartments (asterisk in Fig. 4B and D). These results further suggest that lysosomes and lysosomal membranes are markedly expanded in Fig4 mutant animals.
Figure 4.
Ultrastructural analysis of WT (A) and Fig4Δ1/Δ1 mutant (B–D) larval muscles. Boxed area in (C) is magnified in (D). WT lysosomes are small and electron-dense (arrowhead in A), whereas many Fig4Δ1/Δ1 lysosomes are markedly expanded (arrows in B–D) and contain numerous membranous whorls. Asterisk indicates electron-lucent regions that surround membranous whorls. Scale bars: 2 μm.
Overexpression of Drosophila Fig4 bearing CMT and ALS mutations partially rescues Fig4 null phenotypes
The robust phenotype we observe in Fig4 mutants allowed us to examine the functional effect of FIG4 mutations found in patients with CMT4J and ALS11 (OMIM #612577) (7). The residues I41 and L17, mutated in CMT4J, and D53 mutated in a family with ALS (3, 7), are conserved in Drosophila Fig4 (Supplementary Material, Fig. S1). We mutated these residues to the corresponding pathogenic amino acids to generate Drosophila Fig4I41T, Fig4L17P and Fig4D53Y cDNA rescue constructs. Multiple transgenic lines for each of these Myc-tagged constructs were generated, and a set of transgenic lines showing similar expression of Fig4 mRNA (Fig. 5A) and Myc-tagged protein (Fig. 5B) was chosen for further analysis. Importantly, driving ubiquitous overexpression of these UAS-Myc-Fig4 transgenic lines overexpresses Fig4 mRNA at >100× the level of endogenous Fig4, consistent with modENCODE data that CG17840 transcript is normally expressed at very low levels (35). These Myc-tagged WT and mutant UAS-transgenes were expressed using the mef2-Gal4 driver in the muscles of Fig4Δ1 mutant larvae, and their ability to substitute for the endogenous Fig4 gene was assessed by staining larval muscles with Lysotracker. Muscle-specific expression of WT Fig4 completely rescues the lysosomal defects observed in Fig4 mutants (Fig. 5C), and overexpression of WT and mutant Myc-FIG4 proteins in a wild-type background does not cause abnormal accumulation of Lysotracker (data not shown). All three disease-related transgenes were able to at least partially rescue the Fig4 mutant phenotype (Fig. 5C and Supplementary Material, Fig. S4B). Based on our assay, the mutation L17P appears to cause the most severe disruption of FIG4 function. These data are consistent with these mutations causing a partial loss of Fig4 function.
Figure 5.
Functional significance of human disease-related Fig4 mutations. (A) Quantitative RT-PCR analysis demonstrates that ubiquitous overexpression of N-terminal Myc-tagged Fig4 transgenes are expressed at >100× endogenous levels. (B) Western blot of whole body lysates from adult flies expressing N-terminal Myc-tagged Drosophila Fig4 transgenes under control of actin-Gal4. (C) Transgenes carrying Drosophila Fig4 with orthologous disease-associated mutations were expressed in muscle cells using Mef2-Gal4 in a Fig4 null background. Whereas wild-type FIG4 protein expression fully rescues the lysosome expansion phenotype (lysosomes labeled with Lyostracker red), CMT4J (I41T and L17P) and D53Y mutants only partially rescue this phenotype. Interestingly, a catalytically inactive mutation (C450S) in the Fig4 phosphatase domain also rescues the lysosome expansion phenotype. Scale bars: 30 μm. **P < 0.01 using pairwise t-test.
We next examined the significance of the phosphatase activity of FIG4 in lysosomal regulation. A conserved CX5R(T/S) motif is required for catalytic activity of FIG4 and other related Sac domain-containing phosphatases (31). We generated a Drosophila UAS-Fig4 mutant transgene in which the key residue (C450) of this motif was mutated to serine that has been shown to abolish phosphatase activity in multiple SAC phosphatases (Fig. 5A and B) (36,37). Surprisingly, the C450S mutant was able to fully rescue the Fig4 mutant phenotype (Fig. 5C and Supplementary Material, Fig. S4B). Interestingly, we noticed that the C450S mutant proteins reproducibly expressed at higher levels, when compared with other Fig4 transgenes (Fig. 5B), suggesting that inactivation of phosphatase activity may stabilize the FIG4 protein. These observations suggest that the phosphatase activity of FIG4 is not required for maintaining lysosomal size.
