Abstract
Non-syndromic (NS) cleft lip with or without cleft palate (CL/P) is a common disorder with a strong genetic underpinning. Genome-wide association studies have detected common variants associated with this disorder, but a large portion of the genetic risk for NSCL/P is conferred by unidentified rare sequence variants. Mutations in IRF6 (Interferon Regulatory Factor 6) and GRHL3 (Grainyhead-like 3) cause Van der Woude syndrome, which includes CL/P. Both genes encode members of a regulatory network governing periderm differentiation in model organisms. Here, we report that Krüppel-like factor 17 (Klf17), like Grhl3, acts downstream of Irf6 in this network in zebrafish periderm. Although Klf17 expression is absent from mammalian oral epithelium, a close homologue, Klf4, is expressed in this tissue and is required for the differentiation of epidermis. Chromosome configuration capture and reporter assays indicated that IRF6 directly regulates an oral-epithelium enhancer of KLF4. To test whether rare missense variants of KLF4 contribute risk for NSCL/P, we sequenced KLF4 in approximately 1000 NSCL/P cases and 300 controls. By one statistical test, missense variants of KLF4 as a group were enriched in cases versus controls. Moreover, two patient-derived KLF4 variants disrupted periderm differentiation upon forced expression in zebrafish embryos, suggesting that they have dominant-negative effect. These results indicate that rare NSCL/P risk variants can be found in members of the gene regulatory network governing periderm differentiation.
Introduction
Non-syndromic cleft lip with or without cleft palate (NSCL/P) affects about one in 700 newborns in the USA and is the second most common structural birth defect (after heart anomalies) (1). The pathogenesis of NSCL/P involves both environmental and genetic risk factors, as is true for other common diseases (2). Genome-wide association studies (GWASs) (3–7), linkage studies (8) and a meta-analysis of these (9) have identified 18 loci in which specific common alleles are associated with NSCL/P. Despite the success of GWASs, a substantial portion of the heritability for NSCL/P remains unexplained. Additional common variants conferring risk for NSCL/P may have escaped detection by GWASs, for instance, because of a low density of genotyped SNPs, as in the vicinity of FOXE1 (8). By analogy to other complex diseases, another portion of missing heritability is likely derived from rare or de novo sequence variants (10). Candidate genes to carry these variants are those pointed to by the GWASs, i.e. the genes whose expression level or function is altered by common variants associated with NSCL/P (11). However, the results of sequencing 13 such genes/loci in over 1000 patient–parent trios were not consistent with coding variants of these genes, as a group, being over-transmitted to patients; it nonetheless remains possible that specific function-altering variants confer risk for NSCL/P (12). In principle, genes encoding members of the same gene regulatory networks (GRNs) as GWAS-indicated genes are also candidates to harbour risk variants (13). In this context, the GRN regulating differentiation of oral periderm is of special interest.
The periderm is the most superficial layer of embryonic epidermis and oral epithelium. Evidence that periderm development is relevant to pathogenesis of cleft palate came from animal models of Van der Woude syndrome (VWS), which includes CL/P and, usually, lip pits. This syndrome is caused most frequently by mutations in IRF6 (14). Moreover, GWASs have revealed that common variants near IRF6 are highly associated with NSCL/P (12). In murine Irf6-null mutant embryos, markers of the oral periderm are absent, and markers of the outermost suprabasal layers of the epidermis are either absent or lower than that in wild-type mice (15,16). The absence of oral periderm leads to pathological adhesions between the palate shelves and the tongue (16). Zebrafish periderm, known as the enveloping layer in gastrula stage embryos, is derived from deep blastomeres and one of the first lineages to become distinct (17). Later, in juvenile zebrafish, the periderm is derived from basal keratinocytes (18). In frog and zebrafish embryos injected with an RNA encoding a dominant-negative variant of Irf6 (dnIrf6), the expression of genes encoding regulatory molecules, including several transcription factors (e.g. grhl3, klf17 and klf2b), and differentiation effectors (e.g. keratin genes, periplakin and plakophilin), is lower than normal (19,20). dnIrf6-injected zebrafish embryos rupture through the animal pole during epiboly (19) possibly because of weakened cell adhesion between periderm cells.
Interestingly, analysis of the zebrafish periderm GRN and the hunt for a second cause of VWS converged on the same molecule. Expression of grhl3, encoding a transcription factor necessary for the development of a permeability barrier in flies (21) and mice (22), is blocked by dnIrf6, and artificially restoring grhl3 expression in dnIrf6-injected zebrafish embryos partially restores the expression of markers of periderm differentiation (20). Mutations in GRHL3 were identified in eight of 45 families with VWS lacking IRF6 mutations, and missense variants of GRHL3 segregating with VWS mimicked the ability of dominant-negative Grhl3 to disrupt zebrafish periderm differentiation (23). These findings highlight the importance of periderm integrity for correct morphogenesis of the face and suggest that mutations in genes encoding additional members of the periderm GRN may influence risk for syndromic and/or non-syndromic forms of CL/P.
In addition to Grhl3 and Irf6, the zebrafish periderm GRN includes Krüppel-like factor 17 (Klf17). The 17 members of KLF superfamily of DNA-binding transcriptional regulators share the structural feature of a C-terminal DNA binding domain (DBD) with three zinc fingers; depending on the family member, the cellular context and the presence of co-factors, they regulate proliferation, differentiation, migration and pluripotency in embryonic development and in cancer (24,25). Among the KLF proteins, KLF2, KLF4 and KLF17 constitute a more-closely related subgroup (25,26). In zebrafish, klf17 [previously annotated as biklf or klf4b (27)] is expressed at high levels in the periderm from 30% epiboly onwards (27,28), whereas klf2a and klf2b are expressed in a mosaic, salt-and-pepper pattern in the periderm at this stage; klf4 is not expressed at gastrula stages (27,28). Kotkamp et al. (28) showed that Klf17, Klf2a and Klf2b function in a regulatory network to promote zebrafish periderm differentiation, acting downstream of both Pou5f1 and an additional transcription factor speculated to be Irf6. They found that many keratin genes are direct transcriptional targets of Klf17 and that promoters of these genes were enriched in motifs matching the preferred binding site of the Klf2/4/17 subgroup (28).
Here we show that, as anticipated, Klf17 acts downstream of Irf6 in the GRN governing zebrafish periderm and that embryos injected with RNA encoding dominant-negative Klf17, which likely inhibits Klf17, Klf2a and Klf2b, recapitulate the phenotype of embryos injected with dnIrf6. Switching to mammalian oral periderm, in which Klf17 is not expressed, we shift focus to a member of the same structural subgroup, Klf4, that is expressed in this tissue (25). Klf4 is essential for differentiation of epidermis in mice (29), and we find human KLF4 functions like zebrafish Klf17 in over-expression assays. We show that IRF6 binds an enhancer of KLF4 that is active in oral epithelium cells, supporting the possibility that KLF4 is directly downstream of IRF6 in a GRN regulating oral periderm differentiation. Finally, we find evidence that rare variants in KLF4 confer risk for NSCL/P.
