Abstract
Mitochondrial anomalies have been previously reported in patients′ brain and peripheral tissue, suggesting their relevance in sporadic Alzheimer's disease (AD). The present work evaluates mitochondrial function and recycling in human fibroblasts and brain biopsies. Functional studies using patients′ skin fibroblasts showed slower mitochondrial membrane potential recovery after a mitochondrial insult together with alterations in lysosomes and autophagy, accompanied by an increase of oxidized and ubiquitinated proteins. Impairment in mitophagy has been proven in these cells due to diminished PARK2 and insufficient vesicle induction, accumulating depolarized mitochondria and PINK1. Augmented Δ1 PINK1 fragment levels suggest an inhibitory effect over PARK2 translocation to the mitochondria, causing the accumulation of activated PINK1. Moreover, the overexpression of PARK2 diminished ubiquitinated proteins accumulation, improves its targeting to mitochondria and potentiates autophagic vesicle synthesis. This allows the reversion of mitophagy failure reflected in the recovery of membrane potential and the decrease of PINK1 and mitochondria accumulation. Sporadic AD fibroblasts exhibited alterations similar to what it could be found in patients’ hippocampal samples at early stages of the disease, where there was an accumulation of PINK1 and Δ1 PINK1 together with abnormally increased mitochondrial content. Our findings indicate that mitophagy alterations can be considered a new hallmark of sporadic AD and validate the use of fibroblasts for modelling this pathology.
Introduction
Alzheimer is a silent neurodegeneration in which amyloid-beta (Aβ) senile plaques, microtubule-associated protein tau (MAPT/Tau) tangles and cellular degeneration appear before the onset of disease symptoms (1). Only a small proportion of Alzheimer's disease (AD) cases can be related to mutations in Amyloid-beta (A4) precursor protein (APP) and Presenilin 1 and 2 (PSEN1 and PSEN2) genes (2), which can be easily detected by a genetic analysis and are classified as familial AD (FAD). Conversely, the majority of the patients cannot be associated with any known mutation, and they are considered sporadic AD (SAD) cases. Therefore, it is necessary to identify new early and easily accessible markers of the disease to be able to identify future patients presymptomatically.
Abnormal mitochondrial dynamics and distribution as well as related oxidative stress together with synaptic degeneration are the earliest and most prominent features in vulnerable neurons in the brain of AD patients (3–7). Reactive oxygen species (ROS) are associated with enhanced Aβ formation (8), as well as MAPT self-assembly into fibrilar polymers (9) and MAPT phosphorylation (10,11). Conversely, increased Aβ may result in mitochondrial dysfunction and augmented ROS levels (12), whereas MAPT accumulation causes mitochondrial distribution deficits (13).
Autophagy is a normal cellular recycling process that involves degradation of unnecessary or dysfunctional cellular components through lysosome. Mitochondrial function is regulated by autophagy in a process called mitophagy, by which damaged mitochondria are engulfed into autophagosomes followed by their subsequent catabolism by lysosomes. The label of dysfunctional mitochondria to be recycled by autophagy involves the Parkin RBR E3 ubiquitin protein ligase (PARK2) (14) and PTEN-induced putative kinase 1 (PINK1) (15). PINK1 precursor translocates to the mitochondria and is processed into two cleaved forms that then move to the cytosol to be degraded by the proteasome (16). After mitochondrial depolarization, full-length PINK1 is stabilized in mitochondrial surface inducing the recruitment of PARK2 (17). PARK2-mediated ubiquitination recruits autophagy adapter proteins such as sequestosome 1 (SQSTM1/p62), which interacts with MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3) mediating the cargo engulfment into autophagosomes (18). After dissipation of mitochondrial membrane potential (ΔΨm), the autophosphorylation of PINK1 is essential for its activation, promoting the recruitment of PARK2 to the mitochondria and its phosphorylation at Ser65 as an initial step of mitophagy (19,20). PARK2 ubiquitinates several mitochondrial proteins, such as voltage-dependent anion channel 1 (21) and mitofusins, that may be involved in the segregation of damaged mitochondria due to the inhibition of mitochondrial fusion events (22). In contrast, it has been recently demonstrated that PINK1 may also play an inhibitory role in the mitophagy process (23). The main cleaved product of PINK1 (Δ1 fragment) is able to physically bind PARK2 in the cytosol, inhibiting its translocation to the mitochondria, therefore, impairing the elimination of damaged mitochondria.
Autophagy is intensively involved in AD-related protein aggregation, being the major degradational pathway after activation of the unfolded protein response, an early event in the affected brain (24,25). Several studies have revealed autophagic vacuole (AV) accumulation that affects both neuronal and peripheral cells in AD (24,26–28). In addition, an increase of autophagic vesicles containing mitochondria is found in pyramidal neurons from AD patients, suggesting a mitophagy alteration (29,30). Furthermore, it has been revealed that functional PSEN1 is required for lysosomal maturation, which is impaired by Alzheimer-related PSEN1 mutations, suggesting a link between FAD and autophagy (28). Conversely, autophagy has been proposed to play an active role in Alzheimer pathogenesis, with autophagic vesicles being an active compartment for Aβ generation (31).
Fibroblasts are abundant in peripheral tissues and can be obtained from a small skin biopsy. Although Alzheimer is a neurodegenerative disease, it has been demonstrated that many disease hallmarks can also be found in fibroblasts such as increased oxidative stress levels and reduced antioxidant defences (32). Fibroblasts from FAD patients related to PSEN1 mutations show autophagy impairment characterized by autophagic vesicle accumulation and inefficient degradation phase due to a deficit in lysosomal acidification (28).
In this work, we have demonstrated a mitophagy failure in SAD fibroblasts in which alterations in mitochondrial function and recycling process have been proven. This mitophagy impairment could be reversed by the increase of PARK2. Similar mitophagy alterations could be found in patients’ hippocampal samples at early stages of the disease, confirming the reliability of the findings and the suitability of this model for the study of the disease.
Results
Mitochondrial anomalies in SAD fibroblasts
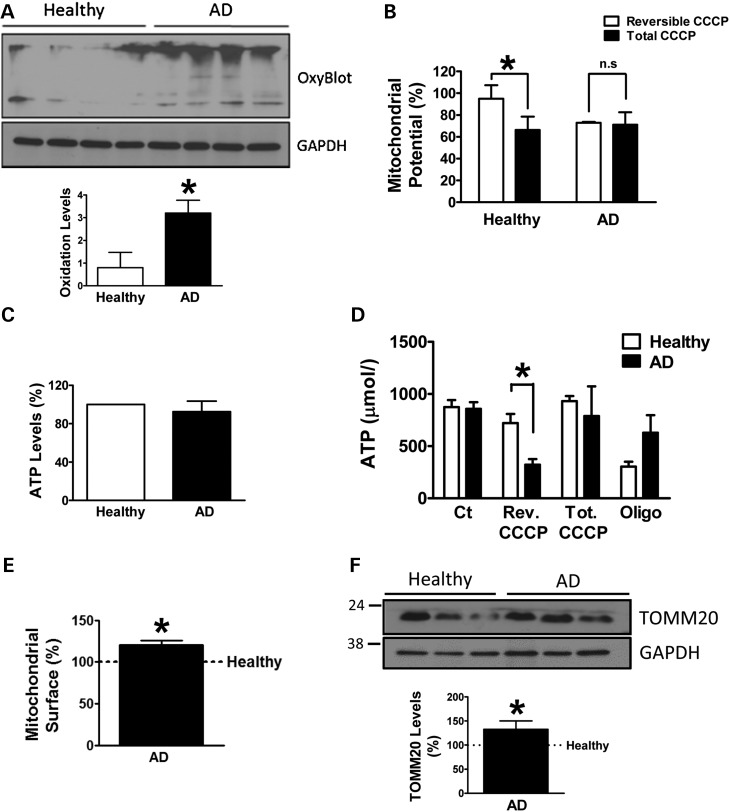
As previous works have demonstrated that some of the AD features mainly due to Aβ pathology can also be found in fibroblasts (33), we wanted to evaluate whether mitochondrial function may also be affected in peripheral tissue. With this aim, we have analysed oxidation of proteins in skin fibroblasts from SAD patients, compared with its correspondent age-matched healthy samples. OxyBlot analysis revealed increased amount of oxidized proteins in SAD cells under basal conditions (Fig. 1A). To determine whether this accumulation may reflect a deficiency in mitochondrial function, we studied ΔΨm after a reversible treatment with a respiratory chain uncoupling agent, carbonyl cyanide m-chlorophenylhydrazone (CCCP). The cells were challenged with CCCP for 6 h and then allowed to recover for 1 h. These conditions induce a reversible depolarization of mitochondria without causing any toxic effect (Supplementary Material, Fig. S1). The recovery of ΔΨm was higher in healthy cells compared with SAD ones, in which there was negligible recovery during the studied period (Fig. 1B). In parallel, we measured adenosine triphosphate (ATP) content of the cells as functional readout of mitochondrial status (Fig. 1C and D). There were no differences in ATP levels between healthy and SAD samples (Fig. 1C). Accordingly, after CCCP challenge for 7 h, similar ATP levels were observed (Fig. 1D, tot. CCCP), indicating that after a long-term CCCP exposure, the cells activate other catalytic mechanisms to recover ATP homeostasis when the mitochondrial function is compromised. Additionally, the recovery of ΔΨm after the reversible treatment is an ATP-demanding process as it can be inferred from the lower ATP levels in this condition in SAD cells (Fig. 1D, rev. CCCP). On the contrary, healthy cells exhibited ATP levels close to the untreated condition, indicating that mitochondrial function was recovered. As a control, we used oligomycin that inhibits mitochondrial H+-ATP-synthase causing the drop of ATP levels, which was more evident in healthy fibroblasts indicating higher dependence on mitochondrial function in these cells to maintain ATP homeostasis (Fig. 1D, Oligo.). In contrast, SAD cells exhibited higher levels of TOMM20, a constitutive mitochondrial protein, by immunofluorescence (see Supplementary Material, Fig. S2 and quantification in Fig. 1E) and western blot (Fig. 1F). The observed accumulation of mitochondria joined to the alterations in mitochondrial potential recovery strongly suggests a possible defect in mitophagy (34–36).