Fab1, Vac14 and trpml mutants phenocopy Fig4 lysosome expansion
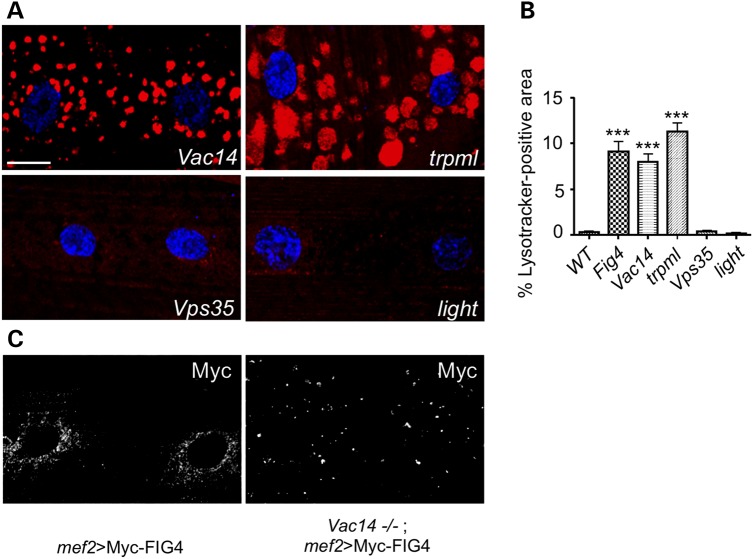
FIG4 forms a ternary complex with the scaffolding protein VAC14 and the kinase FAB1 (17). FAB1 (also called PIKFYVE) catalyzes conversion of PI(3)P to PI(3,5)P2, whereas Fig4 has been proposed to perform the opposite enzymatic reaction. To examine the role of VAC14 and FAB1 in lysosome size, we identified P-element insertions in protein-coding exons of the genes that likely represent null alleles. Vac14 mutants exhibit accumulation of Lysotracker-positive structures in muscles that closely resembles the Fig4 phenotype (Fig. 6A and B). Fab1 larvae are much smaller than control animals and arrest development as late third-instar larvae (38), precluding direct comparison with Fig4 animals at this stage. However, when we perform Lysotracker staining of early third-instar larvae, we find that the lysosomal expansion phenotype of Fab1 animals is at least as severe as that of Fig4 animals (Supplementary Material, Fig. S5).
Figure 6.
FIG4 interactors VAC14 and TRPML also regulate lysosomal size. (A) Larval fillet preps from indicated genotypes were stained with Lysotracker (red) and TO-PRO-3 (blue). (B) Quantification of lysosomal defects observed in indicated genotypes. Data are mean ± s.e.m. ***P < 0.005. (C) Examination of mef2Gal4-driven Myc-FIG4 localization in WT and Vac14 null mutants reveals that VAC14 is required for normal FIG4 localization. n = 30 hemisegments for all experiments. Scale bars: 15 μm in (A) and (C).
Given that Vac14 mutants can phenocopy Fig4 mutants, we examined the role of VAC14 in the recruitment and stabilization of FIG4. To this end, we expressed Myc-tagged FIG4 in WT and Vac14 mutant larval muscles. We observe a marked mislocalization of FIG4 in Vac14 mutants when compared with WT animals (Fig. 6C), consistent with a scaffolding role for VAC14 in regulating the localization of FIG4 (9,16–18).
Given our observations that Fig4 regulates lysosome size, we next determined whether mutations of genes in the lysosome biogenesis pathway show similar defects. Using Lysotracker staining, we examined the phenotype of light, deep orange (dor) and carnation (car) mutants, components of the HOPS complex involved in late endosome–lysosome fusion. None of these mutants exhibit Lysotracker accumulation, suggesting that the enlargement of the acidic compartment observed in Fig4 mutants is not a general consequence of defects in lysosome biogenesis (Fig. 6A and B and data not shown). It has been shown that the transport of the cation-independent mannose 6-phospate receptor to trans-Golgi network, a retromer-dependent process, is defective in FAB1/PIKFYVE-depleted cells (26). This raises the possibility that the lysosomal accumulation observed in Fig4 and Fab1 mutants results from a defect in retromer function (26). To test this hypothesis, we examined mutations in Vps35, an essential component of the retromer complex (39,40). Unlike Fig4 mutants, Vps35 mutants do not exhibit lysosomal enlargement. Thus, our findings suggest that Fig4 function is not required for lysosome biogenesis or recycling from the lysosome via the retromer complex.
We next examined the role of TRPML, a gene mutated in patients with the lysosomal storage disorder mucolipidosis type IV (27). TRPML localizes to lysosomes and directly interacts with PI(3,5)P2 (24). We observe a dramatic accumulation of Lysotracker-positive structures in trpml mutants (Fig. 6A). Lysotracker-positive structures in these mutants are larger in size when compared with Fig4 mutants. Thus, our findings suggest that FIG4, FAB1, VAC14 and TRPML are involved in the regulation of lysosomal size.