Results
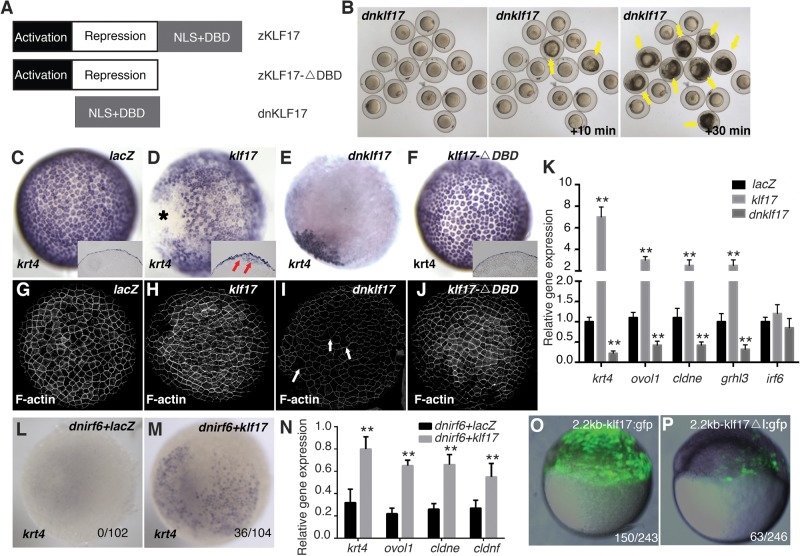
We previously found that klf17 and klf2b expression is strongly downregulated in embryos injected with dnIrf6 (20). Embryos depleted of klf17 exhibit lower-than-normal expression of simple epithelium keratin genes (e.g. krt4, krt5 and krt17), but they live until at least 24 h post fertilization (hpf) (28). The fact that such embryos do not die during gastrulation similar to dnIrf6-injected embryos may reflect the partially redundant activity of Klf2a and Klf2b in promoting periderm differentiation; expression of these genes is upregulated in klf17 knockdown embryos (28). To determine whether activity of the Klf2a/2b/17 network is essential for periderm differentiation and viability during gastrulation, we engineered a cDNA encoding only the Klf17 nuclear localization sequence and its DBD. The promoters of deduced direct targets of all three proteins are enriched for the same binding motif (28), so we predicted that Klf17-DBD would compete with endogenous Klf17, Klf2a and Klf2b for chromatin binding and thereby act in a dominant-negative manner (dnKlf17) (Fig. 1A). We also engineered a truncated Klf17 without the DBD (Klf17-△DBD), anticipating that this would be non-functional (Fig. 1A). For each cDNA construct, we synthesized mRNA in vitro and injected it into wild-type zebrafish embryos at the one-cell stage. Most embryos injected with a control mRNA (lacZ) developed normally, the majority of embryos injected with dnklf17 mRNA ruptured at the animal pole starting at ∼6 hpf (Fig. 1B), recapitulating the phenotype of embryos injected with dnIrf6 mRNA (19). Control RNA-injected embryos had characteristic expression of krt4 in the periderm (Fig. 1C), characteristic polygonal shape and pronounced phalloidin staining, revealing polymerized F-actin, at the cell perimeter (Fig. 1G). In contrast, dnklf17-injected embryos exhibited widespread reduction of krt4 expression (Fig. 1D). In such embryos, periderm cells had weaker-than-normal phalloidin staining and were less regularly shaped (Fig. 1H), again recapitulating the phenotype of dnirf6-injected embryos (19). Embryos injected with klf17-△DBD developed normally and exhibited normal krt4 (Fig. 1E) and phalloidin stain (Fig. 1I); those injected with intact klf17 will be discussed subsequently. By quantitative reverse transcriptase–polymerase chain reaction (RT–PCR), we determined that dnklf17-injected embryos also exhibited reduced expression of several periderm markers (i.e. krt4, cldne, grhl3 and ovol1) in comparison to control RNA-injected embryos (Fig. 1K), whereas the levels of irf6 expression were similar in dnklf17-injected and control mRNA-injected embryos (Fig. 1K). These findings suggest that the Klf2a/2b/17 network is required for periderm differentiation and that it acts downstream of Irf6.
Figure 1.
KLF17 is an effector of IRF6 during differentiation of the embryonic zebrafish periderm. (A) Schematic of wild-type zebrafish Klf17 and of the variants used in this study. (B) Representative images of embryos injected at the 1-2 cell stage with RNA encoding dominant-negative Klf17 (dnklf17) shown at 6 hpf and at the indicated intervals later. Yellow arrows indicate rupture of the embryos. (C–J) Dorsal views of wild-type embryos injected at the one to two cell stage with the indicated mRNA construct, fixed at 5 hpf and processed to reveal (C–F) krt4 expression or (G–J) polymerized actin (phalloidin stain) (C–F, insets): cross-sections revealing that krt4 expression is confined to the superficial cell layer except in (F), where it is also observed in deep cells in a mosaic pattern, arrows. Asterisk in (F) indicates the ventral side of embryos. In (H) dnklf17-injected embryos, the overall level of phalloidin stain is lower than that in other groups, and there are occasional gaps in it, arrowheads. (Embryos dead at 6 hpf over number injected: lacZ, 2/89, dnklf17, 56/85, klf17-△DBD, 4/82, klf17, 6/84.) (K) Quantitation of periderm markers in dnklf17- and klf17-injected embryos (5 hpf), as assessed by qRT–PCR. Asterisks indicate statistical significance compared with the lacZ-injected control groups: *P < 0.05 and **P < 0.01. (L and M) Dorsal views of wild-type embryos injected with RNA encoding dominant-negative IRF6 (L) alone or (M) together with klf17 mRNA. Numbers at bottom right represent the ratio of unruptured/total injected embryos. (N) Quantitation of rescue of periderm markers by klf17 in dnirf6-injected embryos. Asterisks indicate statistical significance compared with the control group: **P < 0.01. (O and P) Representative images of GFP expression in transient transgenic zebrafish embryos injected with (O) the intact klf17 promoter and (P) one that lacks IRF6 binding sites (ΔI). The number at the bottom right represents the ratio of embryos with five or more GFP-periderm positive cells over the total number of injected embryos.
It was previously reported that over-expression of klf17 will induce elevated expression of many markers of the periderm, but the spatial quality of the ectopic expression was not shown (28). To test whether exogenous Klf17 expression is sufficient to induce deep cells to adopt characteristics of periderm cells, we injected embryos with klf17 mRNA. Such embryos exhibited ectopic expression of krt4 in deep cells at 6 hpf (Fig. 1F inset, arrowheads); upregulation of other periderm markers was detected by qRT–PCR, confirming the earlier report (28) (Fig. 1K). Two qualities of the krt4 expression in klf17-injected embryos were surprising. First, the pattern of ectopic krt4 in deep cells was mosaic, which is in contrast to embryos injected with grhl3 mRNA, in which ectopic krt4 expression appears to be contiguous in deep cells (20). Secondly, such embryos exhibited variable loss of endogenous krt4 expression in the ventral periderm (Fig. 1F, asterisk) (the ventral position of the loss was confirmed by processing such embryos to reveal a marker of the shield, data not shown). Notably, in contrast to dnklf17-injected embryos, such embryos had normal phalloidin staining in both the dorsal and the ventral periderm (Fig. 1J) and did not rupture. Although we did not pursue these unexpected observations further, we speculate that they relate to feedback between Klf17 and Klf2 paralogues: over-expression of klf2b induces ectopic expression of krt4 and over-expression of Klf17 inhibits expression of klf2b (28). There are many additional examples in which KLF family members function in a network with mutual repression (30–33). Because expression of klf2a and klf2b is confined to the ventral periderm, it is not unexpected that over-expression of klf17 would have different effects on dorsal and ventral sides. Finally, although embryos injected with dnirf6 mRNA largely lacked krt4 expression (i.e. in the dorsal and ventral periderm), this phenotype was partially rescued in embryos that were co-injected with klf17 and dnirf6 mRNAs (Fig. 1M and N). These results further support a model whereby Klf17 acts downstream of Irf6 in the GRN that governs periderm differentiation.