Figure 1.
Mitochondrial anomalies in SAD fibroblasts. (A) Representative OxyBlot showing oxidized proteins of healthy and SAD fibroblasts under basal conditions and quantification of the whole set of data by using the following healthy/AD sample couples: AG11362/AG05809, AG07803/AG06262, AG05813/AG06263 and AG07310/AG06869. (B) Mitochondrial membrane potential of fibroblasts reversibly treated with CCCP (20 μM) for 6 h and then allowed to recover for 1 h (reversible CCCP) or treated for 7 h with CCCP (total CCCP) using the following cells: AG07310/AG06869, AG11020/AG05810 and AG11362/AG05809. (C) Intracellular ATP measurement of healthy and SAD fibroblasts under basal conditions using the following cells: AG11362/AG05809, AG11020/AG05810, AG05813/AG06263 and AG07310/AG06869. (D) ATP measurement of fibroblasts reversibly treated as in (B) besides the treatment with oligomycin (10 μM) for 7 h. The graph shows representative experiment using AG07310/AG06869 samples performed in duplicate, although similar results were obtained using following cells: AG11362/AG05809 and AG07310/AG06869 (data not shown). (E) Quantification of the mitochondrial surface per cell by measuring the TOMM20 label when the cell limit was determined by phalloidin label in the confocal images as shown in Supplementary Material, Figure S1 using the following cells AG11362/AG05809, AG07803/AG06262 and AG05813/AG06263. (F) Representative western blot and quantification of TOMM20 expression under basal conditions using the cells AG11362/AG05809, AG07310/AG06869 and AG05813/AG06263. All graphs show means and standard deviations of the indicated healthy/AD age-matched couple samples. *P < 0.05.
Autophagy alterations in SAD fibroblasts
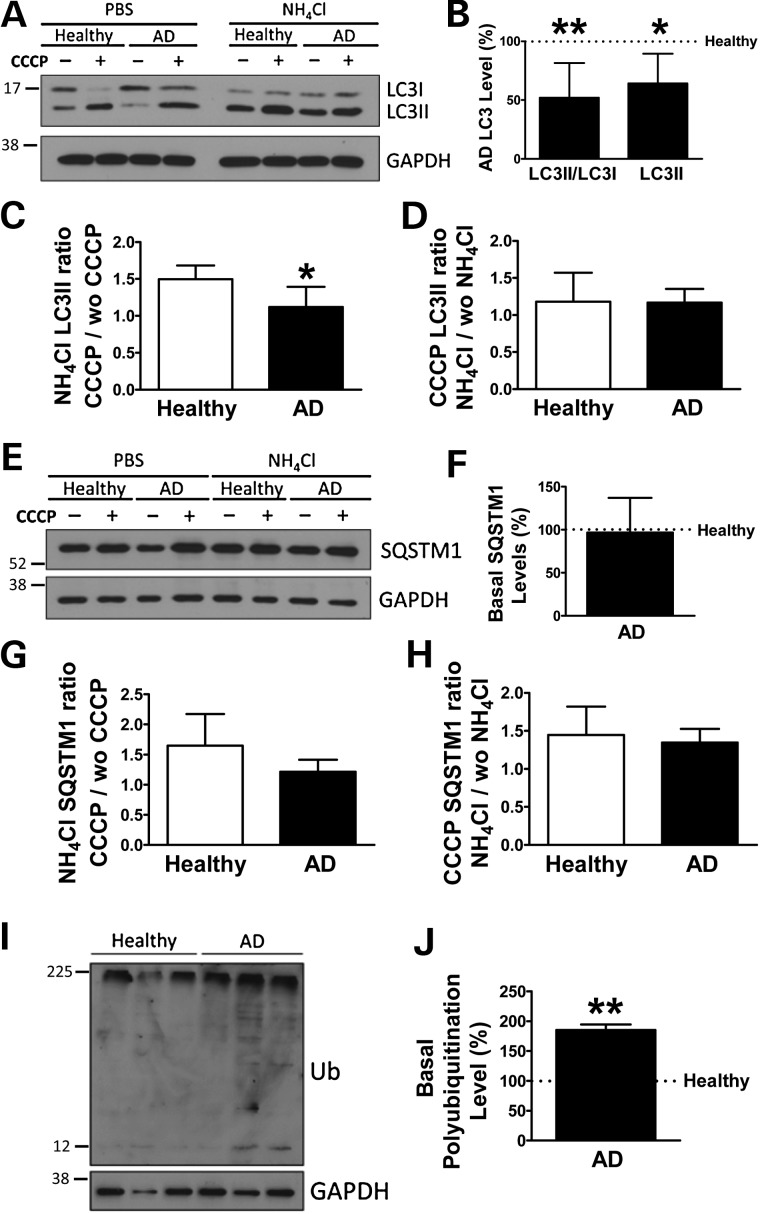
To determine autophagy flux, fibroblasts were treated with CCCP followed by an additional treatment of NH4Cl in the presence of CCCP to block the degradation phase of autophagy. Surprisingly, western blot analysis revealed a significantly diminished amount of AVs in SAD fibroblasts, exhibiting a decrease in LC3II protein levels as well as lower levels of LC3II/LC3I ratio under basal conditions (Fig. 2A and B). Although all samples showed induction of AVs after CCCP treatment, a reduction of autophagosome synthesis in SAD cells was observed, correlating with significantly smaller LC3II accumulation compared with healthy samples after CCCP treatment when vacuole degradation was blocked by NH4Cl (Fig. 2C). In contrast, we do not observe significant differences in LC3II degradation between SAD and healthy fibroblasts (Fig. 2D). These data suggest a decreased autophagy flux mainly due to a deficient AV formation. Additionally, similar levels of SQSTM1 adaptor protein were observed in both SAD and healthy fibroblasts under basal conditions (Fig. 2E and F). Accordingly, accumulation of SQSTM1 was diminished in SAD samples, although differences were not significant (Fig. 2G). Finally, the degradation of SQSTM1 was comparable in both samples, confirming no differences in the degradation phase (Fig. 2H). Conversely, the levels of BECN1 were not altered in SAD fibroblasts when compared with healthy ones under basal conditions (Supplementary Material, Fig. S3A and B), as well as its induction and degradation (Supplementary Material, Fig. S3C and D, respectively) occurring during autophagy. Consistent with these autophagy alterations, we also detected a significant accumulation of ubiquitinated proteins in SAD cells (Fig. 2I and quantification of data in Fig. 2J).
Figure 2.
Autophagy alterations in SAD fibroblasts. (A) Representative western blot of LC3 expression of the cells treated with CCCP (20 μM) followed by an additional treatment of PBS or NH4Cl (15 mM). (B) Quantification of LC3II/LC3I ratio and LC3II levels in the SAD samples with respect to the healthy ones under basal conditions. (C) Quantification of LC3II synthesis ratio represented as the values of the cells treated with CCCP and NH4Cl with respect to the condition without CCCP but maintaining NH4Cl treatment. (D) Quantification of LC3II degradation ratio obtained by the relation between the values of the cells treated with CCCP and NH4Cl and the ones without NH4Cl but maintaining CCCP treatment. (E) Representative western blot of SQSTM1 expression of the cells described in (A). (F) Quantification of the basal levels of SQSTM1 expression. (G) Quantification of SQSTM1 synthesis ratio as described for (C). (H) Quantification of SQSTM1 degradation ratio as defined in (D). (I) Representative western blot and (J) quantification of the basal levels of ubiquitinated proteins. All graphs show means and standard deviations of the five different healthy/AD age-matched couple samples (A–H) or the following three couples AG11362/AG05809, AG07310/AG06869 and AG11020/AG05810 (I). *P < 0.05 and **P < 0.01.