Disruption of Rab7 suppresses the Fig4 mutant phenotype
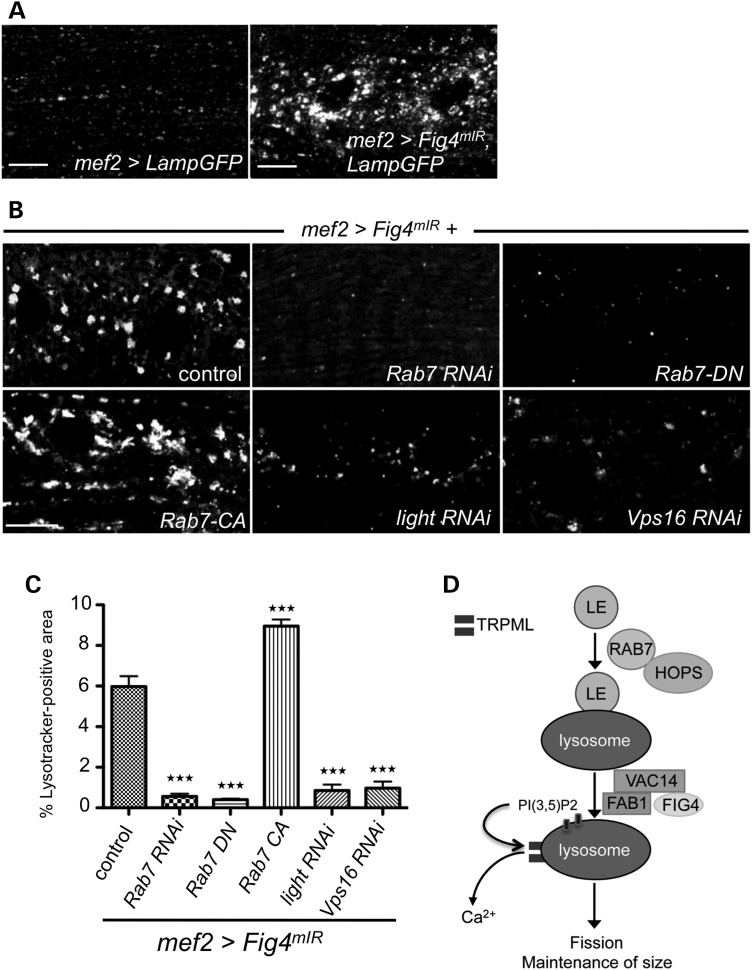
To allow tissue-specific depletion of Fig4 and high-throughput identification of genetic modifiers, we generated a transgenic line harboring two microRNAs (mIRs) in tandem that target two different Fig4 gene sequences to efficiently knockdown Fig4 under the control of the UAS promoter. As expected, the Lamp-GFP and Lysotracker accumulation phenotype is recapitulated in animals expressing this microRNA transgene (Fig4mIR) in muscle cells under control of mef2-GAL4 (Fig. 7A and B). To identify genetic modifiers of the Fig4 mutant phenotype, we examined interactions between Fig4 and genes implicated in endolysosomal trafficking, including genes implicated in CMT and ALS. We depleted these transcripts in larval muscles by expressing RNAi transgenes in combination with Fig4mIR. Interestingly, Rab7 RNAi suppresses the Fig4 loss-of-function Lysotracker accumulation (Fig. 7B and C) and CathepsinL accumulation (Supplementary Material, Fig. S6). Furthermore, expression of dominant-negative Rab7 (T22N) also potently suppresses the Fig4 mutant phenotype, whereas expression of a constitutively active Rab7 (Q67L) enhances the phenotype (Fig. 7B and C). Rab7 mediates endolysosomal trafficking through interactions with two different protein complexes: the HOPS complex and the retromer complex (39,40). We investigated whether disruption of these complexes can also suppress the Fig4 phenotype. RNAi-mediated depletion of light and Vps16, two of the components of the HOPS complex, also suppress the Fig4 phenotype (Fig. 7B and C). In contrast, disruption of the retromer complex through vps26 RNAi does not suppress the Fig4 phenotype (data not shown). Thus, Rab7 and the HOPS complex are necessary for the lysosomal accumulation observed in Fig4 mutant animals, indicating that FIG4 functions downstream of endolysosomal fusion.
Figure 7.
Lysosomal defects in response to disruption of Fig4 can be suppressed by depletion of Rab7 and HOPS complex components. (A) mIR-mediated knockdown of Fig4 in muscles with mef2-GAL4 reproduces the Fig4 null lysosome accumulation phenotype (lysosomes labeled with Lamp-GFP). (B) Candidate-based screen for modifiers of lysosome expansion (Lysotracker-positive punctae) identifies Rab7 and HOPS complex components (light and Vps16). (C) Quantification of suppression of Fig4 phenotype by various transgenes. n = 30 hemisegments. Data are mean ± s.e.m. ***P < 0.005. (D) Model for FIG4 function in lysosomal homeostasis: Late endosome (LE) fusion with the lysosome requires the Rab7/HOPS complex, but not FIG4. The FIG4-VAC14-FAB1 complex is required for PI(3,5)P2 formation, which activates TRPML function; TRPML causes calcium efflux from the lysosome which maintains lysosome size, possibly by activating lysosomal fission. This function of FIG4 requires VAC14, but not phosphatase activity, and is disrupted by mutations that cause CMT4J.
