Abstract
Dental caries is the most common chronic disease worldwide, and exhibits profound disparities in the USA with racial and ethnic minorities experiencing disproportionate disease burden. Though heritable, the specific genes influencing risk of dental caries remain largely unknown. Therefore, we performed genome-wide association scans (GWASs) for dental caries in a population-based cohort of 12 000 Hispanic/Latino participants aged 18–74 years from the HCHS/SOL. Intra-oral examinations were used to generate two common indices of dental caries experience which were tested for association with 27.7 M genotyped or imputed single-nucleotide polymorphisms separately in the six ancestry groups. A mixed-models approach was used, which adjusted for age, sex, recruitment site, five principal components of ancestry and additional features of the sampling design. Meta-analyses were used to combine GWAS results across ancestry groups. Heritability estimates ranged from 20–53% in the six ancestry groups. The most significant association observed via meta-analysis for both phenotypes was in the region of the NAMPT gene (rs190395159; P-value = 6 × 10−10), which is involved in many biological processes including periodontal healing. Another significant association was observed for rs72626594 (P-value = 3 × 10−8) downstream of BMP7, a tooth development gene. Other associations were observed in genes lacking known or plausible roles in dental caries. In conclusion, this was the largest GWAS of dental caries, to date and was the first to target Hispanic/Latino populations. Understanding the factors influencing dental caries susceptibility may lead to improvements in prediction, prevention and disease management, which may ultimately reduce the disparities in oral health across racial, ethnic and socioeconomic strata.
Introduction
Dental caries (i.e. tooth decay) is the most prevalent chronic disease affecting all populations, worldwide, and if left untreated, leads to serious concomitants and comorbidities. In the USA, over 90% of adults are affected by dental caries with over 25% having untreated decay (1). Furthermore, enormous disparities in disease burden persist, with vulnerable populations such as racial/ethnic minorities, low-income groups, and residents of rural areas suffering disproportionate rates of untreated disease and greater barriers to accessing oral health care. These disparities in disease burden persist despite the fact that prevalence of caries (which includes treated decay) is higher in non-Hispanic whites than minority groups (2). In fact, Hispanic and Latino Americans count among the highest-risk populations; for example, the National Health and Nutrition Examination Survey (NHANES) 1999–2004 survey showed that Mexican Americans have nearly twice as many teeth with untreated decay, have more missing teeth due to caries, and are far more likely to report poor perceived dental health as compared with non-Hispanic whites (1). Similar disparities were observed for Hispanic children, adolescents and adults in the NHANES 2011–2012 survey (2,3).
The core disease mechanism leading to dental caries is well-known: the dissolution of the mineralized dental tissues due to acidic byproducts of bacterial metabolism. In the non-disease state, this demineralization is counteracted by natural remineralizing processes. Therefore, whether or not dental caries develops depends on the balance between mineral dissolution and re-precipitation, which is greatly influenced by a number of moderating factors. Among these factors are characteristics of the saliva (including buffering capacity, anti-microbial agents, remineralization-promoting agents and rate of saliva production), characteristics of the teeth (including morphology and position of the teeth, and structure and composition of the enamel), microbial flora, host immunity and exogenous fluoride exposures. These factors are in turn influenced by many environmental exposures (e.g. tobacco, diet and medications), behaviors (e.g. oral hygiene) and socioeconomic conditions (e.g. access to oral health care, cultural attitudes toward oral health). In all, the etiology of dental caries is extremely multifactorial (7).
Many of the factors influencing risk of dental caries are hypothesized to include a genetic component, and indeed, indices of dental caries experience are highly heritable (30–60%) (4–6). Previous genetic studies have identified some variants influencing dental caries. Notably, enamel matrix and related genes have been implicated in candidate gene approaches [as reviewed in Vieira et al. (8) and Opal et al. (9)], and a variety of loci have been nominated via genome-wide association studies (GWASs) (10–14). For example, MPPED2 and ACTN2, were among the top signals from the first GWAS of dental caries, which was limited to children of European ancestry (10), and have subsequently shown evidence of association in some, but not all, independent replication samples (15). Other GWAS studies in adults have yielded significant and ‘suggestive’ associations within or near genes with roles in tooth development and host defense (11–13). The most significant among these are LYZL2, a bacteriolytic agent that may affect oral pathogens, and AJAP1, a gene implicated in independent GWAS studies of both adults (12) and children (14). To date, none of the dental caries loci nominated in GWAS studies in adults have been followed up in fine-mapping or replication studies. Altogether, previously identified genetic variants explain only a fraction of the disease heritability suggesting that additional genetic contributors have yet to be identified. Moreover, given the complexity of caries etiology, and the spectrum of contributing environmental factors, we speculate the effects of some genetic variants may vary across different populations.
No GWAS studies of dental caries have been performed in non-white populations. Therefore, it remains unknown whether the same or different variants are important across ancestry groups. Here, we report the first GWAS of dental caries in samples from six Hispanic and Latino US populations as part of the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) initiative. The aim of the present study, which is the largest GWAS of dental caries to date, was to nominate genetic loci in this understudied and growing population. Our ultimate goal of understanding the genetic contributors to dental caries may lead to insights into preventive or intervention strategies for alleviating disparities in disease burden and improving dental health.
Results
Characteristics of the HCHS/SOL sample are given in Table 1. In general, dental caries prevalence, and two indices of dental caries experience, decayed, missing and filled tooth surfaces (DMFS) and decayed, missing and filled teeth (DMFT) (see the ‘Materials and Methods’ section for phenotype definition), were higher than national averages, which was expected given the large proportion of foreign-born immigrants and their status as ethnic minorities with relatively low socioeconomic status (14). Differences across ancestry groups were observed, with Mexicans and Central Americans having lower caries prevalence and lower DMFT and DMFS indices than other ancestry groups [due in large part to lower rates of missing teeth (17)]. Cuban and Dominican groups had higher numbers of missing teeth.
Table 1.