Lysosomal alterations in SAD fibroblasts
Lysosomal function is fundamental for the degradative phase of autophagy; therefore, we wanted to evaluate lysosomal properties in SAD cases. The pH of lysosomes was analysed by using LysoTracker assay with and without bafilomycin, a specific inhibitor of vacuolar proton ATPase (37), which prevents the acidification of lysosomes (38). Under basal conditions, SAD fibroblasts exhibited a lower capacity to retain LysoTracker probe, suggesting an increase in pH (Fig. 3A). However, when we compared the LysoTracker label under basal conditions with respect to bafilomycin, SAD fibroblasts showed a higher ratio (Fig. 3B). These apparently contradictory results suggest that SAD fibroblasts had a small number of lysosomes, but the pH of each lysosome is more acidic, giving higher ratio between basal and bafilomycin condition. We confirmed this hypothesis by studying the level of lysosomal-associated membrane protein 1 (LAMP1), which was markedly lower in SAD cells, consistent with a diminished amount of lysosomes (Fig. 3C). In contrast, the cysteine protease cathepsin B (CTSB) levels were decreased in SAD cells (Fig. 3D). Conversely, CTSB activity was significantly increased in SAD fibroblasts compared with controls, especially at short time points (Fig. 3E and Supplementary Material, Fig. S4). A lower amount of CTSB protein but increased activity confirmed our hypothesis of a lower amount of lysosomes, but having each one lower pH. Despite these alterations, lysosomal function does not appear to be affected because, as previously stated, there were no differences in the degradation phase of autophagy (Fig. 2D and H, and Supplementary Material, Fig. S3D). Moreover, when lysosomal function was blocked, we could observe the accumulation of p62/SQSTM1 which is normally degraded by autophagy (39) in both healthy and SAD samples, leading us to conclude that lysosomal was active in both cases (Fig. 3F). SQSTM1 accumulation was higher in healthy cells (Fig. 3F) probably due to a major autophagy induction in these samples (Fig. 2C and G). Accordingly, autophagolysosomes formation that depends on lysosomal function was similar in healthy and SAD cells (Fig. 3G and H).
Figure 3.
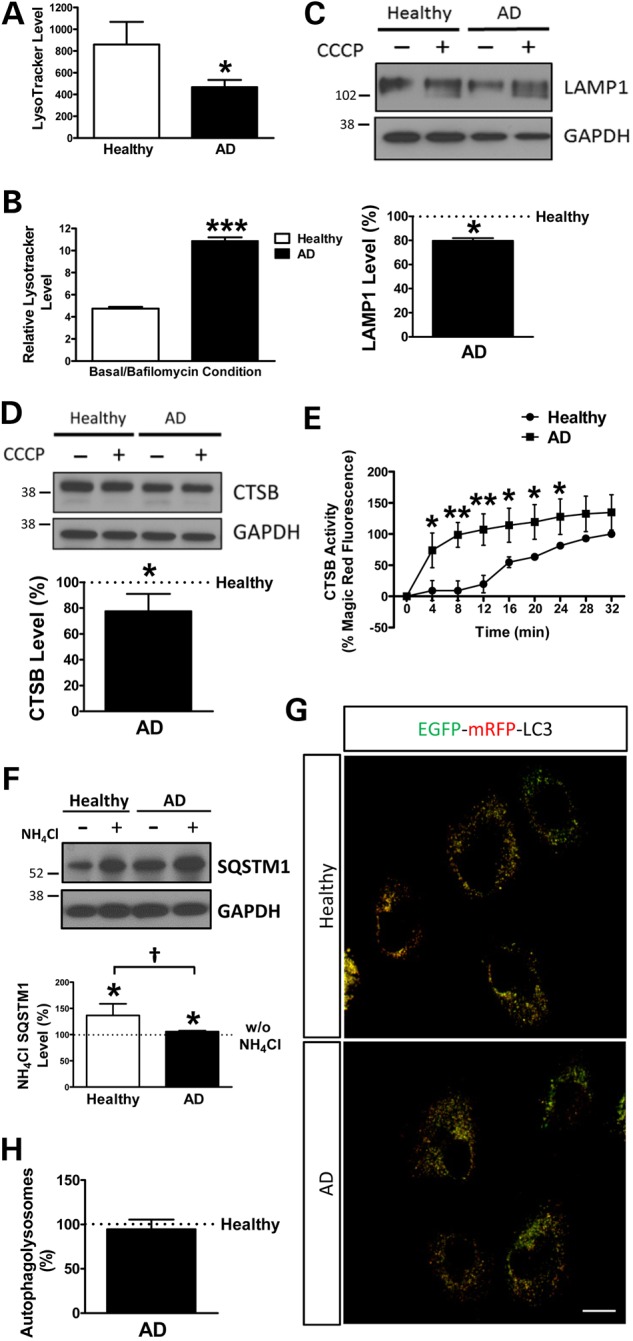
Lysosomal alterations in SAD fibroblasts. (A) Quantification of the amount of lysosomes per cell. (B) Quantification of the lysosomal acidity represented by the ratio between untreated and bafilomycin (100 nM)-treated cells. The following healthy/AD age-matched samples were used: AG1136/AG05809, AG07803/AG06262 and AG05813/AG06263. Every measurement was performed in triplicate, and the graphs represent the means and standard deviations of the average of every cell line. (C) Representative western blot of LAMP1 protein in the absence or presence of CCCP (20 μM) and quantification of the levels under basal conditions. The following couples were used: AG11362/AG05809, AG05813/AG06263 and AG07310/AG06869. (D) Representative western blot analysis of the protein levels of Cathepsin B/CTSB and quantification of the protein levels of CTSB under basal conditions. The following four couples were used in this assay: AG11362/AG05809, AG07803/AG06262, AG05813/AG06263 and AG07310/AG06869. (E) CTSB activity along time (representative images are shown in Supplementary Material, Fig. S3). The activity was normalized by the maximum fluorescence detected in the last time point of the healthy sample. The couples used for this experiment were: AG11362/AG05809, AG05813/AG06263 and AG07310/AG06869. Every measurement was performed in triplicate, and the average obtained for each cell line was used for the graph. (F) Representative western blot analysis of SQSTM1 and quantification of the levels in the presence of NH4Cl with respect to the absence. The following couples were used: AG11362/AG05809, AG07310/AG06869 and AG07803/AG06262. (G) Representative confocal microscopy images of healthy and AD fibroblasts expressing mRFP-EGFP-LC3. (H) Quantification of autophagolysosomes per cell corresponding to the percentage of only red structures. The couples used for this experiment were: AG11362/AG05809, AG05813/AG06263 and AG07310/AG06869. All graphs show means and standard deviations of the mentioned healthy/AD age-matched couple samples. †P < 0.08, *P < 0.05, **P < 0.01 and ***P < 0.001. Scale bar: 40 μm.
Mitophagy failure in SAD fibroblasts
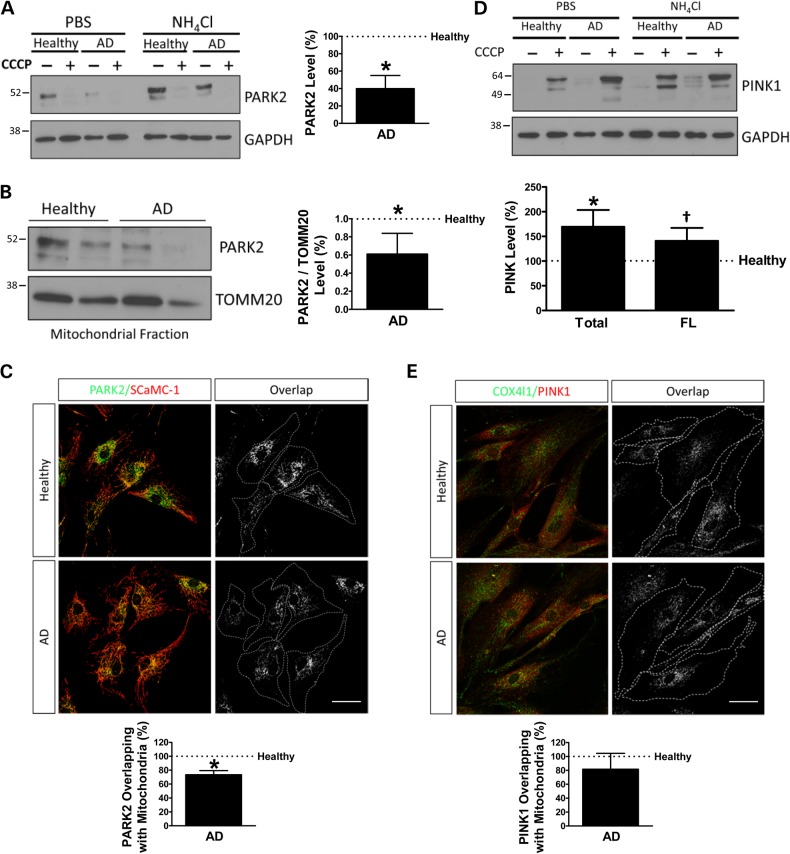
The levels of PARK2 were substantially diminished in SAD cells (Fig. 4A) as well as in a mitochondria-enriched fraction (Fig. 4B). Degradation of PARK2 under resting conditions was mediated by autophagy and proteasome in healthy cells, but, after mitophagy induction by CCCP, degradation was mainly by autophagy as it would be expected (Supplementary Material, Fig S5). In SAD samples, there was a significant accumulation of PARK2 after 3 h of CCCP challenge, which correlates with the previously described failure in autophagy. Accordingly, in these cells, there was no significant degradation of PARK2 mediated by lysosome. The recruitment of PARK2 to the mitochondria after CCCP treatment was significantly decreased in SAD fibroblasts, suggesting a defective labelling of damaged mitochondria to be recycled by mitophagy (Fig. 4C and Supplementary Material, Fig. S6). Additionally, both full-length and all isoforms of PINK1 were significantly increased in SAD cells after a mitochondrial insult (Fig. 4D). However, immunofluorescence analysis of the stabilization of PINK1 in the mitochondria after CCCP challenge showed that there were no significant differences between SAD and healthy fibroblasts (Fig. 4E). Moreover, there was similar colocalization of PARK2 and PINK1 (Supplementary Material, Fig. S7), suggesting that PINK1 was able to recruit PARK2 in these conditions.