Discussion
FIG4 regulates lysosome size
FIG4 has been implicated in regulating vesicle trafficking through different vesicle compartments including early endosomes, late endosomes, lysosomes and autophagosomes in different organisms (2,15,16,21,31). In Drosophila Fig4 mutants, we observe a marked accumulation of acidic organelles that colocalize with lysosomal markers (lysosome-associated membrane protein (LAMP) and cathepsin L protease), but not with markers of early or late endosomes or autophagosomes. Furthermore, electron microscopy reveals whorls of membranous structures (Fig. 4), characteristic of lysosome storage diseases. These data indicate that FIG4 is critical for maintenance of lysosomal size in neurons and muscle cells, and in its absence, lysosomes enlarge and accumulate membranes. Similar findings of enlarged lysosomes have been made in mouse Fig4−/− neurons and glial cells (7,15), although lysosome abnormalities have not been observed in mouse Fig4−/− muscles (41).
CMT and ALS mutations cause partial loss of FIG4 function
Drosophila, yeast and mouse are all powerful experimental systems to assess the functional significance of genetic variants discovered in patients. In yeast studies, I41T and D53Y FIG4 mutants partially rescue the vacuole expansion phenotype of FIG4−/− cells treated with hyperosmolar stress (2,7). In mouse, I41T overexpression also rescues the Fig4 null phenotype (9). Similarly, in our study we find that Drosophila Fig4 transgenes containing I41T and D53Y partially rescue the lysosome expansion phenotype. Interestingly, the functional significance of L17P has not been assessed in any model organism, and among the three disease mutations (L17P, I41T and D53Y) we assessed, L17P appears to have the most deleterious impact on FIG4 function. We propose that Drosophila may be an ideal model system for investigating the functional significance of novel FIG4 variants identified in whole exome sequencing, as the effects of mutations can be quickly assessed in different cell types in a multicellular organism.
Biochemical studies suggest that the I41T Fig4 mutation disrupts its interaction with VAC14 which mislocalizes and destabilizes the protein (8,9,17). The loss of VAC14 in the mouse leads to loss of FIG4 protein as well (17). Similarly, in Drosophila, the absence of Vac14 causes marked mislocalization of FIG4 (Fig. 6C). Our findings are consistent with a hypomorphic function of I41T, as high-level expression of FIG4[I41T] can significantly rescue the Fig4 null phenotype, as has been seen in mice (9). In contrast, our data for L17P suggest that this mutation likely disrupts FIG4 scaffolding function, because at similar levels of expression to WT, this mutant shows a compromised ability to rescue Fig4 function.
FIG4 enzymatic activity is not required for maintenance of lysosome size
FIG4 forms a ternary complex with two proteins: VAC14 and FAB1 (16,17). We investigated whether its function in muscles is also dependent on its binding partners. Vac14 mutants phenocopy Fig4 mutants, suggesting that the scaffolding function of VAC14 is necessary for FIG4 function. In line with previous studies, our results show that in Vac14 mutants, FIG4 protein is markedly mislocalized (17). Together, our data support the view that VAC14 stabilizes and/or localizes FIG4 protein to membranes and is required for FIG4 function.
One of the conundrums in the field has been the observation that even though these enzymes have opposing enzymatic activities, both Fab1 and Fig4 yeast mutants exhibit a similar vacuole enlargement phenotype and reduced PI(3,5)P2 biosynthetic activity (42). Similarly, we also observe accumulation of lysosomes in both Fig4 and Fab1 mutants. To explain these observations, it has been proposed that in addition to its phosphatase activity, FIG4 also functions as a scaffolding protein, necessary for the integrity of the FAB1-VAC14-FIG4 complex (18). To test this hypothesis, we analyzed a phosphatase catalytically inactive form of FIG4 (C450S), and indeed, this mutant protein can largely substitute for wild-type FIG4. In a cosubmitted manuscript by Lenk and colleagues, (43) these authors also find that catalytically inactive FIG4 rescues vacuolization and neurodegeneration of Fig4−/− mice. These data support the hypothesis that the lysosome expansion phenotype arises due to a loss of a noncatalytic function of FIG4. Thus, the role of FIG4 phosphatase activity remains unclear and is not required to prevent lysosomal storage and neurodegeneration.
Model of FIG4 function in lysosomal homeostasis
In principle, expansion of lysosomes can occur through several mechanisms including: (i) increased delivery of organelles to the lysosome, (ii) impaired recycling of membranes from the lysosome, and (iii) impaired lysosomal fission. The retromer complex is involved in retrograde trafficking from late endosomes and lysosomes to Golgi, and retromer-dependent recycling is defective in Fab1-depleted cells (26). Thus, it is plausible that the lysosomal enlargement observed in Fig4 mutants is due to defective retromer-mediated retrograde trafficking from lysosomes. Our data argue against this possibility, as we did not observe lysosomal accumulation in mutants that disrupt retromer function. This suggests that at least in Drosophila muscles, lysosomal defects observed in Fig4 mutants do not arise from a defect in retromer function.