Based on the aforementioned findings, we asked whether klf17 is a direct target of Irf6 in zebrafish periderm. With JASPAR (34), we found two canonical Irf6 binding sites within 2.2 kb upstream of the klf17 transcription start site, suggesting that Irf6 directly regulates klf17 expression. We engineered this promoter element, either intact or after deleting the two Irf6 binding sites, into a plasmid upstream of the gene that encodes green fluorescent protein (GFP). In embryos injected with the plasmid containing the intact klf17 promoter, GFP expression was present in the periderm; in embryos injected with the promoter construct lacking the two Irf6 binding sites, GFP expression in periderm was much lower (Fig. 1G) and was detected in fewer animals (Fig. 1). To confirm that this promoter responds to IRF6, we engineered the intact and binding-site-deleted versions (separately) into a luciferase reporter plasmid and then transfected the resulting constructs into a human embryonic kidney cell line that lacks endogenous IRF6 expression. Whereas there was modest luciferase expression in cells transfected with either the intact or Irf6-binding-site-deleted versions of the reporter construct, co-transfection with a plasmid encoding zebrafish Irf6 elevated reporter expression in the cells transfected with former but not with the latter version (Supplementary Material, Fig. S1). These findings are consistent with the possibility that Irf6 directly regulates Klf17 expression in zebrafish periderm.
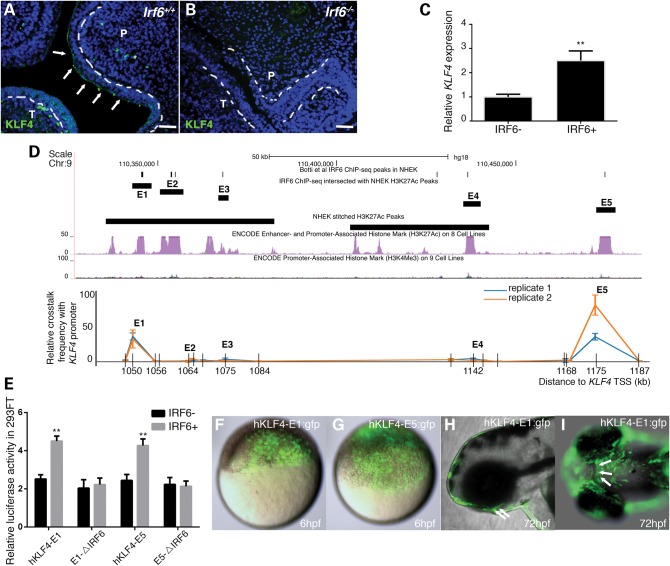
We wondered whether this aspect of the periderm GRN is conserved to human keratinocytes. However, in mouse, Klf17 is mainly expressed in the central nervous system (35) and urothelium (36). In an atlas of murine gene expression patterns, Klf17 expression appears to be absent from oral epithelium (www.jax.informatics.org, date last accessed, August 30, 2015). In contrast, Klf4, the paralogue with highest sequence similarity to Klf17 (25), is expressed in the murine epidermis and is essential for its differentiation (29), and transgenic mice with epithelium-specific over-expression of Klf4 exhibit a cleft palate (37). We therefore pursued the possibility that KLF4 functions downstream of IRF6 in a mammalian oral epithelium GRN. Supporting this possibility, knockdown of IRF6 expression in primary human keratinocyte reduced KLF4 expression (38). In wild-type mouse embryos at E14.5, we detected anti-KLF4 immunoreactivity in the outer surface of the palate and tongue (i.e. oral periderm), whereas in the Irf6-null mutant mice such immunoreactivity was undetectable (Fig. 2A). We also found that over-expression of full-length human IRF6 in a human oral epithelium cell line (i.e. GMSM-K) led to the upregulation of KLF4 RNA expression (Fig. 2B). These results place IRF6 upstream of KLF4 expression in mammalian oral epithelium.
Figure 2.
IRF6 directly promotes the expression of KLF4 through binding to super-enhancers of KLF4. (A and B) Representative image of anti-KLF4 immunofluorescence (green) and nuclear DNA (DAPI, blue) in coronal section of mouse E14.5 head of (A) wild-type (Irf6+/+) and (B) Irf6-null mice (Irf6−/−). Arrowheads, immunoreactivity in oral periderm in wild-types, absent from mutants. P, palate shelf; T, tongue. Bar, 50 µm. (C) Quantitation of KLF4 mRNA in oral epithelial cell line without and with over-expressed human IRF6. Asterisks indicate statistical significance compared with control plasmid-transfected group: **P < 0.01. (D) Quantitative chromosome configuration capture (3C) experiment with the anchoring (i.e. bait) primer in the promoter of KLF4. Upper panel: UCSC Genome Browser view of hKLF4 E1-E5, peaks of IRF6 binding according to ChIP-seq and positions of two NHEK super-enhancers 1000 kb downstream of the KLF4 TSS. Lower panel: results of chromatin conformation capture experiment in human oral epithelial cell line confirming physical crosstalk between KLF4-E1 and -E5 with KLF4 promoter. Grids in x-axis represent each EcoRI restriction site. (E) Dual luciferase reporter assays for KLF4 activity in 293FT cells validate that IRF6 can bind to the putative IRF6 binding sites in hKLF4-E1 and -E5 and increase their activities. Asterisks indicate statistical significance compared with the luciferase activity of empty luciferase reporter co-transfected with IRF6 plasmid: **P < 0.01. (F–H) Lateral or (I) ventral epifluorescence views of live, wild-type zebrafish embryos at the indicated stage, previously injected with the indicated reporter construct. Arrowheads, GFP expression in internal, oral epithelium cells.
To test the possibility that IRF6 directly regulates KLF4 expression, we examined published catalogues of peaks of chromatin immunoprecipitation with anti-IRF6 and anti-H3K27Ac, an indicator of active chromatin, detected in primary normal human epidermal keratinocytes (NHEKs) (38,39). Whereas there were no overlapping peaks near the KLF4 promoter, there were five present nearly 1 Mb downstream from KLF4, with no intervening genes (Fig. 2C and Supplementary Material, Fig. S2). The sequences of these elements are evolutionarily conserved among mammals (www.genome.ucsc.edu, date last accessed, August 30, 2015), supporting the notion that they are enhancers; hereafter, we refer to them as KLF4 candidate enhancers 1–5 (KLF4-E1 to -E5). To test whether they are indeed enhancers of KLF4 expression, we conducted chromatin conformation capture (3C) in a human oral keratinocyte cell line (i.e. GMSM-K), anchored with a primer in the KLF4 promoter, and found that KLF4-E1 and KLF4-E5 interact with the KLF4 promoter (Fig. 2C). Notably, four of the five putative KLF4 enhancers are located within an extended segment (>12 kb) of chromatin marked with the H3K27Ac in NHEKs. This is a feature of so-called ‘super-enhancers’, which are proposed to regulate the expression of key cell-identity genes (40,41) (Fig. 2C and Supplementary Material, Fig. S2 and Table S1). We again performed 3C, with the anchor primer in KLF4-E1, and found that KLF4-E1 interacts with the other four elements in NHEK cells (Supplementary Material, Fig. S2). A published ChIA-PET analysis of MCF7 breast epithelium cells supports the findings of our 3C studies (42).
We next cloned KLF4-E1 and KLF4-E5 elements (individually) into a luciferase reporter vector with a minimal promoter. We found that both elements have enhancer activity in 293FT cell line that is elevated when the cells are co-transfected with a plasmid encoding IRF6 and that the responsiveness to exogenous IRF6 was lost when conserved IRF6 binding sites were deleted (Fig. 2D). We then engineered the intact elements into a GFP reporter vector and found that they have enhancer activity in the zebrafish periderm and oral epithelium (Fig. 2E and Supplementary Material, Fig. S3). Together, the 3C and reporter studies suggest that, in mammals, IRF6 directly drives KLF4 expression in the oral epithelium and in skin and that it does so by via distal enhancers that may function in a single chromatin complex.