Figure 4.
Imbalance of PARK2 and PINK1 pattern in SAD fibroblasts. (A) Representative western blot of PARK2 expression in cells treated with 20 μM CCCP followed by an additional treatment of PBS or 15 mM NH4Cl. Quantification of PARK2 expression under basal conditions using the following couples: AG11362/AG05809, AG07310/AG06869 and AG11020/AG05810. (B) Representative western blot of the mitochondrial fraction of the cells treated with CCCP as indicated in (A) corrected by the TOMM20 expression as a mitochondrial housekeeping gene. Quantification of three healthy/AD age-matched samples using the following couples: AG11362/AG05809, AG07803/AG06262 and AG05813/AG06263. (C) Representative confocal microscopy immunofluorescence images showing PARK2 in green and SCaMC-1 as a mitochondria constitutive marker in red of fibroblasts treated with 20 μM CCCP for 1 h. On the right, binary image representing the colocalization of both labels and dotted line delimits cytoplasm of each cell. Quantification of the colocalization between PARK2 and SCaMC-1 expressed as area occupied by the overlapping elements per cell. The following healthy/AD samples were used for this assay: AG11362/AG05809, AG07310/AG06869 and AG05813/AG06263. (D) Representative western blot of the expression of PINK1 in fibroblasts treated as in (A). Quantification of the levels of all isoforms of PINK1 (total) or only full-length isoform (FL) in SAD fibroblasts in the presence of CCCP using the following couples: AG11362/AG05809, AG07803/AG06262, AG05813/AG06263 and AG07310/AG06869. (E) Representative confocal microscopy immunofluorescence images as in (C) showing PINK1 in red and COX4I1 as a mitochondrial constitutive marker. Quantification of the colocalization between PINK1 and COX4I1 using the following couples: AG11362/AG05809, AG07803/AG06262 and AG05813/AG06263. All graphs show means and standard deviations of the mentioned healthy/AD age-matched couple samples. †P < 0.08 and *P < 0.05. Scale bar: 40 μm.
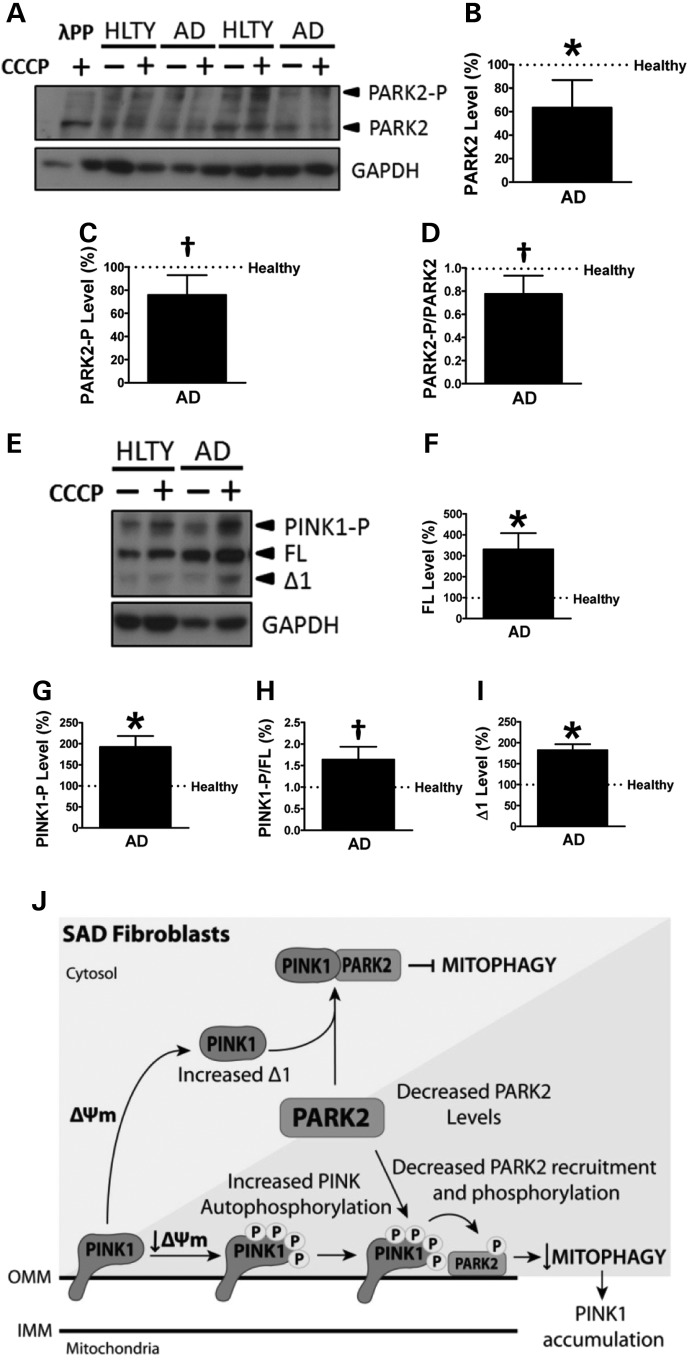
Accordingly with our previous results, the analysis of protein phosphorylation by Phos-tag assays revealed that non-phosphorylated PARK2 was significantly decreased in SAD cells (Fig. 5A and B). Additionally, although there were no differences in PARK2 phosphorylation under basal conditions (data not shown), after CCCP treatment, SAD fibroblasts exhibited a decreased PARK2 phosphorylation as well as a diminished phosphorylation ratio (Fig. 5C and D, respectively). In contrast, after CCCP challenge, non-phosphorylated full-length PINK1 isoform appeared to be significantly increased in SAD fibroblasts as we observed by conventional western blot (Fig. 5E and F). Moreover, in these conditions, we detected an increased PINK1 phosphorylation and phosphorylation ratio in SAD cells (Fig. 5G and H, respectively). Together, these results strongly suggest that there is a disruption of the crosstalk between PINK1 and PARK2. Remarkably, SAD cells exhibited significantly higher levels of Δ1 PINK1 (Fig. 5I), the main cleaved product of PINK1, which is known to bind and inhibit PARK2 in the cytosol (23), suggesting an additional inhibitory effect over mitophagy (see model in Fig. 5J).
Figure 5.
Mitophagy failure in SAD. (A) Representative Phos-tag western blot showing PARK2 and GAPDH pattern of fibroblasts treated or not with 20 μM CCCP for 1 h. Phosphorylated bands of PARK2 were confirmed by their disappearance after lambda protein phosphatase (λPP) treatment. (B) Quantification of non-phosphorylated PARK2 levels in the SAD samples with respect to the healthy (HLTY) ones under basal conditions. (C) Quantification of PARK2 phosphorylation in cells treated with CCCP for 1 h. (D) Quantification of PARK2 phosphorylation ratio in the presence of CCCP represented as the relation of phospho-PARK2 with respect to non-phosphorylated PARK2 protein levels. (E) Representative Phos-tag western blot of PINK1 of fibroblasts treated as described in (A). (F) Quantification of non-phosphorylated full-length PINK1 levels in fibroblasts treated with CCCP. (G) Similar quantification of PINK1 phosphorylation levels in cells treated with CCCP. (H) Quantification of PINK1 phosphorylation ratio of cells treated with CCCP represented as the relation between phospho-PINK1 and non-phosphorylated PINK1 protein levels. (I) Quantification of Δ1 PINK1 fragment under basal conditions. All graphs represent values corrected by GAPDH signal and normalized by each age-matched healthy sample using the following couples AG11362/AG05809, AG07310/AG06869 and AG01020/AG05810. (J) Schematic model representing the mitophagy failure in SAD. PARK2 can follow two different pathways in a mitochondrial membrane potential-dependent manner. †P < 0.08 and *P < 0.05.