Another gene that plays an important role in lysosomal regulation is TRPML, which is mutated in the lysosomal storage disease mucolipidosis IV. TRPML interacts with PI(3,5)P2 on lysosomes, where it regulates calcium homeostasis (24,25) (Fig. 7D). Considering the role of FIG4 in PI(3,5)P2 synthesis, we examined lysosomal morphology in Drosophila trpml mutants (44). Indeed, similar to Fig4 mutants, we observe a dramatic accumulation of lysosomes in trpml mutants. Thus our study suggests that TRPML and Fig4 may act in the same molecular pathway to govern lysosomal size as has recently been proposed (25).
Genetic modifiers of Fig4-related lysosomal defects
To identify functional interactors of Fig4, we performed a candidate-based screen for suppressors of the Fig4 mutant phenotype. Such an approach may prove useful in identifying novel therapeutic strategies for patients with mutations in FIG4 and potentially other lysosomal storage diseases. We found that disruption of Rab7 function, either through knockdown with RNAi or using a dominant-negative approach, suppresses the Fig4 phenotype. Rab7 is required for late endosome-to-lysosome fusion through its interaction with the HOPS complex (39) (Fig. 7D). Similarly, depletion of components of the HOPS complex, a Rab7 effector, also suppresses the Fig4 phenotype. Future studies in mice or human cells are required to determine whether inhibiting endosome-to-lysosome fusion may be a valid therapeutic approach for these forms of CMT.
In summary, we have shown that Drosophila Fig4 and its binding partners play a pivotal role in regulating lysosomal size. This function of FIG4 is disrupted by mutations that cause CMT4J, but not by inactivation of the phosphatase domain. Our data support a model for FIG4 function in maintenance of lysosome size downstream of endosome-to-lysosome function.
Materials and Methods
Drosophila genetics
Fig4 (CG17840) deletion mutants were generated by imprecise excision of the P-element Fig4P(EP)G3648 inserted upstream of the first Fig4 exon. We generated multiple imprecise excisions; two of these were selected for further analysis: Fig4Δ1 and Fig4Δ2 delete 322 bp (genomic region 2L:4465214–4465535) and 3194 bp (genomic region 2L:4465214–4468407), respectively, downstream of the Fig4P(EP)G3648 P-element. Fig4Δ1 deletes the first exon that includes the translation initiation site and part of the second exon, introducing a frameshift mutation in the remaining Fig4 ORF. Fig4Δ2 deletes the entire Fig4 gene along with part of the adjacent gene inebriated. The following transgenic lines from Bloomington Stock Center were used: Rab8EYFP, UAS-YFP.Rab5, UAS-YFP.Rab8, UAS-YFP.Rab7.T22N, UAS-YFP.Rab7.Q67L, UAS-Rab7 RNAi, UAS-Vps16 RNAi, UAS-light RNAi, lt1, Mi(ET1)Fab1MB05024 and vac14 mutant- P(XP)CG560805045. The Fab1MB05024 was generated by a Minos insertion (45) into exon5 of the Fab1 gene (ET023947.1), and would be predicted to insert multiple stop codons after amino acid 1047 of FAB1 protein. Thus, this allele would be predicted to truncate the FAB1 protein before the start of the kinase domain (amino acids 1413–1797) and cause a null; however, we cannot exclude the possibility that an amino-terminal truncated protein is expressed and retains some function. vps35MH20 was a gift from Konrad Basler. UAS-Lamp.GFP was obtained from Helmut Kramer and trpml1 mutant was obtained from Craig Montell.
Fig4 microRNA constructs targeting the sequences cacctggtctacactattaagg and atcgatgcctactcaaactatc in the Fig4 transcripts were generated as described (46). Briefly, while keeping the stem-loop backbone and surrounding sequences unaltered, the Drosophila Mir6.1 gene-targeting microRNA sequence was replaced with the 22 bps complementary to the Fig4 transcript. Two microRNA constructs targeting distinct Fig4 sequences were inserted in tandem into the pUAST vector and used to generate UAS-Fig4mIR transgenic flies. For rescue experiments, the Drosophila Fig4 transgene (either wild type or I41T, L17P, D53Y, C450S generated by site-directed mutagenesis) was cloned into pUAST containing an amino-terminal MYC tag. Transgenic lines expressing similar amounts of protein were selected, and MYC-FIG4 protein expressed under the control of mef2-Gal4 driver in a Fig4 null background.