On the basis of the results from the model systems, we asked whether mutations in KLF4 predispose individuals to cleft lip with or without cleft palate. As mentioned earlier, dominant-negative variants of GRHL3, encoding another member of the periderm GRN, are present in a subset of VWS patients who lack IRF6 mutations (23). We sequenced KLF4 exons in 40 individuals who had been diagnosed with VWS and lacked mutations in either IRF6 or GRHL3. However, we detected no non-synonymous mutations, indicating that KLF4 mutations are not a common cause of VWS.
Next, to test hypothesis that common sequence variants near KLF4 elevate risk for NSCL/P, we genotyped common variants near KLF4 exons in 184 multiplex families from the Philippines. We found no association with NSCL/P (the lowest P-value was P = 0.228 for rs149109998, Supplementary Material, Table S4). Similarly, we did not detect association of common variants near the distal enhancer with NSCL/P (E.L., unpublished data). This argues that common variants near KLF4, at least within the linkage disequilibrium units that we analysed, are unlikely to contribute risk for NSCL/P.
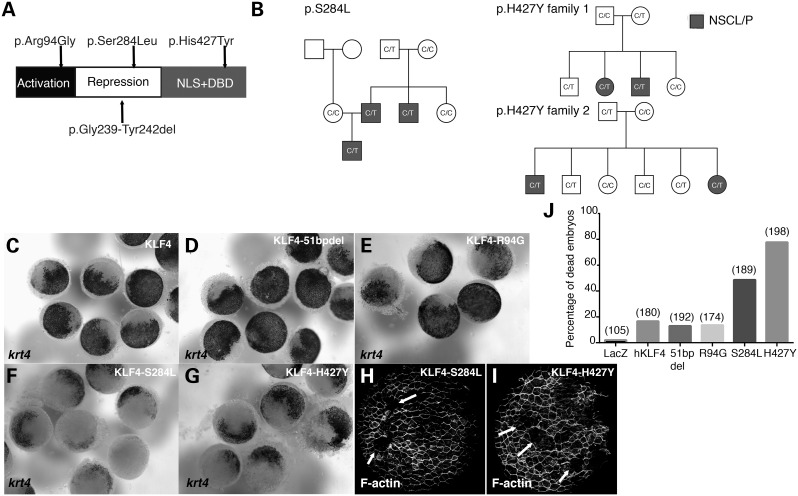
Finally, we identified KLF4 missense variants and tested for their association with NSCL/P using several statistical methods. We sequenced KLF4 exons in 1034 patients with NSCL/P from the Philippines, USA and Ethiopia and 338 controls from the Philippines (not all exons were sequenced in every sample, for details see Supplementary Material, Table S2). We detected one common coding variant, previously submitted to dbSNP (rs45520942/ ss70352771, see Materials and Methods), in two US cases. This variant, a 51 bp deletion resulting in the loss of 17 amino acids from the repression domain of KLF4, is present at a frequency of 2.6% in Western European samples (i.e. CEPH) catalogued at HapMap. Thus, it is unlikely to have a strong pathogenic effect. We also detected five novel missense variants: three in the Filipinos, p.S284L, p.H427Y (in two different families) and p.R94G, all predicted to be ‘probably damaging’ by Polyphen and ‘deleterious’ by SIFT; and two in the American cases, p.R309P and p.V261M, both predicted to be probably damaging by Polyphen but ‘tolerated’ by SIFT (Supplementary Material, Table S2). In three families, we sequenced all affected and unaffected individuals and found that the respective KLF4 variant was present in all affected individuals (Fig. 3B). In each family, the missense variant was inherited from an unaffected parent, as opposed to being de novo. Next we employed two burden tests: the cohort allelic sums test (CAST) (43) and the variable threshold (VT) method (44). These tests revealed an apparent trend towards association that did not reach formal significance (i.e. CAST, P = 0.058 and VT, P = 0.071). Of note, burden tests assume that all variants in the analysis have the same effect (i.e. all increase risk of disease) and have limited power when there are variants with opposite effects. We also applied two statistical analyses that do not make this assumption: the replication-based test (RBT) (45) and the C-alpha test (46). One but not the other revealed association that reached nominal significance (i.e. RBT, P = 0.026 and C-alpha, P = 0.362). These statistical analyses constitute mild support for the possibility that missense KLF4 variants contribute to NSCL/P pathogenesis.
Figure 3.
Rare NSCL/P alleles of KLF4 disrupt development of the zebrafish periderm. (A) Schematic representation of KLF4 protein. Positions of the rare, patient-derived variants predicted to be damaging by PolyPhen and SIFT are indicated above, and that of the common variant is indicated below. (B) Pedigrees of families with KLF4 p.S284L and p.H427Y variants. (C–I) Dorsal views of embryos, injected with the indicated mRNA and fixed at 5 hpf and processed to reveal (C–G) krt4 expression or (H and I) phalloidin stain. Arrows indicate periderm lacking phalloidin stain. (J) Quantification of embryos that died around 6 hpf when injected with the indicated mRNA. The numbers in parentheses indicate the total number of embryos for each group.
Next, to test for potential dominant-negative activity of patient-derived KLF4 variants, we synthesized KLF4 mRNA that matched the reference sequence, the common missense variant (i.e. 51 bp deletion) or the variants predicted to be probably damaging by Polyphen and deleterious by SIFT (i.e. R94G, S284L and H427Y). As expected, >90% of the embryos injected with the reference allele KLF4 survived through 24 hpf, and none ruptured during gastrulation. Similarly, the frequency of survival for embryos injected with the 51 bp deletion and R94G variants was comparable to that for those injected with KLF4. In contrast, >30% of the embryos injected with the S284L variant and >70% of those injected with H427Y exhibited widespread reduction of krt4 expression in the periderm at 5 hpf and ruptured at ∼6 hpf (Fig. 3C and D). These observations support the possibility that S284L and H427Y variants are pathogenic for NSCL/P. The fact that they were inherited from unaffected parents suggests that at normal expression levels they are not as severely damaging as when over-expressed.
Discussion
To identify candidates for genes that when mutated cause CL/P, we first employed the zebrafish gastrula-stage periderm as a model of human oral periderm, then pursued the results from zebrafish with functional tests in an oral epithelium cell line and finally tested a promising candidate gene by sequencing its exons in patients with NSCL/P. Previously, we found that in comparison to control embryos, embryos injected with dnIrf6 exhibit reduced expression of several transcription-factor-encoding genes, including klf17 (20), in addition to genes encoding periderm differentiation effectors such as keratin genes. Here, we found that forcing expression of klf17 could partially reverse the effects of dnIrf6, restoring expression of keratin genes. A promoter analysis suggested that this gene is directly regulated by Irf6. We found no published evidence that Klf17 is expressed in mammalian periderm or epidermis. In contrast, KLF4 is a close homologue of KLF17 and is known to be necessary for epidermal differentiation (29). Referring to published IRF6-ChIP-SEQ (47) data, we found that IRF6 binds several distal elements. We established that these elements interact with the KLF4 promoter and that they have epidermal enhancer activity that depends on the presence of the IRF6 binding sites. Sequencing within all exons for KLF4 in more than 1000 CL/P probands revealed five rare coding variants presenting in different CL/P pedigrees, and with zebrafish periderm as in vivo model, two of them were functionally identified exhibiting dominant-negative effect. Together with earlier work, this study reveals that mutations in genes encoding three elements of the oral periderm GRN—i.e. IRF6, GRHL3 and KLF4—can all predispose individuals to cleft lip with or without cleft palate.