PARK2 overexpression compensates autophagy deficiency and restores mitophagy
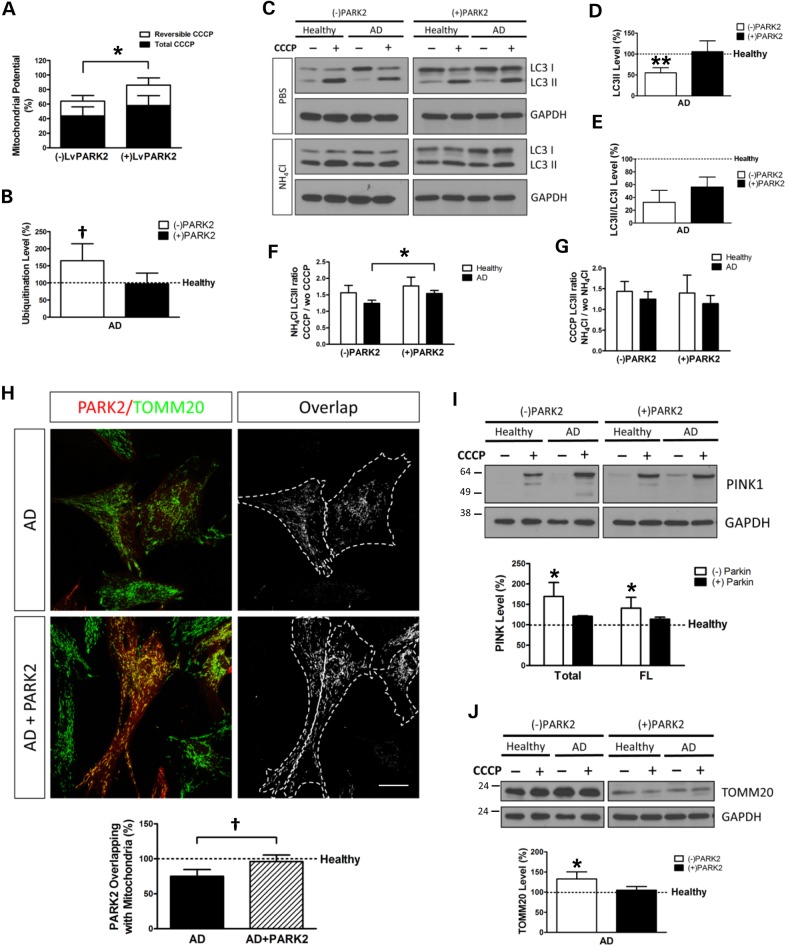
In order to investigate whether increasing the levels of PARK2 would be able to modulate the observed autophagy defect in SAD cells, we induced the overexpression of PARK2 by a lentivector encoding WT PARK2 in healthy and SAD fibroblasts. PARK2 enhancement allowed a significant mitochondrial function improvement indicated by the ΔΨm recovery in SAD cells after a reversible CCCP challenge (as described for Fig. 1B), which was negligible when they were uninfected (Fig. 6A). Degradation of polyubiquitinated proteins was mainly mediated by proteasome and, only in healthy cells, there was a slight contribution of autophagy (Supplementary Material, Fig. S8A–C). Proteasome-mediated degradation flux is higher in SAD samples, probably due to compensatory mechanism to counteract autophagy impairment in these samples (Supplementary Material, Fig. S8C). Surprisingly, although PARK2 is a ubiquitin ligase, its overexpression in SAD fibroblasts, far from increasing ubiquitination levels, decreased them to the levels found in healthy samples (Fig. 6B). This can be explained because PARK2 overexpression favoured ubiquitinated proteins degradation by autophagy and proteasome, especially in SAD samples (Supplementary Material, Fig. S8D). Furthermore, PARK2 overexpression recovered the basal levels of AVs in SAD samples which was shown by the increase of LC3II protein levels up to the ones observed in healthy samples (Fig. 6C and D). However, the induction of AVs represented by the LC3II/LC3I ratio was not significantly increased (Fig. 6E). Noteworthy, we also could observe a significant improvement of AV synthesis (Fig. 6F). This also correlates with the increased degradation of polyubiquitinated proteins via autophagy observed (Supplementary Material, Fig. S8D, AD samples). On the contrary, the autophagosome degradation after PARK2 overexpression maintains levels similar to the ones found in the absence of PARK2 in both SAD and healthy fibroblasts (Fig. 6G), highlighting that the improvement of autophagy flux is due to an increase in AV generation.
Figure 6.
PARK2 overexpression compensates mitophagy deficiency. (A) Mitochondrial membrane potential of SAD fibroblasts treated with CCCP with 20 μM for 6 h and allowed them to recover for 1 h (reversible) or treated for 7 h with CCCP (total) when the cells were infected or not with a lentivector encoding WT PARK2. The following healthy/AD couples were used: AG11362/AG05809, AG07310/AG06869 and AG05813/AG06263. Every measurement was performed in triplicate for each cell line, and the average was used to calculate the percentages with respect to the untreated samples. (B) Protein ubiquitination levels of SAD samples when they are infected or not with a lentivector encoding PARK2. Results are normalized with respect to the levels found in the correspondent age-matched healthy fibroblasts using the following couples: AG11362/AG05809, AG07803/AG06262 and AG05813/AG06263. (C) Representative western blot of LC3 expression in fibroblasts infected with a lentivector encoding WT PARK2. Then, cells were treated with CCCP (20 μM) followed by an additional treatment of PBS or NH4Cl (15 mM). (D–E) Quantification of the western blot showing the levels of LC3II (D) or LC3II/LC3I ratio (E) of SAD fibroblasts infected or not with a lentivector encoding PARK2 under basal conditions. (F) Quantification of LC3II synthesis ratio represented as the relation between the cells treated with CCCP and NH4Cl and the cells without CCCP but maintaining NH4Cl treatment. (G) Quantification of LC3II degradation ratio expressed as the relation between the cells treated with CCCP and NH4Cl and the cells without NH4Cl but maintaining CCCP treatment. (D–G) The following healthy/AD samples were used: AG11362/AG05809, AG05813/AG06263 and AG07310/AG06869. (H) Representative confocal microscopy immunofluorescence images showing PARK2 in red and TOMM20 as a mitochondria constitutive marker in green of fibroblasts infected with a lentivector encoding PARK2 treated with CCCP (20 μM) for 1 h. On the right, binary images representing the colocalization of both labels and dotted line delimit the cytoplasm of each cell. Quantification of the colocalization between PARK2 and TOMM20 expressed as area occupied by the overlapping elements per cell of the fibroblasts infected or not with a lentivirus encoding PARK2 of the following healthy/AD couples: AG11362/AG05809, AG07310/AG06869 and AG05813/AG06263. (I) Representative western blot of PINK1 expression in fibroblasts treated with CCCP for 24 h, when indicated, and when these cells are infected or not with a lentivirus encoding PARK2. Quantification of the levels of total and full-length (FL) PINK1 in the cells treated with CCCP with respect to the correspondent healthy sample treated with CCCP using the following couples: AG11362/AG05809, AG07803/AG06262, AG05813/AG06263 and AG07310/AG06869. (J) Representative western blot of TOMM20 levels in fibroblasts treated as described in (I). Quantification of the levels of TOMM20 of SAD fibroblasts infected or not with a lentivector encoding PARK2 with respect to the healthy fibroblasts under basal conditions using the couples AG11362/AG05809, AG07310/AG06869 and AG05813/AG06263. All graphs show means and standard deviations of the mentioned healthy/AD age-matched couple samples. †P < 0.08, *P < 0.05 and **P < 0.01. Scale bar: 40 μm.
PARK2 transduction improved the targeting of PARK2 to the mitochondria after CCCP treatment in SAD fibroblasts (Fig. 6H and Supplementary Material, Fig. S9). Additionally, the accumulation of either total or full-length isoforms of PINK1 in SAD samples was reduced after PARK2 overexpression to levels closer to the ones observed in healthy cells (Fig. 6I). Finally, we could observe that the accumulation of TOMM20 label in SAD fibroblasts under basal conditions was restored by PARK2 overexpression up to healthy levels (Fig. 6J). Moreover, in normal conditions or after CCCP treatment, there was a clear recycling of mitochondria by lysosomal degradation in healthy samples that was not observed AD samples where there was an accumulation of mitochondria (Supplementary Material, Fig. S10). But PARK2 overexpression dramatically improved mitochondrial recycling bypassing the impairment due to NH4Cl treatment. Overall, these data indicate that the impairment of mitophagy in SAD cells can be restored by PARK2 overexpression.