Quantitative RT–PCR
For each genotype, mRNA was collected from 10 whole flies using the TRIzol reagent (Life Technologies) following the manufacturer's protocol. Reverse transcription was performed using SuperScript III First-Strand synthesis kit (Life Technologies) following the manufacturer's protocol. Quantitative PCR was performed using SYBR Green PCR system (Applied Biosystem) on a 7900 HT fast Real-Time PCR system (Applied Biosystem). The following primers were used. For actin: forward 5′-GCGCGGTTACTCTTTCACCA-3′, reverse 5′-ATGTCACGGACGATTTCACG-3′. For Fig4: forward 5′-GCATCAACTTGTTCTTGGGCATA-3′, reverse 5′-GTAGTCCGTCTGTAGCTCCCAAA-3′. The primers were designed to span the second intron-exon junction of Fig4 (see Fig. 1).
Lysotracker staining of larval fillet preparations
Fillet preparations of wandering 3rd instar larvae were incubated in Lysotracker Red DND-99 (1:1000, Thermo Fisher Scientific) in Schneider media at RT for 5 min (47). Dissected larvae were fixed in 4% paraformaldehyde for 2 min at RT, and washed four times with PBS over 10 min. The partially fixed larvae were mounted in 70% glycerol and immediately imaged using a Zeiss LSM 510 confocal microscope. In all experiments, muscles 6 and 7 were imaged and quantitation of the lysosomal accumulation phenotype was determined by measuring the percentage area occupied by Lysotracker-positive structures using Image J.
Immunostaining and fluorescence microscopy
Dissected larval fillet preps were fixed in 4% paraformaldehyde for 20 min and immunostaining was performed as previously described (48). The following antibodies were used: mouse anti-Myc at 1:1000 (Invitrogen), rabbit anti-GFP at 1:500 (Invitrogen), rabbit anti-Rab7 at 1:100 and rabbit anti-Cathepsin L at 1:200 (gifts from P.J. Dolph). TO-PRO-3 (Thermo Fisher Scientific) was used to label nuclei. Images were acquired using a Zeiss LSM 510 confocal microscope in the JHU MPI Imaging core.
Electron microscopy
Drosophila 3rd instar larval fillets from Fig4Δ1/Δ1 mutants and controls were processed as described with minor changes (49). Larval fillets were fixed with 1% glutaraldehyde and 4% paraformaldehyde in cacodylate buffer at 4°C overnight, stained first with 2% osmium tetroxide at 4°C for an hour and then with 2% uranyl acetate for 30 min. Samples were dehydrated by incubating with increasing concentrations of ethanol and equilibrated in varying concentrations of propylene oxide and Epon resin. Larval fillets were pinned to embedding molds using insect pins and embedded in Epon resin. Ultrathin sections of the larval fillet were cut using an ultramicrotome. Sections were subsequently stained with uranyl acetate and lead citrate; TEM was performed using a Hitachi H-7000 transmission electron microscope.
Supplementary Material
Funding
The MPI Imaging Core is funded by an NINDS Core Center Grant (P30 NS050274: “JHU Center for Neuroscience Research”). This work was supported by an NIH/NINDS R01NS082563 to T.E.L. and an ALSA fellowship to K.Z.
Supplementary Material
Acknowledgements
We thank Bloomington Stock Center, Hellmut Kramer, Konrad Basler and Craig Montell for Drosophila lines and Patrick Dolph for Cathepsin and Rab7 antibodies. We thank Guy Lenk, Miriam Meisler and Mark Wu for helpful discussion and comments on this manuscript. We thank Megan Fauci, Sarah Collins, James Machamer and Brian Woolums for technical assistance.
Conflict of Interest statement: None declared.
References
- 1.Di Paolo G., De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature, 443, 651–657. [DOI] [PubMed] [Google Scholar]
- 2.Chow C.Y., Zhang Y., Dowling J.J., Jin N., Adamska M., Shiga K., Szigeti K., Shy M.E., Li J., Zhang X. et al. (2007) Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature, 448, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson G., Lenk G.M., Reddel S.W., Grant A.E., Towne C.F., Ferguson C.J., Simpson E., Scheuerle A., Yasick M., Hoffman S. et al. (2011) Distinctive genetic and clinical features of CMT4J: a severe neuropathy caused by mutations in the PI(3,5)P(2) phosphatase FIG4. Brain, 134, 1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campeau P.M., Lenk G.M., Lu J.T., Bae Y., Burrage L., Turnpenny P., Roman Corona-Rivera J., Morandi L., Mora M., Reutter H. et al. (2013) Yunis-Varon syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am J Hum Genet, 92, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakajima J., Okamoto N., Shiraishi J., Nishimura G., Nakashima M., Tsurusaki Y., Saitsu H., Kawashima H., Matsumoto N., Miyake N. (2013) Novel FIG4 mutations in Yunis-Varon syndrome. J Hum Genet, 58, 822–824. [DOI] [PubMed] [Google Scholar]