Demographic characteristics and dental caries phenotypes in the HCHS/SOL sample and sub-samplesa
| Sample | N | Female | Age | Caries prevalence | DMFT |
DMFS |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| % | Mean (range) | % | Mean (range) | Heritability (95% CI) | λ | Mean (range) | Heritability (95% CI) | λ | ||
| Cuban | 1950 | 52.8 | 47.6 (18, 75) | 96.9 | 11.54 (0, 26) | 0.32 (0.09, 0.56) | 0.976 | 39.12 (0, 117) | 0.37 (0.13, 0.61) | 0.975 |
| Dominican | 1083 | 64.3 | 45.1 (18, 75) | 96.0 | 10.77 (0, 25) | 0.47 (0.16, 0.77) | 0.999 | 34.7 (0, 109) | 0.45 (0.14, 0.76) | 0.995 |
| Mexican | 4578 | 60.3 | 44.4 (18, 76) | 92.7 | 8.79 (0, 26) | 0.20 (0.10, 0.30) | 1.009 | 26.72 (0, 127) | 0.28 (0.18, 0.39) | 1.026 |
| Puerto Rican | 2006 | 57.2 | 47.0 (18, 75) | 95.4 | 10.3 (0, 26) | 0.34 (0.14, 0.55) | 0.994 | 34.59 (0, 118) | 0.33 (0.13, 0.54) | 0.985 |
| Central American | 1294 | 59 | 44.1 (18, 74) | 93.1 | 9.42 (0, 26) | 0.53 (0.25, 0.82) | 0.999 | 30.51 (0, 122) | 0.46 (0.18, 0.74) | 0.998 |
| South American | 843 | 59.4 | 46.3 (18, 76) | 96.9 | 11.63 (0, 26) | 0.44 (0.09, 0.79) | 0.993 | 38.95 (0, 117) | 0.42 (0.08, 0.76) | 0.996 |
| Total | 11754 | 58.7 | 45.5 (18, 76) | 94.5 | 9.96 (0, 26) | 0.16b (0.11, 0.22) | 1.006c | 32.15 (0, 127) | 0.20b (0.14, 0.25) | 1.017c |
aDescriptive statistics are shown for the sample of participants included in the GWAS (rather than target population estimates).
bHeritability for the total sample was estimated via ‘mega-analysis’ of all participants combined.
cGenomic inflation calculated from the genome-wide meta-analysis of the six ancestry groups.
The heritability (i.e. proportion of variation attributable to the genetically determined kinship) of DMFT ranged from 20% in the Mexican group to 53% in the Central American group. The heritability of DMFS was similar (ranging from 28% in the Mexican group to 46% in the Central American group). These values are comparable with traditional heritability estimates of DMFT (6) and DMFS (5) in the permanent dentition using family-based study designs. The range in values is expected because heritability estimates are population-specific, and may reflect differences across ancestry groups in either the cumulative effects of genetic factors, or the total variance (including environmental sources of variance) or both (18). Across all ancestry groups combined, the proportion of variation in DMFT and DMFS attributable to the genetically determined kinship was somewhat lower, 16 and 20%, respectively, which is expected given the increased total variance when combining groups. Overall, these estimates indicate that genetics has an important role in dental caries experience, and reinforces our view that gene-mapping approaches such as GWAS are suitable for identifying specific variants influencing disease.
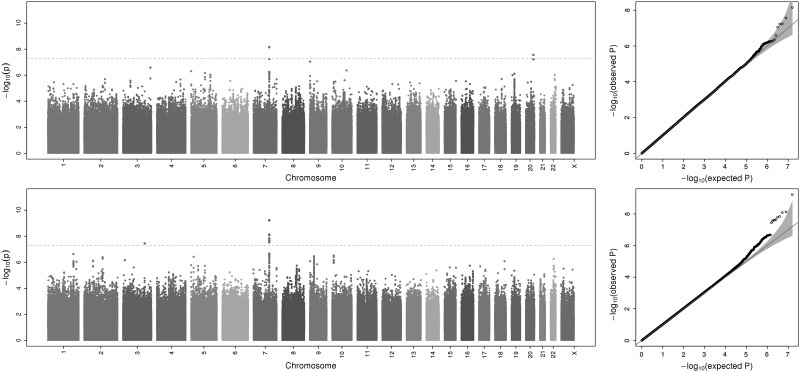
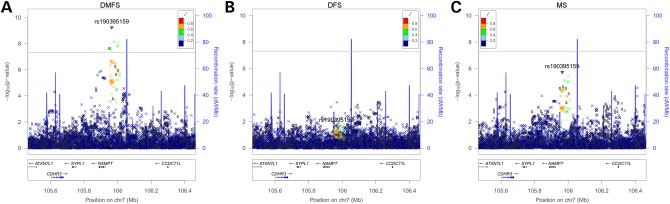
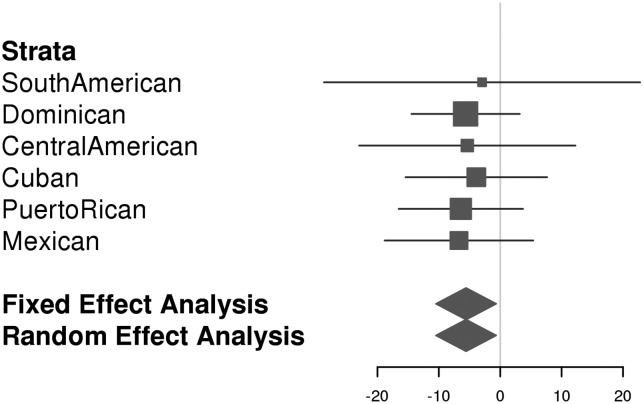
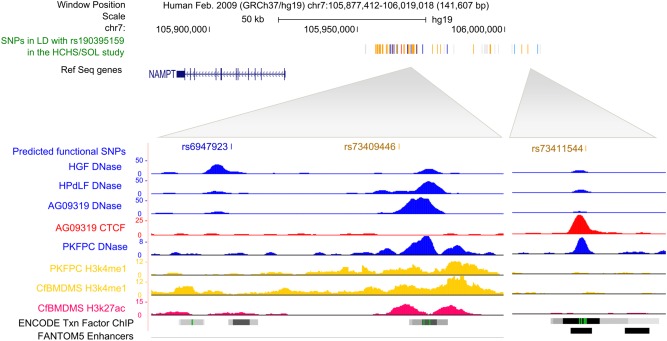
GWAS scans were performed for each ancestry group and meta-analysis was used to combine results across the six samples. Overall, GWAS scans for DMFT and DMFS yielded multiple associations meeting genome-wide significance (i.e. P-values <5 × 10−8; Fig. 1), and no evidence of genomic inflation was observed [see quantile– quantile (Q–Q) plots in Fig. 1; λ = 1.006 and 1.017, respectively, for DMFT and DMFS] indicating that model assumptions were adequately satisfied. For both dental caries phenotypes, the strongest association signal was observed for the single-nucleotide polymorphism (SNP) rs190395159 just upstream of the gene NAMPT (P-value = 7.14 × 10−9 for DMFT and P-value = 5.97 × 10−10 for DMFS; Fig. 2A). This SNP (A/G) has a minor allele (G) frequency of 0.7% in HCHS/SOL sample, whereas in the 1000 Genomes Project the G allele is absent from Asian and South Asian populations, and occurs at frequencies of 19, 2 and ∼0% in African, American and European populations, respectively. Effect sizes for rs190395159 were similar within each of the six ancestry groups (Fig. 3), and there was no evidence of heterogeneity (Cochran's Q P-value = 0.9). Multiple intronic SNPs within NAMPT, which were uncorrelated with the top SNP rs190395159, showed associations having P-values in the 1 × 10−7 to 5 × 10−6 range; however, no associations with coding variants were observed. Of note, data from large-scale efforts to identify functional non-coding elements [ENCODE (19), Roadmap Epigenomics (20) and FANTOM5 (21) projects] show that three SNPs (rs6947923, rs73409446 and rs73411544) in high-linkage disequilibrium (r2 ≥ 0.7) with rs190395159 occur within putative enhancers upstream of NAMPT (Fig. 4).