Mitophagy alterations in SAD brain
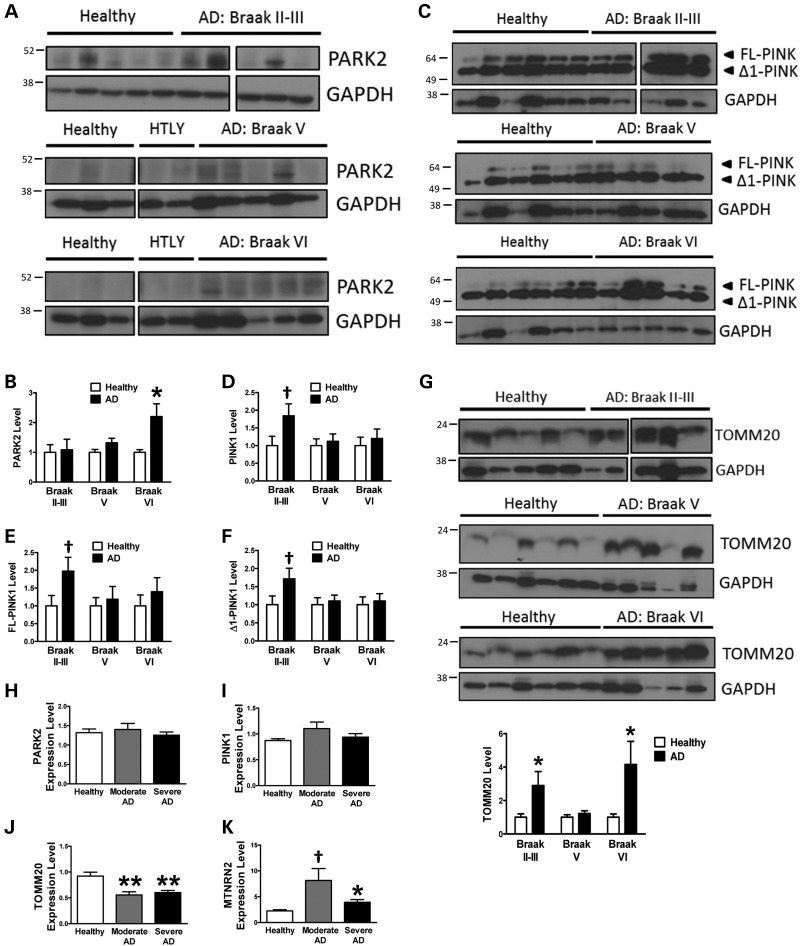
It is well known that one of the brain structures mainly affected in SAD patients is the hippocampus (40), so we focused our study in this region. In contrast to what we observed in fibroblasts (Fig. 4A and B), we could not detect significant differences in PARK2 levels in the hippocampus at early stages (Braak II and III), but at later stages (Braak V and VI) of the disease there was an increase of PARK2, being significant at Braak VI (Fig. 7A and B). On the contrary, the study of PINK1 showed an accumulation of this protein at the first stages of the disease (Braak II and III) (Fig. 7C–E). Accordingly to what we have described for fibroblasts (Fig. 5E and I), we could also find an increase of Δ1 PINK1 fragment at early stages of the disease (Braak II and III) (Fig. 7F). These data strongly indicate that a similar inhibitory mechanism over PARK2 translocation may be taking place in the brain. Moreover, the study of TOMM20 showed an increase in mitochondrial content (Fig. 7G), according to what we demonstrated for fibroblasts under basal conditions (Fig. 1E and F). The analysis of PARK2 expression levels by reverse transcription quantitative real-time PCR (RT–qPCR) showed similar RNA levels in AD and healthy brain (Fig. 7H). However, when a larger set of brain samples was used, such in the Hisayama study (41), a reduction in PARK2 RNA levels could be observed (Supplementary Material, Fig. S11), correlating with decreased PARK2 protein levels exhibited by our fibroblasts. The absence of differences in PINK1 RNA levels observed in our samples (Fig. 7I) or the reduction observed in the Hisayama study (Supplementary Material, Fig. S11) indicates that the protein accumulation of PINK1 was due to an impaired degradation rather than an excess of synthesis. Finally, although TOMM20 RNA levels were significantly diminished in our brain samples (Fig. 7J) and in the Hisayama study (Supplementary Material, Fig. S11), the levels of 16S/MT-NRN2 RNA (Fig. 7K), a mitochondrial DNA-encoded constitutive protein which should be proportional to the amount of mitochondria, were markedly increased. This clearly indicates higher mitochondrial content due to an accumulation rather than an increased synthesis. All these evidences together suggest that in brain samples, there is a strong mitophagy alteration at early stages of the disease that may cause the accumulation of dysfunctional mitochondria, similar to what we have described for fibroblasts.
Figure 7.
Mitophagy alterations in SAD brain. (A) Representative western blot of PARK2 in human hippocampal samples divided accordingly to Braak stage when compared with the same group of healthy donors. (B) Quantification of the data shown in (A). (C) Study of PINK1 as described for (A) and quantification of the levels of PINK1 (D). (E) Quantification of FL-PINK1 levels. (F) Quantification of Δ1 PINK1 levels. (G) Representative western blot and quantification of the levels of TOMM20 protein in human hippocampal samples. (H–K) Expression levels of PARK2 (H), PINK1 (I), TOMM20 (J) and MT-NRN2 (K) measured by RT–qPCR in brain samples. All graphs show means and standard deviations of the indicated healthy/AD patients′ samples. †P < 0.08, *P < 0.05 and **P < 0.01.
Discussion
The most commonly used models of AD involve mutations in APP, PSEN1 or PSEN2 genes found in familial cases, despite the fact that the majority of the cases of AD are sporadic and not related to these mutations. Therefore, we have focused our study in sporadic cases of the disease using unmodified peripheral and brain tissues. Due to the vast variability of SAD, it is necessary to generate cell models that must be easy to obtain and handle. Fibroblasts offer us an accessible cell source that can be obtained by a small skin biopsy from living patients. However, it is imperative to demonstrate that fibroblasts retain certain hallmarks of the disease, and, being the skin—a tissue that is not affected in AD pathology—this is not a trivial issue. Several works have previously demonstrated that fibroblasts exhibit oxidized proteins accumulation and mitochondrial anomalies that can also be found in AD brain (42). Moreover, accumulation of Aβ has been also demonstrated in these cells (33).
In this work, the mitochondrial respiratory chain uncoupling agent CCCP has been used to induce mitochondrial depolarization in order to potentiate the possible differences between SAD and healthy fibroblasts. We have demonstrated that mitochondrial function was compromised due to the lack of membrane potential and ATP homeostasis recovery after this insult, suggesting an inefficient recycling of damaged mitochondria by autophagy. Accordingly, we have found lower levels of basal autophagy represented in a diminished amount of autophagic vesicles (LC3), correlating with the accumulation of oxidized and ubiquitinated proteins. Additionally, after the treatment with CCCP, there was a deficiency in autophagy induction shown in lower levels of autophagic vesicle formation. The absence of significant differences in BECN1 levels in SAD samples may be explained because autophagy can also occur independently of BECN1 (43,44). Lysosomal anomalies have also been found in our SAD samples. We assume a reduction in the amount of lysosomes reflected in lower LysoTracker label under basal conditions as well as diminished levels of lysosomal proteins such as LAMP1 and CTSB. However, we hypothesize that these lysosomes have a more acidic pH as confirmed by the increased ratio in LysoTracker label between the basal conditions and the bafilomycin-treated ones, which correlates with enhanced CTSB activity. In fact, it was previously described that lysosomal CTSB exopeptidase activity is increased as the pH decreases (45). Despite these anomalies, the lack of significant differences in the degradative phase of autophagy together with the active degradation of proteins mediated by autophagy such as SQSTM1 as well as similar rates of fusion of autophagosomes with lysosomes to form autophagolysosomes indicates that these lysosomes may be functional and suggests that the altered properties may be a consequence of autophagy failure. Lysosomal alterations have been previously described for AD fibroblasts associated with PSEN mutations; however, in this case, lysosomal function was compromised by a deficit in lysosomal acidification, causing a defect in the degradative phase of autophagy (28). These divergent phenotypes observed in SAD and PSEN1-associated FAD may suggest different mechanisms for a common final failure in the autophagy process.
The study of mitophagy after a mitochondrial insult indicates that this process is severely affected in SAD fibroblasts. We hypothesize that under basal conditions the increased levels of Δ1 fragment of PINK1 sequester cytosolic PARK2 (23), which is in turn diminished in SAD samples, inhibiting its activation. This defect in mitochondrial recycling may explain the accumulation of oxidized proteins observed. Additionally, upon mitochondrial depolarization, although there is a correct stabilization and autophosphorylation of PINK1, there is not a proper PARK2 activation. This may be due to the lower levels of PARK2 and its inhibition mediated by Δ1 PINK1. This causes the accumulation of damaged mitochondria, reflected in the increase of full-length and phosphorylated PINK1 and diminished phosphorylated PARK2, emphasizing the deficiency in the crosstalk between PINK1 and PARK2 (see model in Fig. 5J). The situation observed in these cells may be similar to what can be found in the hippocampus at early stages of the disease where we observed accumulation of both full-length and Δ1 PINK1 accompanied by abnormally increased mitochondrial content due to an impaired degradation rather than an increased synthesis. This strongly suggests that mitophagy failure may play an important role in the onset of the disease. At latest stages of the disease, the increase of PARK2 may correlate with recently published results, in which accumulation of insoluble PARK2 with intraneuronal Aβ and phospho-MAPT was detected in autophagosomes in AD brains, reflecting an autophagy failure (46). Moreover, the upregulation of PARK2 in these patients′ brain has been suggested as a defence mechanism to counteract stress-induced damage in AD pathogenesis (47).