- 6.Baulac S., Lenk G.M., Dufresnois B., Ouled Amar Bencheikh B., Couarch P., Renard J., Larson P.A., Ferguson C.J., Noe E., Poirier K. et al. (2014) Role of the phosphoinositide phosphatase FIG4 gene in familial epilepsy with polymicrogyria. Neurology, 82, 1068–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chow C.Y., Landers J.E., Bergren S.K., Sapp P.C., Grant A.E., Jones J.M., Everett L., Lenk G.M., McKenna-Yasek D.M., Weisman L.S. et al. (2009) Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet, 84, 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikonomov O.C., Sbrissa D., Fligger J., Delvecchio K., Shisheva A. (2010) ArPIKfyve regulates Sac3 protein abundance and turnover: disruption of the mechanism by Sac3I41T mutation causing Charcot-Marie-Tooth 4J disorder. J Biol Chem, 285, 26760–26764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenk G.M., Ferguson C.J., Chow C.Y., Jin N., Jones J.M., Grant A.E., Zolov S.N., Winters J.J., Giger R.J., Dowling J.J. et al. (2011) Pathogenic mechanism of the FIG4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet, 7, e1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson C.J., Lenk G.M., Jones J.M., Grant A.E., Winters J.J., Dowling J.J., Giger R.J., Meisler M.H. (2012) Neuronal expression of Fig4 is both necessary and sufficient to prevent spongiform neurodegeneration. Hum Mol Genet, 21, 3525–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaccari I., Carbone A., Previtali S.C., Mironova Y.A., Alberizzi V., Noseda R., Rivellini C., Bianchi F., Del Carro U., D'Antonio M. et al. (2014) Loss of Fig4 in both Schwann cells and motor neurons contributes to CMT4J neuropathy. Hum Mol Genet, 24, 383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varghese P., Collins N., Warner G., Leitch J., Ho E., Crock P. (2014) Yunis-varon syndrome: further delineation of cardiovascular and endocrine outcome. Am J Med Genet A, 164A, 1213–1217. [DOI] [PubMed] [Google Scholar]
- 13.Ho C.Y., Alghamdi T.A., Botelho R.J. (2012) Phosphatidylinositol-3,5-bisphosphate: no longer the poor PIP2. Traffic, 13, 1–8. [DOI] [PubMed] [Google Scholar]
- 14.Efe J.A., Botelho R.J., Emr S.D. (2005) The Fab1 phosphatidylinositol kinase pathway in the regulation of vacuole morphology. Curr Opin Cell Biol, 17, 402–408. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson C.J., Lenk G.M., Meisler M.H. (2009) Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum Mol Genet, 18, 4868–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudge S.A., Anderson D.M., Emr S.D. (2004) Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol Biol Cell, 15, 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin N., Chow C.Y., Liu L., Zolov S.N., Bronson R., Davisson M., Petersen J.L., Zhang Y., Park S., Duex J.E. et al. (2008) VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J, 27, 3221–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Botelho R.J., Efe J.A., Teis D., Emr S.D. (2008) Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol Biol Cell, 19, 4273–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikonomov O.C., Sbrissa D., Shisheva A. (2001) Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J Biol Chem, 276, 26141–26147. [DOI] [PubMed] [Google Scholar]
- 20.Nicot A.S., Fares H., Payrastre B., Chisholm A.D., Labouesse M., Laporte J. (2006) The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans. Mol Biol Cell, 17, 3062–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katona I., Zhang X., Bai Y., Shy M.E., Guo J., Yan Q., Hatfield J., Kupsky W.J., Li J. (2011) Distinct pathogenic processes between Fig4-deficient motor and sensory neurons. Eur J Neurosci, 33, 1401–1410. [DOI] [PubMed] [Google Scholar]
- 22.Martyn C., Li J. (2013) Fig4 deficiency: a newly emerged lysosomal storage disorder? Prog Neurobiol, 101–102, 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo J., Ma Y.H., Yan Q., Wang L., Zeng Y.S., Wu J.L., Li J. (2012) Fig4 expression in the rodent nervous system and its potential role in preventing abnormal lysosomal accumulation. J Neuropathol Exp Neurol, 71, 28–39. [DOI] [PubMed] [Google Scholar]
- 24.Dong X.P., Shen D., Wang X., Dawson T., Li X., Zhang Q., Cheng X., Zhang Y., Weisman L.S., Delling M. et al. (2010) PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat Commun, 1, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou J., Hu B., Arpag S., Yan Q., Hamilton A., Zeng Y.S., Vanoye C.G., Li J. (2015) Reactivation of Lysosomal Ca2+ Efflux Rescues Abnormal Lysosomal Storage in FIG4-Deficient Cells. J Neurosci, 35, 6801–6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Lartigue J., Polson H., Feldman M., Shokat K., Tooze S.A., Urbe S., Clague M.J. (2009) PIKfyve regulation of endosome-linked pathways. Traffic, 10, 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun M., Goldin E., Stahl S., Falardeau J.L., Kennedy J.C., Acierno J.S. Jr., Bove C., Kaneski C.R., Nagle J., Bromley M.C. et al. (2000) Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum Mol Genet, 9, 2471–2478. [DOI] [PubMed] [Google Scholar]