GWASs (3–7), linkage studies (8) and a meta-analysis of these (9) have identified at least 18 loci in which specific common alleles are associated with NSCL/P at genome-wide significance. The high ratio of the number of risk-associated loci identified versus the number of samples sequenced makes NSCL/P one of the most successful applications of GWASs; together, the loci account for more than half of the overall population-attributable risk for this disorder (48). The remaining ‘missing heritability’ may reside in rare variants of large effect size, among other possibilities (e.g. gene interactions, structural variation and misdiagnosis (49)]. Supporting this view, two rare KLF4 variants we identified in NSCL/P cases have the potential to have a large effect size, based on the in silico analysis and on zebrafish-based functional studies. The fact that they were inherited from unaffected parents indicates they are not sufficient to cause disease on their own, consistent with the diagnosis of NSCL/P in the patients. Additional rare variants pathogenic for NSCL/P may be found in other candidate loci with strong biological support either through direct sequencing (as we have done here) or through exome and whole-genome sequencing efforts.
This work indicates that fish Klf17 and mammalian Klf4 share the function of promoting differentiation of superficial epithelia. Phylogenic analysis shows that klf17 groups with klf2 and klf4 (25,26). In zebrafish, klf2a, klf2b and klf17 function in a network to promote periderm differentiation (28). Our work extends the earlier study to show that excess klf17 has the potential to repress expression of some periderm markers on the ventral side, but not to disrupt cell adhesion in the periderm as is induced by dnklf17, perhaps by repressing klf2b expression (28). Zebrafish klf4 is not expressed in gastrula-stage embryos (28), but embryos depleted of klf4 exhibit abnormal differentiation of intestinal epithelium (50), as do murine Klf4 mutants (51–53). Indeed, in mouse, Klf4 has an essential role in the differentiation of many epithelia, including epidermis, by stimulating expression of CDKN1A (25). We found that a human Klf4 enhancer drives a periderm expression pattern that resembles zebrafish klf17, not zebrafish klf4, and that human Klf4 functioned equivalently to zebrafish klf17 in an over-expression assay. A parsimonious evolutionary scenario consistent with these data is that an ancestral gene, encoding a KLF orthologue that promoted periderm differentiation, was duplicated, giving rise to KLF2, KLF4 and KLF17. Differential loss of regulatory elements or changes in protein function (both forms of subfunctionalization) (54) in different taxa resulted in retention of periderm differentiation function in distinct paralogues in distinct taxa. Supporting this model, in the mammalian but not in the fish lineage, KLF17 has evolved rapidly, consistent with protein subfunctionalization (25,26). It has been proposed that Klf family members are especially prone to evolvability by virtue of their quality of being co-regulated by the same upstream regulators (28).
Exploring the zebrafish periderm GRN has led us to two genes that appear to contribute risk for orofacial clefting (i.e. GRHL3 and KLF4); we anticipate that other members of this GRN will contain the rare variants that contribute to the missing heritability of NSCL/P. It is noteworthy that among the 18 genes presumed to be associated with NSCL/P in genome-wide studies, at least six are expressed in the oral epithelium (i.e. ARHGAP29, FOXE1, IRF6, MAFB, NOG1, and SPRY2). VAX1, also identified in such studies, is expressed in the brain but is essential for normal patterning of oral epithelium (55). Zebrafish embryonic periderm is a tractable model for deducing the architecture of the regulatory networks containing these proteins, and for testing the function of patient-derived variants. Combined with ChIP and analyses of chromatin interactions in cell lines or primary periderm cells, it should be possible to assemble the periderm GRN, including the direct regulatory interactions, at high resolution.
Materials and Methods
Human samples and study approval
Patients with NSCL/P were recruited from sites in the Philippines, Ethiopia and the USA. Individuals with additional congenital malformations or developmental delays suggestive of a syndrome were excluded. Patients in the US cohort with self-reported race/ethnicity as Asian, Hispanic or Black were also excluded. Controls with no known history of craniofacial anomalies were recruited from the Philippines. Informed consent was obtained for all participants, and the study was approved by the Institutional Review Board at the University of Iowa (approval numbers 199804080, 199804081 and 201102785).
Animal models
Breeding Danio rerio were maintained as described (56) in the University of Iowa Animal Care Facility. Zebrafish embryos were staged according to Kimmel et al. (57) at 28.5°C by hours or days post fertilization (hpf or dpf).
The Irf6 mutant allele (Irf6−/−), which was maintained in a C57BL/6 background, has been described previously (58). C57BL/6 mice were used as wild-type control. Murine embryos were obtained from pregnant female mice at 14.5 days after conception (E14.5). Animal use protocols were approved by the Institutional Animal Care and Use Committees at the University of Iowa.
Zebrafish KLF17 and human KLF4 mRNA injection and in situ hybridization
Full-length, wild-type zebrafish klf17 cDNA (RefSeq: NM_131723.1) was cloned from zebrafish 6 hpf cDNA and inserted into pCS2+ plasmid. Different functional domains of Klf17 were determined by comparing its amino acid sequence with human KLF4 (UniProtKB: O43474). Different truncated forms of zebrafish KLF17 were generated by overlapping PCR. Sanger sequencing confirmed that no variants were present in the clones. Full-length, wild-type and p.S284L mutant human KLF4 cDNAs (RefSeq: NM_004235.4) were obtained from Stratagene company. Other mutations from NSCL/P patients were generated via PCR-mediated mutagenesis. All the KLF4 variants were engineered into pCS2+ plasmid.
Capped mRNA was synthesized in vitro (mMESSAGE mMACHINE SP6 kit, Ambion) and purified with RNeasy® mini kit (Qiagen, Valencia, CA, USA), and ∼1 ng of mRNA was injected into wild-type zebrafish embryos at one-cell stage. Whole-mount in situ hybridization for krt4 was performed as described before (19).
RNA isolation and quantitative RT–PCR
Total RNA was isolated using RNeasy mini kit (Qiagen) and treated with RNease-free DNaseI (Promega, Madison, WI, USA). RT was performed by High Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA, USA). Real-time PCR was performed in CFX96 Touch™ Real-Time PCR Detection System. For zebrafish and human mRNA expression assays, beta-2-microglobulin and glyceraldehyde 3-phosphate dehydrogenase were used as an internal control, respectively.
Cell culture, luciferase reporter constructs, transfections and dual luciferase assay
The 293FT cell line (human embryonic kidney cell line) was purchased from Invitrogen (Carlsbad, CA, USA) and was maintained in Dulbecco's modified Eagle's medium (Life Technologies, Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (Life Technologies) and 1% antibiotic-antimycotic (Life Technologies). GMSM-K human embryonic oral epithelial cell line (a kind gift from Dr Daniel Grenier) (59) was maintained in keratinocyte serum-free medium (Life Technologies), supplemented with epidermal growth factor human recombinant (Life Technologies) and bovine pituitary extract (Life Technologies). All cells were incubated at 37°C in 5% CO2.