Figure 1.
Manhattan (left) and Q–Q (right) plots showing meta-analysis results (log10-transformed P-values) for DMFT (top) and DMFS (bottom). Genomic inflation (λ) was 1.006 and 1.017, respectively, for DMFT and DMFS, indicating minimal evidence of model misspecification.
Figure 2.
Genetic association for NAMPT and (A) DMFS, (B) DFS and (C) MS. LocusZoom plots show the association (left y-axis; log10-transformed P-values) with dental caries-related phenotypes of SNPs in the region around NAMPT. The purple triangle represents the top SNP, rs190395159, from the genome-wide scans (which was imputed with information quality score = 0.918 and R2 = 0.947). Genotyped SNPs are indicated by circles and imputed SNPs are indicated by Xs. The blue overlay represents recombination rate (right y-axis) from reference data. Positions of genes are indicated below the plot. Linkage disequilibrium (r2) values were calculated from the HCHS/SOL sample.
Figure 3.
Forest plot for rs190395159. β-Values for DMFS models are shown for each ancestry group as squares proportional to the sample size. Error bars indicate the 99.999995% (i.e. genome-wide significant) confidence intervals. Meta-analyses effect sizes and confidence intervals are conveyed by the position and width of diamonds.
Figure 4.
Three SNPs, rs6947923, rs73409446 and rs73411544, in high-linkage disequilibrium (r2 ≥ 0.7) with the lead SNP (rs190395159) overlap with putative enhancers. The rug plot shows positions relative to NAMPT of SNPs in linkage disequilibrium (indicated by color) with the lead SNP: red (r2 = 1), blue (r2 = 0.8), gold (r2 = 0.7), light blue (r2 = 0.6) and gray (r2 = 0.5–0.4). Signal tracks are shown for biochemical signatures typical of upstream enhancer elements: DNase I hypersensitivity (22,23) (an indicator of chromatin accessibility; blue), transcription factor CTCF binding (red), Histone mark H3k4me1 (yellow) and Histone mark H3K27ac (magenta). The ENCODE (19) Txn factor ChIP track shows transcription factor-binding sites as gray boxes where the darkness of the shading is proportional to the maximum value seen in any ENCODE cell line in that region. A green highlighted region indicates the highest scoring site of a Factorbook-identified (24) canonical motif for the corresponding factor. The black boxes shown in the FANTOM5 enhancers track represent in vivo-transcribed enhancers identified using cap analysis gene expression in the FANTOM5 (21) project. HGF: human gingival fibroblasts; HPdLF: normal human periodontal ligament fibroblasts; AG09319: gum tissue fibroblasts from apparently healthy 24-year old; PKFPC: penis foreskin fibroblast primary cells; CfMdMS: chondrocytes from bone marrow-derived mesenchymal stem cell cultured cells.
NAMPT is thought to be a pro-inflammatory adipokine, and increased serum levels of NAMPT have been observed for a number of diseases including obesity, metabolic syndrome, type 2 diabetes and cardiovascular diseases (25). Though there is currently no direct evidence for the role of NAMPT in dental caries, previous studies have implicated NAMPT in supporting tissues. For example, gingival fibroblasts have been shown to constitutively produce NAMPT, and to increase synthesis in response to oral bacteria (26,27). Increased NAMPT levels have also been observed in gingival biopsies from patients with periodontal disease (26), and in vitro work has shown NAMPT inhibits periodontal healing (28). NAMPT has also been shown to mediate osteoarthritic cartilage destruction in a mouse model by regulating matrix-degrading enzymes [matrix metalloproteinases (MMPs)] (29); MMPs, in turn, are known to be important for the progression of tissue damage in both dental caries and periodontitis (30). Overall, the confluence of several lines of evidence suggests that NAMPT is expressed in the periodontal tissue, up-regulated in response to oral bacteria and in periodontitis patients, and operates through pro-inflammatory and pro-tissue destructive mechanisms. How NAMPT may affect dental caries is currently unknown, although its critical roles in epithelial-mesenchymal interaction and inflammatory response in a variety of biological contexts supports its candidacy as a susceptibility gene.
Its possible role in dental caries notwithstanding, given that NAMPT is a putative gene for periodontitis, which also ultimately leads to tooth loss, we tested the association of this locus with two additional caries-related phenotypes: DFS, which included only current decay and restorations, but not missing teeth, and MS, which included only missing teeth (reportedly lost due to dental caries or periodontitis). The purpose of testing these additional phenotypes was to explore whether tooth loss was driving the observed association. Indeed, MS showed strong evidence of association with this locus, whereas DFS did not (Fig. 2B–C), suggesting that the missing teeth component of DMFT and DMFS is largely responsible for the association. Because there is uncertainty regarding the cause of tooth loss we explored the possibility that the association was due to periodontitis rather than dental caries by testing three measures of periodontal disease, per se: (i) mean attachment loss, a quantitative phenotype, (ii) moderate periodontal disease, a dichotomous affection status defined by the Centers for Disease Control (CDC) and (iii) severe periodontal disease, a dichotomous affection status defined by the CDC. None of these three periodontal disease phenotypes showed association with the NAMPT locus (results not shown).
Additional loci showing genome-wide significant evidence of association are summarized in Table 2. One notable association was observed via meta-analysis between DMFT and rs72626594 (P-value = 2.75E−8), downstream of BMP7. BMP7 is a known tooth development gene (31,32), the deletion of which causes craniofacial manifestations including teeth and salivary gland defects in mice (33). In humans, expression of BMP7 was down-regulated in dental pulp of human third molars that were experimentally injured in vivo (in patients having the teeth extracted for orthodontic reasons within 15 days of injury) (34). BMP7 has not previously been implicated in dental caries, but its role in promoting growth and repair of mineralized tissues is well-known. Indeed, BMP7 protein currently is used in orthopedic care, and its therapeutic potential for tooth repair/re-growth has been investigated, with multiple animal studies showing that BMP7 (or crude BMP extract) stimulates dentin formation (35,36). It is currently unknown whether BMP7 is involved in dental caries, although plausible mechanisms by which genetic variation in BMP7 may influence risk of dental caries include its role in tooth development or possible role in remineralizing pre-cavitated dental lesions.