Considering the fact mentioned earlier, we wanted to evaluate whether PARK2 enhancement would be able to reverse the observed mitophagy deficiency in fibroblast. The overexpression of wild-type PARK2 was able to improve autophagy shown in the recovery of autophagic vesicle level (LC3II) under basal conditions up to the levels found in healthy samples. This improvement was reflected in the elimination of ubiquitinated proteins and the decrease of abnormal accumulated mitochondria (TOMM20) under basal conditions or after CCCP treatment. Additionally, PARK2 enhancement would exceed the inhibitory effect of Δ1 PINK1 and, upon CCCP treatment, PARK2 would be recruited by full-length PINK1 to damaged mitochondria, leading them to be degraded by mitophagy. Consequently, the levels of PINK1 were diminished and the ΔΨm was recovered by increasing of autophagy vesicle synthesis. We may thus assume that PARK2 overexpression restored autophagy and was able to compensate mitochondrial anomalies found in SAD fibroblasts. The previous work has demonstrated that the overexpression of PARK2 mediated the clearance of Aβ and phospho-MAPT by enhancing autophagy, and it was able to increase tricarboxylic acid cycle activity, suggesting an improvement of mitochondrial function as well as a restored synaptic function in a triple transgenic AD mice model (46,48). Taking all these evidences together, we may conclude that promoting autophagy by PARK2 overexpression restores mitochondrial recycling and function, induces Aβ and phospho-MAPT clearance and improves synaptic function. The most important point is that the therapeutic capacity of PARK2 enhancement that we have demonstrated in sporadic cases highlights the similarities not only with FAD but also with other neuropathies such as Parkinson's disease, where the role of PARK2 has been widely demonstrated (14).
Further studies comparing fibroblast samples from different stages of the disease and brain tissue from the same patient will be very interesting in order to investigate whether mitophagy failure in peripheral tissue may be used as an indicator of the disease progression. In particular, the accumulation of PINK1 and Δ1 PINK1 in peripheral tissue may serve as an early marker of the disease. Therefore, non-invasive methods for the detection of this protein would be a powerful tool to detect these patients presymptomatically.
Mitophagy alterations have been defined as a new hallmark of SAD. The mitochondrial and autophagy defects found in fibroblasts resemble the pathology observed in patients’ brain at early Braak stages, demonstrating the feasibility of this model for the study of the disease. Additionally, the accumulation of PINK1 both in brain and in peripheral tissue demonstrates its suitability to be used as a biomarker of this pathology. Finally, the therapeutic potential of improving mitophagy by the enhancement of PARK2 signalling was established.
Materials and Methods
Primary cells and culture conditions
Primary skin fibroblasts were obtained from Coriell Institute for Medical Research (NJ, USA). Five fibroblast cell lines from SAD patients and five correspondent apparently healthy sex- and age-matched samples have been used (see Table 1 for details about age, sex and stage of the disease). Human fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS), 2 mm glutamine, 10 U/ml penicillin, 10 μg/ml streptomycin, in 5% CO2 in a humid incubator at 37°C. The use of fibroblasts has been restricted to a maximum of 10 cell passages to avoid replicative senescence, and cultures were always kept below confluence.
Table 1.
Core set of fibroblast lines
| Line | Age/sex | Clinical diagnosis |
|---|---|---|
| AG11362 | 63/F | Non-affected |
| AG05813 | 67/F | Non-affected |
| AG07803 | 66/M | Non-affected |
| AG07310 | 60/F | Non-affected |
| AG11020 | 79/F | Non-affected |
| AG05809 | 63/F | Moderate dementia |
| AG06263 | 67/F | Moderate dementia |
| AG06262 | 66/M | Moderate dementia |
| AG06869 | 60/F | Moderate dementia |
| AG05810 | 79/F | Severe dementia |
Coriell Institute for Medical Research.
Translocase of outer mitochondrial membrane 20 homologue (TOMM20).
Antibodies
The primary antibodies used were: TOMM20 (sc-11415, Santa Cruz Biotechnology, Santa Cruz, CA, USA), COX4I1 (complex IV) (459600, Invitrogen, Eugene, OR, USA), LC3 (B7931, Sigma, St. Louis, MO, USA, for WB), LC3 (M152-3, MBL, Woburn, MA, USA, for immunostaining), SQSTM1 (610832, BD Biosciences, San Jose, CA, USA), BECN1 (sc-11427, Santa Cruz), Ubiquitin (Z0458, Dako, Glostrup, Denmark), LAMP1 (611042, BD Biosciences), CTSB (AB4064, Chemicon, Darmstadt, Germany), GAPDH (ab8245, Abcam, Cambridge, UK), PARK2 (sc-32282, Santa Cruz), PINK1 (BC100-494, Novus, Cambridge, UK) and SCaMC-1 [short calcium-binding mitochondrial carrier 1, kindly given by Dr Araceli del Arco (49)]. The secondary antibodies for western blot studies were horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgGs (P0448 and P0161, DAKO) and for immunofluorescence were anti-mouse or anti-rabbit IgGs alexa-488 or -555 labelled (Molecular Probes, Millipore, Waltham, MA).
MTT assay
Cells were grown in 96-well plates and treated as indicated. Metabolic status was determined by 200 μg/ml MTT (3-[4,5-dimethylthiazo-2-yl]-2,5-diphenyltetrazolium bromide) (Sigma-Aldrich, St. Louis, MO, USA), as described previously (50).
Western blot analysis
The cells and tissue samples were homogenized in lysis buffer [50 mM, pH 7.5 HCl–Tris, 300 mM NaCl, 0.5% sodium dodecyl sulphate (SDS) and 1% Triton X-100] and incubated for 15 min at 95°C. Protein concentration of the extracts was measured using the Dc protein assay kit (500-0111, Bio-Rad, Hercules, CA, USA). Equal amounts of total protein extract from healthy and SAD cells were resolved by SDS–polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes (G9917809, Amersham, Germany). Western blot and immunoreactive proteins were developed using an enhanced chemiluminescence detection kit (NEL105001EA, Perkin Elmer, Waltham, MA, USA), following instructions of the supplier. Quantification was performed by densitometry of the obtained bands in each lane with respect to the correspondent housekeeping protein in each experiment (Quantity One software, Bio-Rad). Data were normalized with respect to the values obtained from each correspondent age-matched healthy sample.
Lentivirus production
Pseudotyped lentivectors were produced using reagents and protocols from Didier Trono with the following modifications: 293T cells were transiently co-transfected with 5 μg of the corresponding lentivector plasmid, 5 μg of the packaging plasmid pCMVdR8.74 (Addgene plasmid 22036) and 2 μg of the VSV G envelope protein plasmid pMD2G (Addgene plasmid 12259) using Lipofectamine and Plus reagents, following instructions of the supplier (18324 and 11514, respectively, Life Techonologies, Carlsbad, CA, USA). The lentivector used encodes wild-type PARK2 and was a gift from Dr Andrew B. West (University of Alabama) (51).
Mitochondrial potential measurement
Cells were treated with 20 μM CCCP (C2759, Sigma) for 7 h and, in the reversible condition, CCCP was removed and the medium was replaced for DMEM 10% FBS during the last 60 min. Then, fibroblasts were incubated with 50 nM DiOC6(3) (D273, Molecular Probes, Waltham, MA, USA) for 30 min at 37°C and analysed by flow cytometry (FACSCalibur, BD Biosciences). Mitochondrial membrane potential for each condition was represented as the percentage of the fluorescence intensity with respect to the fluorescence intensity exhibited when these cells remained untreated.
Protein oxidation detection
OxyBlot Protein Oxidation Detection Kit (S7150, Millipore) was used to detect oxidized proteins in cell extracts using the supplier guidelines. Briefly stated, carbonyl groups of cell extract proteins were derivatized with 2,4-dinitrophenylhydrazine. Equal amounts of total protein extract (20 μg) from healthy and SAD were loaded, and western blot detection was performed as described previously. Quantification was performed by densitometry of the obtained bands in each lane (Quantity One software, Bio-Rad) with respect to the GAPDH signal, and every result was normalized with respect to the values obtained from each correspondent age-matched healthy sample.
ATP quantification
ATP was quantified using ATP determination kit (Molecular Probes, Eugene, OR, USA). After correspondent treatments, cell extracts were prepared in phosphate-buffered saline (PBS) 0.5% Triton X-100; following the manufacturer's instructions, ATP content was determined by luminescence using a FLUOstar OPTIMA Plate Reader (BMG Labtech, Ortenberg, Germany) at 28°C, using Costar 96-well black plates (Fisher Scientific, Pittsburgh, PA, USA). As a control, we used Oligomycin (O4876, Sigma).
Ubiquitination levels detection
A conventional western blot was performed as described earlier by using the primary antibody against ubiquitin motive (Z0458, Dako). Quantification was performed by densitometry of each lane (Quantity One software, Bio-Rad) with respect to the correspondent GADPH signal, and every result was normalized with respect to the correspondent age-matched healthy sample.