For zebrafish klf17 promoter assay, about 2.2 kb upstream of zebrafish klf17 (GRCz10, chr2:33 341 076–33 343 238) was cloned directly from zebrafish genomic DNA, two IRF6 binding motifs were deleted subsequently using overlapping PCR as described previously (60). For human KLF4 potential enhancers reporter assay, H3K27Ac ChIP-seq peaks containing IRF6 ChIP-seq peaks were cloned (KLF4-E1: NCBI36/hg18, chr9:110 344 050 –110 346 209; KLF4-E2: NCBI36/hg18, chr9:110 352 710–110 356 644; KLF4-E3: NCBI36/hg18, chr9:110 367 137–110 368 773; KLF4-E4: NCBI36/hg18, chr9:110 435 915–110 438 546; KLF4-E5: NCBI36/hg18, chr9:110 472 889–110 476 694). Truncations of IRF6 binding sites in KLF4-E1 and KLF4-E5 were performed with overlapping PCR. All the cloned elements were engineered into pTol2-cFos-FLuc as reported before (12). To verify whether these elements are IRF6-dependent, we co-transfected these reporter plasmids with full-length zebrafish or human IRF6 over-expression plasmids. pCS2+ plasmid which harbours these insertions was used as empty plasmid control. Transient transfection in 293FT cells was performed using Lipofectamine® 3000 (Life Technologies), according to manufacturer's instructions. Transient transfection in GMSM-K cells was performed using Nucleofector™ II system (Lonza, Cologne, Germany), according to manufacturer's instructions.
For each reporter construct, three independent transfections were carried out with Renilla luciferase plasmid (pTol2-cFos-RLuc) co-transfection. The Dual-Luciferase Reporter Assay System (Promega) and 20/20n Luminometer (Turner Biosystems, Sunnyvale, CA, USA) were used to measure luciferase activity in cell lysates. Relative luciferase activities were calculated by the ratio between the read for firefly and renilla. Three measurements were made on every three biological replicates. All quantified results are presented as mean ± SEM. One-way analysis of variance test was used to determine statistical significance.
Immunofluorescent staining
Zebrafish embryos were fixed at dome stage in 4% paraformaldehyde/phosphate-buffered saline (PBS) at 4°C overnight. For F-actin staining, 0.165 m Phalloidin Alexa 546 (University of Iowa Central Microscopy Research Facility, Iowa City, IA, USA) was used as described previously (19). Confocal images of periderm cells in Phalloidin-stained embryos were collected with a 20×/0.5 NA objective lens (Carl Zeiss).
Immunodetection for mouse Klf4 was performed on coronal sections of 4% formaldehyde-fixed, paraffin-embedded E14.5 mouse heads. After blocking with 3% goat serum (Vector Laboratories, Burlingame, CA, USA), sections were incubated with rabbit anti-human KLF4 (Abcam, Cambridge, MA, USA, ab72543), washed in PBS and incubated in goat anti-rabbit Alexa-Fluor 488. DAPI (LifeTechnologies) was used as a nuclear stain. Images were viewed with a Nikon Eclipse E800 (Melville, NY, USA) and acquired with a SPOT RT Slider Diagnostic Instruments CCD camera using Spot Advanced software (Diagnostic Instruments, Sterling Heights, MI, USA). Black and white images were pseudocolourized and merged.
Chromatin conformation capture (3C)
The 3C-qPCR assay was performed as described (61) for at least two replicates. The 1×107 GMSM-K oral epithelial cells were crosslinked with 1% formaldehyde at room temperature for 10 min, followed by glycine quenching and cell lysis. Nuclei were resuspended in 500 µl 1× EcoRI Buffer and then lysed with 0.2% sodium dodecyl sulphate (SDS), followed by SDS sequestration with 1.2% Triton X-100. Lysates were digested overnight at 37°C with EcoRI (NEB, Ipswich, MA, USA) and the restriction enzyme then inactivated for 1 h at 65°C with 1.6% SDS. Ligation was performed with NEB T4 ligase in 1.15× T4 Ligase buffer at 16°C for 4 h. Finally, the ligation products were purified with phenol-extraction protocol. 3C ligation products were quantified with three replicates by SYBR Green qPCR. The 3C signals at the each locus were normalized first to a loading control (primers located in the GAPDH gene and that do not flank any EcoRI digestion site), and second to a random collision control, a region 8.1kb from the KLF4 TSS, chosen because it lacks chromatin marks in NHEK). Primers were designed using Primer3 Plus online software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and tested against EcoRI-digested, ligated BAC templates (RP11-150J11 and RP11-589C8) by serial dilution and melt-curve analysis to ensure specific and linear amplification. Digestion efficiencies were determined with SYBR Green qPCR by comparison of aliquots taken pre- and post-digestion using primer pairs that amplified a region in each interrogated fragment spanning an EcoRI digestion site.
Genomic interval manipulation
Identification of super-enhancers of NHEK cells was performed according to the algorithm described before (41). Briefly, constituent enhancers in NHEKs were defined as ChIP-seq peaks of H3K27ac in NHEK (39), with exclusion of 3 kb in the vicinity of TSS for each gene. H3K27ac ChIP-seq raw reads were sorted and indexed using SAMtools (v. 0.1.18) (62). About 1470 super-enhancers in NHEKs were defined. Intersections between NHEK super-enhancers and IRF6 ChIP-seq peaks (38) were carried out in Bedtools (v. 2.24.0) (63), and the positive intersection flanking KLF4 was visualized in UCSC Genome Browser.
Zebrafish enhancer in vivo reporter assay
Human KLF4 potential enhancers KLF4-E1 to KLF4-E5 were cloned as described earlier and engineered into the zebrafish enhancer detection (ZED) vector (64). ZED vectors (25–30 ng/μl) were injected along with the tol2 mRNA (20–30 ng/μl) into 200 one-cell stage embryos. The embryos were screened at 6 and 72 hpf for enhancer activity in periderm and oral epithelium, respectively. A consistent GFP expression pattern in a minimum of 10% of injected fish (i.e. F0) was the criterion for tissue-specific enhancer activity, as this usually predicts a given element's expression pattern in F1 animals. Embryos with periderm enhancer activity were photographed in bright field, epi-fluorescent illumination on a Leica DMRA2 compound microscope with a colour 12 bit ‘QIClick’ camera (QImaging). Embryos with oral epithelium enhancer activity were photographed in a Zeiss 700 confocal microscope with a 20×/0.5 NA objective lens (Carl Zeiss).
Human sample genotyping
Three common SNPs (rs2236599, rs149109998 and rs77275043) were genotyped using Taqman SNP Genotyping Assays (Life Technologies, Grand Island, NY, USA) on the ABI Prism 7900HT and analysed with SDS 2.4 software (Applied Biosystems). Families with Mendelian errors were excluded from analyses.
Human sample sequencing
Primers were designed with Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi) to cover all exons of KLF4 (Supplementary Material, Table S1). For technical reasons, only the 375 bp of the 5′-UTR upstream of the start codon and the first 500 bp of the 3′-UTR were sequenced. PCR products were Sanger-sequenced using ABI BigDye v3.1 on an ABI 3730×l instrument (Functional Biosciences, Inc., Madison, WI, USA). The chromatograms were transferred to a Unix workstation, base-called with PHRED (v. 0.961028), assembled with PHRAP (v. 0.960731), scanned by POLYPHRED (v. 0.970312) and visualized using CONSED program (v. 4.0). We used the Variant Effect Predictor tool from Ensemble (http://www.ensembl.org/tools.html) to predict the functional effects of missense variants (65).
The rs45520942 variant was deleted from the dbSNP database because of mapping or clustering errors. The original dbSNP submission can be found with the submission identifier ss70352771. It has also been found in the exomes of the Exome Aggregation Consortium, although it has been flagged as a lower quality site. This variant appears to be a polymorphism, although the frequency is unclear, which may be due to difficulty mapping this region of the genome.
Genetic statistical analysis
Association with rs2236599, rs149109998 and rs77275043 in the Filipino families was tested using the family-based association test (FBAT) (66). We used the -e option in FBAT, which allows for the adjustment for multiple affected individuals or multiple nuclear families within a pedigree by adjusting the variance used in the Z-score calculation.