Table 2.
Genetic loci significantly (P < 5 × 10−8) associated with dental caries indices
| Sample | Phenotype | Chr. | SNP | BP | Allele | MAC | β | SE | P-value | Nearby gene(s)a |
|---|---|---|---|---|---|---|---|---|---|---|
| Metab | DMFT | 7 | rs190395159 | 105 964 857 | A | 162 | −1.24 | 0.21 | 7.14E−9 | SYPL1, NAMPT |
| DMFT | 20 | rs72626594 | 55 535 521 | G | 65 | 2.61 | 0.47 | 2.75E−8 | BMP7, MIR4325, SPO11 | |
| DMFS | 3 | rs138769355 | 151 367 491 | A | 36 | −12.07 | 2.19 | 3.59E−8 | IGSF10, MIR5186, MIR548H2, AADACL2 | |
| DMFS | 7 | rs190395159 | 105 964 857 | A | 162 | −5.56 | 0.90 | 5.97E−10 | SYPL1, NAMPT | |
| Mexican | DMFS | 1 | rs138642966 | 235 695 611 | T | 99 | −11.01 | 1.96 | 1.94E−8 | GNG4, LYST, B3GALNT2, TBCE, GGPS1, ARIB4D |
| DMFS | 10 | rs116717469 | 62 237 360 | C | 39 | −17.01 | 3.08 | 3.23E−8 | ANK3, CDK1, RHOBTB1 | |
| DMFS | 17 | rs71381322 | 48 900 824 | G | 53 | −15.15 | 2.75 | 3.72E−8 | CACNA1G, ABCC3, ANKRD40, LUC7L3, MIR8059, WFIKKN2, TOB1, SPAG9 | |
| DMFS | 18 | rs16946661 | 26 946 870 | T | 31 | −19.23 | 3.50 | 4.02E−8 | None | |
| Puerto Rican | DMFT | X | rs141563584 | 23 757 150 | G | 36 | −2.90 | 0.53 | 3.89E−8 | ACOT9, PRDX4, SAT1, APOO |
aNearby genes were determined based on physical proximity (<400 kb) to the associated SNP while simultaneously considering the linkage disequilibrium structure in the genomic region.
bMeta-analysis of all six sub-samples combined. Chr.: chromosome; BP: base pair position; allele: the effect allele in the model; MAC: minor allele count; β: the beta-coefficient representing the per allele effect on the caries index; SE: standard error of the β-coefficient; DMFT/S: decayed, missing and filled teeth/tooth surfaces.
Another associated locus observed in the meta-analysis of DMFS was the region on chromosome 3 spanning IGSF10, MIR5186, MIR548H2 and AADACL2 (leading SNP rs138769355; P-value = 3.59E−8). None of these genes/miRNAs have known functions that may impact dental caries. In the Mexican ancestry subsample, a few significant loci were observed, notably ANK3 (leading SNP rs116717469, P-value = 3.23E−8), a gene associated with bipolar disorder (37), which is in turn a risk factor for dental caries (38). Likewise, a region on chromosome 17 spanning several genes including CACNA1G was observed (leading SNP rs71381322, P-value = 3.72E−8). Calcium ion channels are important for mineral homeostasis during tooth development, and disruptions of many ion channel genes lead to channelopathies affecting dentition (39). Genetic variants in some ion channels, but not specifically CACNA1G, have been previously shown to be associated with dental caries (Lewis et al. 2014 ASHG abstract; manuscript under review). Moreover, in vitro studies of the effects of BMP2 on ameloblast differentiation showed up-regulation of CACNA1G (40). Likewise, CACNA1G was up-regulated in mouse tooth germ in normal teeth compared with PPARα-associated hypoplastic teeth (41).
Other genome-wide significant associations were observed for Mexican and Puerto Rican groups in loci lacking genes with clear biological roles in oral health or in ‘gene deserts’ (Table 2). Many suggestive associations, some in or near (based on physical proximity) genes with plausible biological roles, were also observed and are reported in the Supplementary Material.
The generalizability of genetic association results and effects of acculturation were addressed by repeating analyses while including years of residency in the USA and its interaction with age as predictors in our model. As indicators of acculturation, these variables were significantly associated with caries indices (P = 0.0086) and improved the fit of our model [change in Akaike's information criterion (AIC) = −5.5]. Strong concordance among genetic association P-values were observed for both DFMT and DMFS for models with and without adjustment for acculturation (see Supplemental Material). Top ranking SNPs and interpretations of results were unchanged.
In addition to the quantitative caries indices, DMFS and DMFT, we also performed GWAS scans of a dichotomous caries phenotype contrasting adults with and without any dental caries. The analysis across all groups recapitulated the association signal on chromosome 20 downstream of BMP7 (lead SNP rs62208680; P-value = 8.20E−7), but not the signals on chromosomes 3 or 7 (see Supplementary Material).
Discussion
In this study, we identified multiple novel genetic loci associated with dental caries. Moreover, this was the first GWAS of dental caries in a Hispanic and Latino sample (and the first GWAS of dental caries in a non-European ancestry sample) as well as the largest GWAS of dental caries, to date. The top association observed for both DMFS and DFMT was in the locus harboring the periodontal health-related gene NAMPT, which appeared to be due to the missing teeth component of the caries indices. Furthermore, the follow-up analyses suggested that this association was not due to periodontitis, but rather due to carious tooth loss. The up-regulation of NAMPT in gingival fibroblasts in response to oral bacterial provides a plausible mechanism by which NAMPT may influence dental caries (27,28). Another significant association was near BMP7, a growth factor important for mineralized tissues. Both of these genes have plausible roles in the host response to carious injury. One could speculate that genes mitigating the progression of active caries through a tooth could impact DMFT and DMFS scores even if their effects are in response to caries rather than in preventing initial decay. Such a mechanism is most plausible for NAMPT and BMP7 based on their known biology.
Previous analyses have shown that the Hispanic and Latino groups included in the HCHS/SOL Study are heterogeneous with respect to their genetic composition (42,43) as well as their oral health status (17) and diet (44). Here, we showed that Hispanic and Latino groups also differ with respect to the cumulative role of genetics on dental caries experience (as indicated by the range of heritability estimates across ancestry groups and the lower heritability estimate in the total sample compared with all individual ancestry groups). While this study focused on the genetic risk factors common to all ancestry groups, the mélange of factors influencing susceptibility to dental caries and their relative effect sizes may differ across groups; additional work contrasting the ancestry groups is needed to extricate these differences.