- 28.Zimprich A., Benet-Pages A., Struhal W., Graf E., Eck S.H., Offman M.N., Haubenberger D., Spielberger S., Schulte E.C., Lichtner P. et al. (2011) A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet, 89, 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilarino-Guell C., Wider C., Ross O.A., Dachsel J.C., Kachergus J.M., Lincoln S.J., Soto-Ortolaza A.I., Cobb S.A., Wilhoite G.J., Bacon J.A. et al. (2011) VPS35 mutations in Parkinson disease. Am J Hum Genet, 89, 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verhoeven K., De Jonghe P., Coen K., Verpoorten N., Auer-Grumbach M., Kwon J.M., FitzPatrick D., Schmedding E., De Vriendt E., Jacobs A. et al. (2003) Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am J Hum Genet, 72, 722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sbrissa D., Ikonomov O.C., Fu Z., Ijuin T., Gruenberg J., Takenawa T., Shisheva A. (2007) Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J Biol Chem, 282, 23878–23891. [DOI] [PubMed] [Google Scholar]
- 32.del Toro D., Alberch J., Lazaro-Dieguez F., Martin-Ibanez R., Xifro X., Egea G., Canals J.M. (2009) Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol Biol Cell, 20, 1478–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunst S., Kazimiers T., von Zadow F., Jambor H., Sagner A., Brankatschk B., Mahmoud A., Spannl S., Tomancak P., Eaton S. et al. (2015) Endogenously tagged rab proteins: a resource to study membrane trafficking in Drosophila. Dev Cell, 33, 351–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neufeld T.P. (2008) Genetic manipulation and monitoring of autophagy in Drosophila. Methods Enzymol, 451, 653–667. [DOI] [PubMed] [Google Scholar]
- 35.modENCODE Consortium, Roy S., Ernst J., Kharchenko P.V., Kheradpour P., Negre N., Eaton M.L., Landolin J.M., Bristow C.A., Ma L. et al. (2010) Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science, 330, 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohde H.M., Cheong F.Y., Konrad G., Paiha K., Mayinger P., Boehmelt G. (2003) The human phosphatidylinositol phosphatase SAC1 interacts with the coatomer I complex. J Biol Chem, 278, 52689–52699. [DOI] [PubMed] [Google Scholar]
- 37.Jackson M.D., Denu J.M. (2001) Molecular reactions of protein phosphatases--insights from structure and chemistry. Chem Rev, 101, 2313–2340. [DOI] [PubMed] [Google Scholar]
- 38.Rusten T.E., Rodahl L.M., Pattni K., Englund C., Samakovlis C., Dove S., Brech A., Stenmark H. (2006) Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors. Mol Biol Cell, 17, 3989–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Epp N., Rethmeier R., Kramer L., Ungermann C. (2011) Membrane dynamics and fusion at late endosomes and vacuoles--Rab regulation, multisubunit tethering complexes and SNAREs. Eur J Cell Biol, 90, 779–785. [DOI] [PubMed] [Google Scholar]
- 40.McGough I.J., Cullen P.J. (2011) Recent advances in retromer biology. Traffic, 12, 963–971. [DOI] [PubMed] [Google Scholar]
- 41.Reifler A., Lenk G.M., Li X., Groom L., Brooks S.V., Wilson D., Bowerson M., Dirksen R.T., Meisler M.H., Dowling J.J. (2013) Murine Fig4 is dispensable for muscle development but required for muscle function. Skeletal Muscle, 3, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duex J.E., Tang F., Weisman L.S. (2006) The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J Cell Biol, 172, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenk G.M., Frei C.M., Miller A.C., Wallen R.C., Mironova Y.A., Giger R.J., Meisler M.H. (2016) Rescue of neurodegeneration in the Fig4 null mouse by a catalytically inactive FIG4 transgene. Hum. Mol. Genet., 25, 340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venkatachalam K., Long A.A., Elsaesser R., Nikolaeva D., Broadie K., Montell C. (2008) Motor deficit in a Drosophila model of mucolipidosis type IV due to defective clearance of apoptotic cells. Cell, 135, 838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Metaxakis A., Oehler S., Klinakis A., Savakis C. (2005) Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics, 171, 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C.H., Huang H., Ward C.M., Su J.T., Schaeffer L.V., Guo M., Hay B.A. (2007) A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science, 316, 597–600. [DOI] [PubMed] [Google Scholar]
- 47.Brent J.R., Werner K.M., McCabe B.D. (2009) Drosophila larval NMJ dissection. J Vis Exp, doi:10.3791/1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bharadwaj R., Roy M., Ohyama T., Sivan-Loukianova E., Delannoy M., Lloyd T.E., Zlatic M., Eberl D.F., Kolodkin A.L. (2013) Cbl-associated protein regulates assembly and function of two tension-sensing structures in Drosophila. Development, 140, 627–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramachandran P., Budnik V. (2010) Electron microscopy of Drosophila larval neuromuscular junctions. Cold Spring Harb Protoc, 2010, pdb prot5474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.