From the sequencing data, we considered a variant ‘novel’ if it was absent from dbSNP137, the 1000 Genomes Project and the NHLBI Exome Variant Server. We performed case–control analyses on two groups of rare variants (MAF <1%): (i) all coding variants and (ii) all missense variants. Association with rare variants was tested using several burden and non-burden statistical methods. Burden tests collapse all of the rare variants in a region into a single ‘score’ and test for association between that score and the phenotype. We employed two burden tests: the CAST (43) and the VT method (44). Burden tests assume that all variants in the analysis have the same effect (i.e. all increase risk of disease) and have limited power when there are variants with opposite effects. We also employed two non-burden tests: the C-alpha test (46) and the RBT (45). All rare variant analyses were implemented in R using the AssotesteR software package.
The Exome Aggregation Consortium catalogue (URL: http://exac.broadinstitute.org) was accessed on July 23, 2015.
The URL for the HapMap project is http://hapmap.ncbi.nlm.nih.gov.
Supplementary Material
Funding
This work was supported by the National Institute of Health grants HD073107 (R.A.C.), K99-DE025060 (E.J.L.), K99/R00 DE022378 (A.B.), DE08559 (J.M.), by the Robert Wood Johnson Foundation grant 72429 (A.B.), by the National Natural Science Foundation of China 81400477 (H.L.), and by Scientific Research Funds for Young Teachers of Sichuan University grant 2015SCU11999 (Z.L.). Funding to pay the Open Access publication charges for this article was provided by a grant from the A.E. and Martha Michelbacher Trust administered by the Marin Community Foundation.
Supplementary Material
Acknowledgement
We are grateful to Gregory Bonde for excellent animal husbandry and technical support of this project.
Conflict of Interest statement. None declared.
References
- 1.Rahimov F., Jugessur A., Murray J.C. (2012) Genetics of nonsyndromic orofacial clefts. Cleft Palate-Craniofac. J., 49, 73–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leslie E.J., Marazita M.L. (2013) Genetics of cleft lip and cleft palate. Am. J. Med. Genet. C: Semin. Med. Genet., 163, 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaty T.H., Murray J.C., Marazita M.L., Munger R.G., Ruczinski I., Hetmanski J.B., Liang K.Y., Wu T., Murray T., Fallin M.D. et al. (2010) A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet., 42, 525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnbaum S., Ludwig K.U., Reutter H., Herms S., Steffens M., Rubini M., Baluardo C., Ferrian M., Almeida de Assis N., Alblas M.A. et al. (2009) Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat. Genet., 41, 473–477. [DOI] [PubMed] [Google Scholar]
- 5.Grant S.F., Wang K., Zhang H., Glaberson W., Annaiah K., Kim C.E., Bradfield J.P., Glessner J.T., Thomas K.A., Garris M. et al. (2009) A genome-wide association study identifies a locus for nonsyndromic cleft lip with or without cleft palate on 8q24. J. Pediat., 155, 909–913. [DOI] [PubMed] [Google Scholar]
- 6.Mangold E., Ludwig K.U., Birnbaum S., Baluardo C., Ferrian M., Herms S., Reutter H., de Assis N.A., Chawa T.A., Mattheisen M. et al. (2010) Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet., 42, 24–26. [DOI] [PubMed] [Google Scholar]
- 7.Sun Y., Huang Y., Yin A., Pan Y., Wang Y., Wang C., Du Y., Wang M., Lan F., Hu Z. et al. (2015) Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat. Commun., 6, 6414–6420. [DOI] [PubMed] [Google Scholar]
- 8.Moreno L.M., Mansilla M.A., Bullard S.A., Cooper M.E., Busch T.D., Machida J., Johnson M.K., Brauer D., Krahn K., Daack-Hirsch S. et al. (2009) FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Hum. Mol. Genet., 18, 4879–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludwig K.U., Mangold E., Herms S., Nowak S., Reutter H., Paul A., Becker J., Herberz R., AlChawa T., Nasser E. et al. (2012) Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet., 44, 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A. et al. (2009) Finding the missing heritability of complex diseases. Nature, 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leslie E.J., Murray J.C. (2013) Evaluating rare coding variants as contributing causes to non-syndromic cleft lip and palate. Clin. Genet., 84, 496–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leslie E.J., Taub M.A., Liu H., Steinberg K.M., Koboldt D.C., Zhang Q., Carlson J.C., Hetmanski J.B., Wang H., Larson D.E. et al. (2015) Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. Am. J. Hum. Genet., 96, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kousa Y.A., Schutte B.C. (2015) Toward an orofacial gene regulatory network. Dev. Dyn., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo S., Schutte B.C., Richardson R.J., Bjork B.C., Knight A.S., Watanabe Y., Howard E., de Lima R.L., Daack-Hirsch S., Sander A. et al. (2002) Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat. Genet., 32, 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ingraham C.R., Kinoshita A., Kondo S., Yang B., Sajan S., Trout K.J., Malik M.I., Dunnwald M., Goudy S.L., Lovett M. et al. (2006) Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat. Genet., 38, 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richardson R.J., Hammond N.L., Coulombe P.A., Saloranta C., Nousiainen H.O., Salonen R., Berry A., Hanley N., Headon D., Karikoski R. et al. (2014) Periderm prevents pathological epithelial adhesions during embryogenesis. J. Clin. Invest., 124, 3891–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimmel C.B., Warga R.M., Schilling T.F. (1990) Origin and organization of the zebrafish fate map. Development, 108, 581–594. [DOI] [PubMed] [Google Scholar]
- 18.Lee R.T.H., Asharani P.V., Carney T.J. (2014) Basal keratinocytes contribute to all strata of the adult zebrafish epidermis. PLoS ONE, 9, e84858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabel J.L., d'Alencon C., O'Brien E.K., Van Otterloo E., Lutz K., Cuykendall T.N., Schutte B.C., Houston D.W., Cornell R.A. (2009) Maternal interferon regulatory factor 6 is required for the differentiation of primary superficial epithelia in Danio and Xenopus embryos. Dev. Biol., 325, 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Garza G., Schleiffarth J.R., Dunnwald M., Mankad A., Weirather J.L., Bonde G., Butcher S., Mansour T.A., Kousa Y.A., Fukazawa C.F. et al. (2013) Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of Grainyhead-like 3. J. Invest. Dermatol., 133, 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bray S.J., Kafatos F.C. (1991) Developmental function of Elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes Dev., 5, 1672–1683. [DOI] [PubMed] [Google Scholar]
- 22.Ting S.B., Caddy J., Hislop N. (2005) A homolog of Drosophila Grainy head is essential for epidermal integrity in mice. Science, 308, 411–413. [DOI] [PubMed] [Google Scholar]
- 23.Peyrard-Janvid M., Leslie E.J., Kousa Y.A., Smith T.L., Dunnwald M., Magnusson M., Lentz B.A., Unneberg P., Fransson I., Koillinen H.K. et al. (2014) Dominant mutations in GRHL3 cause Van der Woude Syndrome and disrupt oral periderm development. Am. J. Hum. Genet., 94, 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McConnell B.B., Yang V.W. (2010) Mammalian Krüppel-like factors in health and diseases. Physiol. Rev., 90, 1337–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tetreault M.P., Yang Y., Katz J.P. (2013) Kruppel-like factors in cancer. Nat. Rev. Cancer, 13, 701–713. [DOI] [PubMed] [Google Scholar]
- 26.Xue Y., Gao S., Liu F. (2015) Genome-wide analysis of the zebrafish Klf family identifies two genes important for erythroid maturation. Dev. Biol., 403, 115–127. [DOI] [PubMed] [Google Scholar]
- 27.Kawahara A., Dawid I.B. (2000) Expression of the Kruppel-like zinc finger gene biklf during zebrafish development. Mech. Dev., 97, 173–176. [DOI] [PubMed] [Google Scholar]