To assess the role of acculturation on our analyses and determine whether results were generalizable to other US Hispanic and Latino Americans, we repeated our analysis while including acculturation-related variables (i.e. number of years of residency in the USA and its interaction with age) in our model. Acculturation was indeed associated with dental caries indices, although genetic association results were robust to these effects. This suggests that genetic associations identified in our study may be generalizable to other Hispanic and Latino populations. Given the admixed nature of our sample, genetic associations identified here may not be generalizable to other ancestry groups. For example, the minor allele of the top SNP rs190395159 upstream of NAMPT is absent or very rare in individuals of European, Asian, and Southeast Asian ancestry and therefore its role in dental caries is precluded from these groups.
Strong a priori candidate genes such as the enamel matrix and related genes, as well as associated loci identified in previous GWAS studies, were notably absent from the list of associations observed in the HCHS/SOL samples. Moreover, associations identified herein were not observed in previous GWAS (11–13). Explanations for this inconsistency include low power to detect weak genetic effects, heterogeneity across populations due to differences in genetic composition or environment and the possibility of false-positive results from previous candidate gene and GWAS studies. Lack of replication is not entirely unexpected, as previously identified associations are likely impacted by the ‘winner's curse’ (e.g. the effect sizes were inflated in the data set in which they were discovered, which facilitated their discovery in the first place and explains subsequent failure to replicate similar effects in other data sets). In fact, one of the challenges of the GWAS approach is the extensive multiple testing and need to accommodate for this by setting a very low P-value threshold for claiming statistical significance. Therefore, we anticipate identifying some number of false positives, and missing a large number of true associations (i.e. many false negatives). Multi-marker tests and Bayesian methods that incorporate outside information about biological functions of genes may be useful for mitigating these issues in the follow-up analyses. Studies of other diseases have also benefited from meta-analyses across large consortia, including orders of magnitude more participants than can be collected in any single study. Such an approach may also be possible for dental caries as genetic information is collected for more cohorts with dental phenotypes.
Another challenge in identifying the risk factors for dental caries is measuring the state of disease and defining a meaningful phenotype to analyze. Traditional DMFT and DMFS indices combine evidence of both active and past decay without distinguishing between the two, which is reasonable for a genetics study. However, DMFT does not consider multiple lesions per tooth, and DMFS may misrepresent the number of carious surfaces if tooth extraction is used as a treatment option for otherwise restorable lesions. Moreover, the missing tooth/surfaces components of these scores may conflate caries and periodontitis due to uncertainty in the reason for extraction. Therefore, both caries indices may misrepresent the true disease state to some degree. Another issue is that access to oral healthcare can counter-intuitively lead to increased DMFT and DMFS scores through aggressive restorations of pre-cavitated lesions and two-surface restorations of interproximal lesions. Indeed, population surveys have shown that majority racial groups and those with higher socio-economic status or greater access to dental care often exhibit greater DMFT and DMFS indices than do minority racial or low SES groups. This is due to the greater number of restorations in the privileged groups, whereas disadvantaged groups may have lower overall DMFT and DMFS, although have more untreated decay and missing teeth (1). These and other sources of noise in the phenotype could reduce statistical power.
Conclusions
In the first GWAS of dental caries in Hispanic and Latino participants, we discovered several novel associations including multiple loci containing genes with compelling biological stories. This study demonstrated the utility of the GWAS approach, although additional work is required to fully identify and understand the genetic contributors of this multifactorial disease. Moreover, we showed evidence of heterogeneity across Hispanic/Latino groups, which suggests that future efforts investigating the differences among the groups, in addition to their similarities addressed herein, may benefit gene-mapping efforts for dental caries. Ultimately, understanding the factors influencing dental caries susceptibility may lead to improvements in prediction, prevention and disease management.
Materials and Methods
Participant recruitment and data collection
The HCHS/SOL is a multicenter prospective cohort study designed to investigate numerous indices of health and disease in the US Hispanic/Latino population. HCHS/SOL was funded by the National Heart, Lung and Blood Institute with support from six additional institutes including the National Institute of Dental and Craniofacial Research. Institutional Review Boards of all participating entities approved this study. All participants provided informed written consent. The present study reports cross-sectional analyses of the dental assessments collected at baseline.
Study recruitment followed a two-stage census block group- and household-based design in four US cities (Bronx borough of NYC, Chicago, Miami and San Diego) chosen based on the geographical distribution and the place of origin of their respective Hispanic and Latino residents. Details regarding study design and recruitment of participants are available elsewhere (45,46). In all, 16 415 participants were recruited.
Intra-oral examinations by trained and calibrated examiners were performed for 15 848 participants based on the protocols used by the NHANES (1). In brief, dental examiners performed tooth-level and tooth surface-level assessments which included determining the presence or absence of each tooth, the reported cause of any missing teeth and the evidence of coronal decay and restorations of each tooth surface. Agreement among examiners was high for missing teeth (98%), decayed surfaces (99%) and decayed or filled surfaces (86%) (17). Based on these assessments, several dental caries phenotypes were derived including indices that correspond to the counts of decayed, missing due to decay or periodontal disease and restored (i.e. filled) Teeth, and tooth surfaces. DMFT and DMFS are the two most widely used indices of dental caries, capturing both current and past evidence of decay. To follow-up our results, we also considered the DFS index, corresponding to the count of decayed and restored surfaces, and the MS index, corresponding to the count of missing tooth surfaces due to decay or periodontitis. All dental caries indices were calculated while excluding the third molars, and indices were not calculated for edentulous participants (4.1% of the sample). In addition to quantitative dental caries phenotypes, we also considered a dichotomous measure of lifetime caries experience where participants with DMFS ≥1 were defined as affected by caries, and participants with DMFS = 0 were defined as caries-free. For this caries presence/absence phenotype, edentulous individuals were considered affected.
Genotyping, imputation and ancestry
A total of 12 803 participants providing consent for genetic analyses were genotyped for over 2.5 million single-nucleotide polymorphisms (SNPs) using a custom Illumina (San Diego) array consisting of the HumanOmni2.5-8v1-1 array content along with a panel of ∼150 000 investigator-chosen SNPs. The custom content was selected to include ancestry-informative markers, variants distinctive of Amerindian populations and candidate polymorphisms. Genotyping was performed by Illumina Microarray Services and all genotype data were extensively cleaned and quality-checked by Illumina Microarray Services, LA Biomed, and the SOL Genetic Analysis Center (GAC) at the University of Washington using analysis pipelines developed by the GAC (47). These analyses were standard for the field and included scrutinizing the participant samples for genetic sex, chromosomal anomalies, relatedness, population structure, Mendelian errors among relatives, concordance among duplicates, batch effects and genotyping call rates. SNP probes were scrutinized for poor performance in inter-sample comparisons (such as checks of Mendelian errors, concordance among sample duplicates etc.), missing call rates, separation of clusters during genotype calling, deviations from Hardy–Weinberg equilibrium and genotype concordance among duplicate probes. Recommended SNP filters were established by the GAC based on quality control analyses.
Imputation of un-typed SNPs was performed as implemented in the IMPUTE2 software (48) using haplotypes observed in the 1000 Genomes Project Phase 1 reference panel. In total, genotypes for >25.5 million additional SNPs were imputed, bringing the total number SNPs with available data to 27.7 million. Masked SNPs (i.e. genotyped SNPs that were imputed in order to assess imputation quality) showed 99.7 and 98.8% concordance for SNPs with minor allele frequencies <0.05 and ≥0.05, respectively.
An iterative procedure was used to disentangle population structure from the family structure given the presence of cryptic biological relatives within the study sample (42). First, the KING-robust method was used to estimate the kinship coefficient (KC; a measure of relatedness) between all pairs of participants using genomic data (49). The sample was then partitioned into a primary subset of mutually unrelated participants (N = 10 861) and a secondary subset comprised of the biological relatives of the unrelated set (N = 2204). Principal components analysis (PCA) was used to model the genetic ancestry of participants within the unrelated subset; the principal components (PCs) of ancestry were then projected onto the subsample of relatives (50). KCs were then re-estimated taking ancestry into account by using the PCs to estimate individual-specific allele frequencies (51). PCA was repeated and KCs were estimated yet again. This process yielded components of ancestry that reflect the population structure free from the influence of the family structure and KC estimates adjusted for ancestry.
Genetic analysis groups were defined using the self-identified Hispanic heritage group and the first five PCs of ancestry in a multivariate outlier detection procedure based on the minimum covariant determinant estimation (42,52). The concordance between self-reported ancestry and genetic ancestry was high (94–98% for each of the six ancestry groups). Nineteen participants with genetically determined Asian ancestry and 37 Central American outliers with unusual ancestry were excluded from analyses. Genetic ancestry was used to stratify participants for genetic association analyses. The top five principal components of ancestry were included in genetic association models.
Statistical analyses
Overview
GWAS scans of DMFT and DMFS were performed separately for six genetic analysis groups: Cuban (N = 1950), Dominican (N = 1083), Puerto Rican (N = 2006), Mexican (N = 4578), Central American (N = 1294) and South American (N = 843). Results from the six ancestry groups were combined via inverse variance-weighted meta-analysis. Cochran's Q was used to test for heterogeneity in SNP effects across samples. SNPs with minor allele counts of ≥30 were included in analyses. The threshold for determining statistical significance was P-value <5 × 10−8. Cluster plots (of allele intensity data used for genotype calling) were visually scrutinized for all significant SNPs to confirm reliable genotyping. Given the strong correlational structure of the genome, this significance threshold may be considered conservative. Therefore, we also noted ‘suggestive’ loci having P-values <1 × 10−7 in the Supplemental Material.
Association models
The two caries indices, DMFT and DMFS, approximate continuous variables and were analyzed as such. A mixed-models approach was used to interrogate each SNP for evidence of association while adjusting for fixed effects, including sex, age, recruitment center, sampling weights and five PCs of ancestry (calculated separately for each group), and random effects, including census block group within each recruitment site, household and kinship. The distributions of DMFT and DMFS are non-normal; however, model residuals were approximately normal, indicating that the phenotypes were reasonably well-suited for our analytic framework. Environmental factors that may explain some phenotype variance (e.g. tobacco use, diet etc.) were not included in the models. Note that such factors cannot confound the analyses because they do not alter constitutional genetic variants; however, by not modeling environmental effects, our GWAS scans may detect associations that are mediated through environments (e.g. genes that influence diet and therefore could impact dental caries). This analysis strategy was chosen to cast the widest net possible given the multifactorial nature of the disease. Genomic inflation was estimated and visualized using Q–Q plots. Analyses were performed in the R statistical environment (R Foundation for Statistical Computing, Vienna, Austria). Associated loci were visualized using LocusZoom (53).
Heritability estimation
Heritability was defined as the proportion of phenotype variance attributable to the observed kinship component from the ‘base’ mixed-model, which accounts for the genetic relatedness among all participants in the sample or subsample. This is similar to heritability estimates using the variance components approach in families, except here we estimate kinship from genomic data rather than using expected sharing due to known familial relationships.
Functional annotation
Identification of biochemical signals associated with non-coding functional elements was performed with the UCSC Genome Browser (https://genome.ucsc.edu/) using the GRCh37/hg19 assembly (54).
Sensitivity analysis and effect of acculturation
To explore the generalizability of results and address the possible role of acculturation on our analyses, we considered several variables related to nativity and acculturation [short acculturation scale (55), MESA nativity subscore (56), first versus second generation immigrant, the USA versus foreign born and years of residence in the USA] by adding them individually to the association model described above. Based on change in AIC, we chose the number of years of residence in the USA and its interaction with age as indices of acculturation. Genetic association analyses were repeated while adjusting for these acculturation variables.
Supplementary Material
Funding
The baseline examination of the Hispanic Community Health Study/Study of Latinos was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236) and San Diego State University (N01-HC65237). The following Institutes/Centers/Offices contribute to the HCHS/SOL through a transfer of funds to the NHLBI: National Institute on Minority Health and Health Disparities, National Institute on Deafness and Other Communication Disorders, National Institute of Dental and Craniofacial Research, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Neurological Disorders and Stroke, NIH Institution-Office of Dietary Supplements. The Genetic Analysis Center at the University of Washington was supported by NHLBI and NIDCR contracts (HHSN268201300005C AM03 and MOD03). Investigators website: http://www.cscc.unc.edu/hchs/
Supplementary Material
Acknowledgements
We express our gratitude to the participants and staff of the HCHS/SOL study for their contribution to our goals of identifying the risk factors and protective factors impacting health and disease in US Hispanic and Latino populations. We thank Marston Youngblood of the University of North Carolina at Chapel Hill for his role in managing and facilitating access to the data, Drs Jane Atkinson and Emily Harris of the National Institute for Dental and Craniofacial Research for their involvement in facilitating this work and Dr Bruce Weir of the University of Washington for his role in overseeing the Genetic Analysis Center and the development of the analysis pipelines utilized in this study.
Conflict of Interest statement. None declared.