- 28.Kotkamp K., Mossner R., Allen A., Onichtchouk D., Driever W. (2014) A Pou5f1/Oct4 dependent Klf2a, Klf2b, and Klf17 regulatory sub-network contributes to EVL and ectoderm development during zebrafish embryogenesis. Dev. Biol., 385, 433–447. [DOI] [PubMed] [Google Scholar]
- 29.Segre J.A., Bauer C., Fuchs E. (1999) Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet., 22, 356–360. [DOI] [PubMed] [Google Scholar]
- 30.Eaton S.A., Funnell A.P., Sue N., Nicholas H., Pearson R.C., Crossley M. (2008) A network of Kruppel-like factors (Klfs). Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J. Biol. Chem., 283, 26937–26947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang C.J., Lemsaddek W., Alhashem Y.N., Bondzi C., Redmond L.C., Ah-Son N., Dumur C.I., Archer K.J., Haar J.L., Lloyd J.A. et al. (2012) Kruppel-like factor 1 (KLF1), KLF2, and Myc control a regulatory network essential for embryonic erythropoiesis. Mol. Cell. Biol., 32, 2628–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simmen F.A., Su Y., Xiao R., Zeng Z., Simmen R.C. (2008) The Kruppel-like factor 9 (KLF9) network in HEC-1-A endometrial carcinoma cells suggests the carcinogenic potential of dys-regulated KLF9 expression. Reprod. Biol. Endocrinol., 6, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner J., Crossley M. (1999) Basic Kruppel-like factor functions within a network of interacting haematopoietic transcription factors. Int. J. Biochem. Cell Biol., 31, 1169–1174. [DOI] [PubMed] [Google Scholar]
- 34.Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H. et al. (2014) JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res., 42, D142–D147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray P.A., Fu H., Luo P., Zhao Q., Yu J., Ferrari A., Tenzen T., Yuk D.-i., Tsung E.F., Cai Z. et al. (2004) Mouse brain organization revealed through direct genome-scale TF expression analysis. Science, 306, 2255–2257. [DOI] [PubMed] [Google Scholar]
- 36.Harding S.D., Armit C., Armstrong J., Brennan J., Cheng Y., Haggarty B., Houghton D., Lloyd-MacGilp S., Pi X., Roochun Y. et al. (2011) The GUDMAP database—an online resource for genitourinary research. Development, 138, 2845–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaubert J., Cheng J., Segre J.A. (2003) Ectopic expression of kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development, 130, 2767–2777. [DOI] [PubMed] [Google Scholar]
- 38.Botti E., Spallone G., Moretti F., Marinari B., Pinetti V., Galanti S., De Meo P.D., De Nicola F., Ganci F., Castrignano T. et al. (2011) Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc. Natl Acad. Sci. USA, 108, 13710–13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium E.P. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adam R.C., Yang H., Rockowitz S., Larsen S.B., Nikolova M., Oristian D.S., Polak L., Kadaja M., Asare A., Zheng D. et al. (2015) Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature, 521, 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. (0000) Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell, 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J. et al. (2012) Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell, 148, 84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgenthaler S., Thilly W.G. (2007) A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST). Mutat. Res., 615, 28–56. [DOI] [PubMed] [Google Scholar]
- 44.Price A.L., Kryukov G.V., de Bakker P.I., Purcell S.M., Staples J., Wei L.J., Sunyaev S.R. (2010) Pooled association tests for rare variants in exon-resequencing studies. Am. J. Hum. Genet., 86, 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ionita-Laza I., Buxbaum J.D., Laird N.M., Lange C. (2011) A new testing strategy to identify rare variants with either risk or protective effect on disease. PLoS Genet., 7, e1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neale B.M., Rivas M.A., Voight B.F., Altshuler D., Devlin B., Orho-Melander M., Kathiresan S., Purcell S.M., Roeder K., Daly M.J. (2011) Testing for an unusual distribution of rare variants. PLoS Genet., 7, e1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Botti E., Spallone G., Moretti F. (2011) Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc. Natl Acad. Sci. USA, 108, 13710–13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marazita M.L. (2012) The evolution of human genetic studies of cleft lip and cleft palate. Annu. Rev Genom. Hum. G, 13, 263–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maher B. (2008) Personal genomes: the case of the missing heritability. Nature, 456, 18–21. [DOI] [PubMed] [Google Scholar]
- 50.Li I.C., Chan C.T., Lu Y.F., Wu Y.T., Chen Y.C., Li G.B., Lin C.Y., Hwang S.P. (2011) Zebrafish kruppel-like factor 4a represses intestinal cell proliferation and promotes differentiation of intestinal cell lineages. PLoS ONE, 6, e20974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katz J.P., Perreault N., Goldstein B.G., Actman L., McNally S.R., Silberg D.G., Furth E.E., Kaestner K.H. (2005) Loss of Klf4 in mice causes altered proliferation and differentiation and precancerous changes in the adult stomach. Gastroenterology, 128, 935–945. [DOI] [PubMed] [Google Scholar]
- 52.Katz J.P., Perreault N., Goldstein B.G., Lee C.S., Labosky P.A., Yang V.W., Kaestner K.H. (2002) The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development, 129, 2619–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y., Goldstein B.G., Chao H.H., Katz J.P. (2005) KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol. Ther., 4, 1216–1221. [DOI] [PubMed] [Google Scholar]
- 54.Van Otterloo E., Cornell R.A., Medeiros D.M., Garnett A.T. (2013) Gene regulatory evolution and the origin of macroevolutionary novelties: insights from the neural crest. Genesis, 51, 457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bharti K., Gasper M., Bertuzzi S., Arnheiter H. (2011) Lack of the ventral anterior homeodomain transcription factor VAX1 leads to induction of a second pituitary. Development, 138, 873–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Westerfield M. (1993) The Zebrafish Book. University of Oregon Press, Eugene, OR. [Google Scholar]
- 57.Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. (1995) Stages of embryonic development of the zebrafish. Dev. Dyn., 203, 253–310. [DOI] [PubMed] [Google Scholar]
- 58.Biggs L.C., Rhea L., Schutte B.C., Dunnwald M. (2012) Interferon regulatory factor 6 is necessary, but not sufficient, for keratinocyte differentiation. J. Invest. Dermatol., 132, 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gilchrist E.P., Moyer M.P., Shillitoe E.J., Clare N., Murrah V.A. (2000) Establishment of a human polyclonal oral epithelial cell line. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodont., 90, 340–347. [DOI] [PubMed] [Google Scholar]
- 60.Heckman K.L., Pease L.R. (2007) Gene splicing and mutagenesis by PCR-driven overlap extension. Nat. Protoc., 2, 924–932. [DOI] [PubMed] [Google Scholar]
- 61.Hagege H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forne T. (2007) Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc., 2, 1722–1733. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol., 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quinlan A.R. (2014) BEDTools: the swiss-army tool for genome feature analysis. Curr. Protoc. Bioinform., 47, 11. 12. 1–11. 12. 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bessa J., Tena J.J., de la Calle-Mustienes E., Fernandez-Minan A., Naranjo S., Fernandez A., Montoliu L., Akalin A., Lenhard B., Casares F. et al. (2009) Zebrafish enhancer detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev. Dyn., 238, 2409–2417. [DOI] [PubMed] [Google Scholar]
- 65.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. (2010) Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics, 26, 2069–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Horvath S., Xu X., Laird N.M. (2001) The family based association test method: strategies for studying general genotype–phenotype associations. Eur. J. Hum. Genet., 9, 301–306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.