References
- 1.Dye B.A., Tan S., Smith V., Lewis B.G., Barker L.K., Thornton-Evans G., Eke P.I., Beltran-Aguilar E.D., Horowitz A.M., Li C.H. (2007) Trends in oral health status: United States, 1988–1994 and 1999–2004. Vital Health Stat.. Series 11, Data from the National Health Survey, 1–92. [PubMed] [Google Scholar]
- 2.Dye B., Thornton-Evans G., Li X., Iafolla T. (2015) Dental caries and tooth loss in adults in the United States, 2011–2012. NCHS Data Brief, 197. [PubMed] [Google Scholar]
- 3.Dye B.A., Thornton-Evans G., Li X., Iafolla T.J. (2015) Dental caries and sealant prevalence in children and adolescents in the United States, 2011–2012. NCHS Data Brief, 1–8. [PubMed] [Google Scholar]
- 4.Boraas J.C., Messer L.B., Till M.J. (1988) A genetic contribution to dental caries, occlusion, and morphology as demonstrated by twins reared apart. J. Dent. Res., 67, 1150–1155. [DOI] [PubMed] [Google Scholar]
- 5.Shaffer J.R., Wang X., Desensi R.S., Wendell S., Weyant R.J., Cuenco K.T., Crout R., McNeil D.W., Marazita M.L. (2012) Genetic susceptibility to dental caries on pit and fissure and smooth surfaces. Caries Res., 46, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X., Shaffer J.R., Weyant R.J., Cuenco K.T., DeSensi R.S., Crout R., McNeil D.W., Marazita M.L. (2010) Genes and their effects on dental caries may differ between primary and permanent dentitions. Caries Res., 44, 277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher-Owens S.A., Gansky S.A., Platt L.J., Weintraub J.A., Soobader M.J., Bramlett M.D., Newacheck P.W. (2007) Influences on children's oral health: a conceptual model. Pediatrics, 120, e510–520. [DOI] [PubMed] [Google Scholar]
- 8.Vieira A.R., Modesto A., Marazita M.L. (2014) Caries: review of human genetics research. Caries Res., 48, 491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Opal S., Garg S., Jain J., Walia I. (2015) Genetic factors affecting dental caries risk. Aust. Dent. J., 60, 2–11. [DOI] [PubMed] [Google Scholar]
- 10.Shaffer J.R., Wang X., Feingold E., Lee M., Begum F., Weeks D.E., Cuenco K.T., Barmada M.M., Wendell S.K., Crosslin D.R. et al. (2011) Genome-wide association scan for childhood caries implicates novel genes. J. Den. Res., 90, 1457–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Shaffer J.R., Zeng Z., Begum F., Vieira A.R., Noel J., Anjomshoaa I., Cuenco K.T., Lee M.K., Beck J. et al. (2012) Genome-wide association scan of dental caries in the permanent dentition. BMC Oral Health, 12, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaffer J.R., Feingold E., Wang X., Lee M., Tcuenco K., Weeks D.E., Weyant R.J., Crout R., McNeil D.W., Marazita M.L. (2013) GWAS of dental caries patterns in the permanent dentition. J. Dent. Res., 92, 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng Z., Shaffer J.R., Wang X., Feingold E., Weeks D.E., Lee M., Cuenco K.T., Wendell S.K., Weyant R.J., Crout R. et al. (2013) Genome-wide association studies of pit-and-fissure- and smooth-surface caries in permanent dentition. J. Dent. Res., 92, 432–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeng Z., Feingold E., Wang X., Weeks D.E., Lee M., Cuenco D.T., Broffitt B., Weyant R.J., Crout R., McNeil D.W. et al. (2014) Genome-wide association study of primary dentition pit-and-fissure and smooth surface caries. Caries Res., 48, 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanley B.O., Feingold E., Cooper M., Vanyukov M.M., Maher B.S., Slayton R.L., Willing M.C., Reis S.E., McNeil D.W., Crout R.J. et al. (2014) Genetic association of MPPED2 and ACTN2 with dental caries. J. Dent. Res., 93, 626–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruz G.D., Chen Y., Salazar C.R., Le Geros R.Z. (2009) The association of immigration and acculturation attributes with oral health among immigrants in New York City. Am. J. Public Health, 99(Suppl. 2):S474–S480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck J.D., Youngblood M. Jr., Atkinson J.C., Mauriello S., Kaste L.M., Badner V.M., Beaver S., Becerra K., Singer R. (2014) The prevalence of caries and tooth loss among participants in the Hispanic Community Health Study/Study of Latinos. J. Am. Dent. Assoc., 145, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visscher P.M., Hill W.G., Wray N.R. (2008) Heritability in the genomics era—concepts and misconceptions. Nat. Rev. Genet., 9, 255–266. [DOI] [PubMed] [Google Scholar]
- 19.Gerstein M.B., Kundaje A., Hariharan M., Landt S.G., Yan K.K., Cheng C., Mu X.J., Khurana E., Rozowsky J., Alexander R. et al. (2012) Architecture of the human regulatory network derived from ENCODE data. Nature, 489, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roadmap Epigenomics C., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al. (2015) Integrative analysis of 111 reference human epigenomes. Nature, 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. et al. (2014) An atlas of active enhancers across human cell types and tissues. Nature, 507, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B. et al. (2012) The accessible chromatin landscape of the human genome. Nature, 489, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.John S., Sabo P.J., Thurman R.E., Sung M.H., Biddie S.C., Johnson T.A., Hager G.L., Stamatoyannopoulos J.A. (2011) Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet., 43, 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J., Zhuang J., Iyer S., Lin X.Y., Greven M.C., Kim B.H., Moore J., Pierce B.G., Dong X., Virgil D. et al. (2013) Factorbook.org: a Wiki-based database for transcription factor-binding data generated by the ENCODE consortium. Nucleic Acids Res., 41, D171–D176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang Y.H., Chang D.M., Lin K.C., Shin S.J., Lee Y.J. (2011) Visfatin in overweight/obesity, type 2 diabetes mellitus, insulin resistance, metabolic syndrome and cardiovascular diseases: a meta-analysis and systemic review. Diabetes Metab. Res. Rev., 27, 515–527. [DOI] [PubMed] [Google Scholar]
- 26.Damanaki A., Nokhbehsaim M., Eick S., Gotz W., Winter J., Wahl G., Jager A., Jepsen S., Deschner J. (2014) Regulation of NAMPT in human gingival fibroblasts and biopsies. Mediat. Inflamm., 2014, 912821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nogueira A.V., Nokhbehsaim M., Eick S., Bourauel C., Jager A., Jepsen S., Cirelli J.A., Deschner J. (2014) Regulation of visfatin by microbial and biomechanical signals in PDL cells. Clin. Oral Invest., 18, 171–178. [DOI] [PubMed] [Google Scholar]
- 28.Nokhbehsaim M., Keser S., Jager A., Jepsen S., Deschner J. (2013) Regulation of regenerative periodontal healing by NAMPT. Mediat. Inflamm., 2013, 202530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang S., Ryu J.H., Oh H., Jeon J., Kwak J.S., Kim J.H., Kim H.A., Chun C.H., Chun J.S. (2015) NAMPT (visfatin), a direct target of hypoxia-inducible factor-2alpha, is an essential catabolic regulator of osteoarthritis. Ann. Rheum. Dis., 74, 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorsa T., Tjaderhane L., Salo T. (2004) Matrix metalloproteinases (MMPs) in oral diseases. Oral Dis., 10, 311–318. [DOI] [PubMed] [Google Scholar]
- 31.Helder M.N., Karg H., Bervoets T.J., Vukicevic S., Burger E.H., D'Souza R.N., Woltgens J.H., Karsenty G., Bronckers A.L. (1998) Bone morphogenetic protein-7 (osteogenic protein-1, OP-1) and tooth development. J. Dent. Res., 77, 545–554. [DOI] [PubMed] [Google Scholar]
- 32.Tasli P.N., Aydin S., Yalvac M.E., Sahin F. (2014) Bmp 2 and bmp 7 induce odonto- and osteogenesis of human tooth germ stem cells. Appl. Biochem. Biotechnol., 172, 3016–3025. [DOI] [PubMed] [Google Scholar]
- 33.Zouvelou V., Luder H.U., Mitsiadis T.A., Graf D. (2009) Deletion of BMP7 affects the development of bones, teeth, and other ectodermal appendages of the orofacial complex. J. Exp. Zool. B Mol. Dev. Evol., 312B, 361–374. [DOI] [PubMed] [Google Scholar]
- 34.Cevallos Gutierrez F.A., Del Portillo Obando P.D.P., Rodriguez Castillo J.G., Tibata Rodriguez V.M. (2005) Expression Patterns of BMP7 of Healthy and Injured Human Teeth. J. Dent. Res., 84A, 0137. [Google Scholar]
- 35.Sloan A.J., Rutherford R.B., Smith A.J. (2000) Stimulation of the rat dentine-pulp complex by bone morphogenetic protein-7 in vitro. Arch. Oral Biol., 45, 173–177. [DOI] [PubMed] [Google Scholar]
- 36.Harichane Y., Dimitrova-Nakov S., Poliard A., Veis A., DenBesten P., Kellermann O., Goldberg M. (2010) In Goldberg M. (ed), Amelogenins: Multifaceted Proteins for Dental and Bone Formation and Repair. Bentham Science Publishers, pp. 174–190. [Google Scholar]
- 37.Ferreira M.A., O'Donovan M.C., Meng Y.A., Jones I.R., Ruderfer D.M., Jones L., Fan J., Kirov G., Perlis R.H., Green E.K. et al. (2008) Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet., 40, 1056–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friedlander A.H., Friedlander I.K., Marder S.R. (2002) Bipolar I disorder: psychopathology, medical management and dental implications. J. Am. Dent. Assoc., 133, 1209–1217. [DOI] [PubMed] [Google Scholar]
- 39.Duan X. (2014) Ion channels, channelopathies, and tooth formation. J. Dent. Res., 93, 117–125. [DOI] [PubMed] [Google Scholar]
- 40.Miyoshi K., Nagata H., Horiguchi T., Abe K., Arie Wahyudi I., Baba Y., Harada H., Noma T. (2008) BMP2-induced gene profiling in dental epithelial cell line. J. Med. Investig., 55, 216–226. [DOI] [PubMed] [Google Scholar]
- 41.Sehic A., Khuu C., Risnes S., Osmundsen H. (2009) Differential gene expression profiling of the molar tooth germ in peroxisome proliferator-activated receptor-alpha (PPAR-alpha) knockout mouse and in wild-type mouse: molar tooth phenotype of PPAR-alpha knockout mouse. Eur. J. Oral Sci., 117, 93–104. [DOI] [PubMed] [Google Scholar]
- 42.Conomos M.P., Laurie C.A., Stilp A.M., Gogarten S.M., McHugh C.P., Nelson S.C., Sofer T., Fernandez-Rhodes L., Justice A.E., Graff M. et al. (2015) Genetic diversity and association studies in U.S. Hispanic/Latino populations: Applications in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet., In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manichaikul A., Palmas W., Rodriguez C.J., Peralta C.A., Divers J., Guo X., Chen W.M., Wong Q., Williams K., Kerr K.F et al. (2012) Population structure of Hispanics in the United States: the multi-ethnic study of atherosclerosis. PLoS Genet., 8, e1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siega-Riz A.M., Sotres-Alvarez D., Ayala G.X., Ginsberg M., Himes J.H., Liu K., Loria C.M., Mossavar-Rahmani Y., Rock C.L., Rodriguez B. et al. (2014) Food-group and nutrient-density intakes by Hispanic and Latino backgrounds in the Hispanic Community Health Study/Study of Latinos. Am. J. Clin. Nutr., 99, 1487–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorlie P.D., Aviles-Santa L.M., Wassertheil-Smoller S., Kaplan R.C., Daviglus M.L., Giachello A.L., Schneiderman N., Raij L., Talavera G., Allison M. et al. (2010) Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann. Epidemiol., 20, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bovbjerg D.H. (2013) Genome-wide analysis of BMI in adolescents and young adults reveals additional insight into the effects of genetic loci over the life course. J. Pain, 22, 3597–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laurie C.C., Doheny K.F., Mirel D.B., Pugh E.W., Bierut L.J., Bhangale T., Boehm F., Caporaso N.E., Cornelis M.C., Edenberg H.J. et al. (2010) Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol., 34, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Howie B.N., Donnelly P., Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. (2010) Robust relationship inference in genome-wide association studies. Bioinformatics, 26, 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conomos M.P., Miller M.B., Thornton T.A. (2015) Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet. Epidemiol., 39, 276–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conomos M.P., Reiner A.P., Weir B.S., Thornton T.A. (2015) Model-free estimation of recent genetic relatedness. Am. J. Hum. Genet., In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rousseeuw P.J., Van Driessen K. (1999) A fast algorithm for the minimum covariance determinant estimator. Technometrics, 41, 121–223. [Google Scholar]
- 53.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marin G., Sabogal F., VanOss Marin B., Otero-Sabogal R., Perez-Stable E.J. (1987) Development of a short acculturation scale for Hispanics. Hispanic J. Behav. Sci., 9, 183–205. [Google Scholar]
- 56.Kandula N.R., Diez-Roux A.V., Chan C., Daviglus M.L., Jackson S.A., Ni H., Schreiner P.J. (2008) Association of acculturation levels and prevalence of diabetes in the multi-ethnic study of atherosclerosis (MESA). Diab. Care, 31, 1621–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.