Immunocytochemistry
Fibroblasts were grown on sterile glass coverslips, treated as described for each experiment, followed by washing with PBS and fixing with 4% paraformaldehyde in PBS for 30 min at room temperature. After blocking with PBS containing 1% bovine serum albumin and permeabilizing with 0.1% Triton X-100 and glycine 1 M for 30 min, cells were washed with PBS and stained by indirect immunofluorescence using the antibodies described before. When indicated, phalloidin (A-12379, Molecular Probes, Life Technologies, Eugene, OR, USA) was added at 1:200 during the second antibody incubation. Samples were mounted with Prolong Gold Antifade (P-36930, Molecular Probes, Life Technologies), and randomly chosen field images were obtained in an Invert Confocal LSM510 (Zeiss, Oberkochen, Germany) fluorescence microscope.
Colocalization assay
Colocalization analysis was performed with ImageJ software (Bethesda, MD, USA), and every cell was manually delimited according to phalloidin staining. The background of different channels was edited with Subtract Background tool with a rolling ball radius of 30 pixels; by a threshold intensity, binary images were obtained. The logical operation AND of the Image Calculator tool was used to generate an image harbouring only overlapping structures of both channels. Colocalization measurement was obtained by quantifying the area occupied by the overlapping elements per cell. At least, 200 cells were measured for each cell line.
Lysosomal function study
Cells were treated with 100 nM bafilomycin A1 (B1793, Sigma) for 2 h or remain untreated. After that, cells were collected and treated with 500 nM LysoTracker Red DND-99 (L-7528, Molecular Probes, Life Technologies) for 20 min. Then, cells were analysed by flow cytometry (FACSCalibur, BD Biosciences). Relative LysoTracker level represents the ratio of the FL-2 fluorescence intensity of the untreated condition and the bafilomycin-treated one.
Cathepsin assay (Magic Red)
Fibroblasts were plated to chambered coverglasses and imaged the following day on FRET in vivo Imaging System (Zeiss, Oberkochen, Germany) coupled to an Axiovert200 microscope (Zeiss). At the start of imaging, Magic Red reagent (937, Immunochemistry Technologies, Bloomington, MN, USA) was diluted 1:365 into PBS, and then this solution was added into the cell culture media at a 1:10 dilution. Images were acquired using a 20X Plan-Apochromat objective (420650-9901-000, Zeiss): numerical aperture of 0.6, at 4 min intervals for a total of 32 min. Image analysis was performed using ImageJ software. Images were thresholded to include only the lysosomal compartment, using the same numerical threshold for all images across a single experiment. The total fluorescence above threshold was normalized to the total area of the lysosomal compartment, measured as the area above threshold at t = 32 min. Graph represents means and standard deviation of normalized fluorescence intensities plotted over time obtained from three Alzheimer and healthy couples.
Mitochondrial isolation
The Mitochondria Isolation Kit for Mammalian Cells (89874, Pierce Protein Biology Products, Rockford, IL, USA) was used to isolate intact mitochondria from healthy and SAD cultured cells using the manufacturer’s protocol.
Autophagolysosomes quantification
Fibroblasts were transfected with mRFP-GFP tandem fluorescent-tagged LC332 using Polyethylenimine (PEI; 408727, Sigma). After 24 h, cells were fixed and processed for analysis in an Invert Confocal LSM510 (Zeiss, Oberkochen, Germany) fluorescence microscope. Quantification of only RFP-positive dots per cell was performed with ImageJ software.
Phos-tag assay
Cells were treated with 20 μM CCCP for 1 h and then lysed in western blot buffer. We use 7.5% polyacrylamide gels containing 50 μM Phos-tag acrylamide AAL-107 (Wako Chemicals, Richmond, VA, USA) and 100 μM MnCl2. After electrophoresis, gels were washed twice with transfer buffer containing 0.01% SDS and 10 mM EDTA for 20 min and then replaced with transfer buffer containing 0.01% SDS for 10 min. Proteins were transferred to PVDF membranes and analysed by conventional immunoblotting. For dephosphorylation, extracts treated with CCCP were incubated for 1 h with 800 U of lambda protein phosphatase (λPP, Sigma), following the guidelines of the supplier.
Brain tissues
Hippocampal brain samples from SAD patients and control subjects were obtained from Banco de Tejidos (Fundación CIEN, Instituto de Salud Carlos III, Madrid, Spain). On the basis of quantitative pathological features, the SAD brain specimens were classified as coming from SAD patients at Braak stages II and III (early AD) (n = 5), Braak stage V (n = 5) and Braak VI (n = 6) (severe AD) and control subjects (n = 6). Written informed consent was obtained from all the patients.
Quantitative real-time PCR assays
Frozen brain tissue was homogenized using a TissueLyser (Retsch MM300, Qiagen, GmbH, Hilden, Germany) run at 30 Hz for 5 min with 5 mm stainless steel beads (69989, Qiagen) in 700 μl of QIAzol Lysis Reagent (79306, Qiagen). RNA was extracted using miRNeasy Mini Kit (217004, Qiagen) with RNase-Free DNase Set (79254, Qiagen), following provider's guidelines. RNA integrity number (RIN) of each sample was calculated using the Agilent 2100 Bioanalyzer system (Agilent Technologies, Santa Clara, CA, USA), and only RNAs with RIN above 5 were used for RT–qPCR. Retrotranscription was performed with Transcriptor First Strand cDNA Synthesis Kit (04379012001, Roche, Mannheim, Germany) using 20 ng/μl of RNA with random primers. Quantitative PCR was performed in a thermocycler LightCycler480 (Roche), using the following conditions: 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. The oligos used for the detection were PARK2-F: CCCTGGGACTAGTGCAGAAT, PARK2-R: TGCGATCAGGTGCAAAGCTA, PINK1-F: CATGCCTACATTGCCCCAGA, PINK1-R: GAACCTGCCGAGATGTTCCA, TOMM20-F: GCTGGGCTTTCCAAGTTACC, TOMM20-R: AGTAACTGCTGTGGCTGTCC, MT-RNR2-F: CGATGGTGCAGCCGCTATTA and MT-RNR2-R: ATCATTTACGGGGGAAGGCG. Gene expression was normalized to GAPDH expression using TaqMan primer human GAPDH (Hs02758991_g1, Applied Biosystems, Darmstadt, Germany).
Microarray analysis
The expression profiling by array (Affymetrix Human Gene 1.0 ST Array) of PARK2, PINK1 and TOMM20 of human brain samples classified into healthy (n = 47) versus AD (n = 32) was retrieved from the Hisayama study (41). Differences in PARK2, PINK1 and TOMM20 expression between healthy and AD were calculated using Student's t-test.
Statistical analysis
The values represented in the graphs were obtained by normalizing every SAD sample data with its correspondent age-matched healthy sample. Randomly chosen AD–healthy couples were used for each experiment without any specific exclusion criteria. The lines used for each experiment have been specified in the correspondent legend to the figure. All graphs represent means and standard deviations of the values obtained from at least three AD–healthy couples. Statistical comparison of the data sets was performed by the Student's t-test. Two-way analysis of variance test was performed to examine the differences between experimental factors and their interaction. A post hoc Bonferroni test was used when more than two experimental groups were compared. When the distribution of the data was not normal, a non-parametric Mann–Whitney U test was used. The differences are given with their corresponding statistical significance or P-value, which is the probability that the difference occurred merely by chance under the null hypothesis.
Supplementary Material
Funding
This work was supported by the Ministerio de Economía y Competitividad (SAF 2011 program, project 24841 and grant BES-2012-055068) from Spain and by a grant from the National Institute on Minority Health and Health Disparities (G12-MD007591) from the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Supplementary Material
Acknowledgements
We would like to specially acknowledge Alberto Rábano from Banco de Tejidos (Fundación CIEN, Instituto de Salud Carlos III, Madrid, Spain) for providing the hippocampus samples. We are grateful for advice and technical aid provided by Veronica Labrador from the Microscopy Service of the Centro de Biología Molecular ‘Severo Ochoa’. We also would like to thank Dr Enrique Gabandé (Barts Cancer Institute, Queen Mary University of London) for his help with CTSB activity assay. We want to acknowledge Dr Jerónimo Jurado (Centro de Biología Molecular Severo Ochoa) for his help with statistical analysis. SCaMC-1 antibody was a kind gift from Dr Araceli de Arco (Universidad de Castilla la Mancha, Toledo, Spain) and PARK2 lentivector from Dr Andrew B. West (UAB School of Medicine, Birmingham, AL, USA).
Conflict of Interest statement. None declared.
References
- 1.Savva G.M., Wharton S.B., Ince P.G., Forster G., Matthews F.E., Brayne C. (2009) Age, neuropathology, and dementia. N. Engl. J. Med., 360, 2302–2309. [DOI] [PubMed] [Google Scholar]
- 2.Price D.L., Tanzi R.E., Borchelt D.R., Sisodia S.S. (1998) Alzheimer's disease: genetic studies and transgenic models. Annu. Rev. Genet., 32, 461–493. [DOI] [PubMed] [Google Scholar]
- 3.Zhu X., Perry G., Smith M.A., Wang X. (2013) Abnormal mitochondrial dynamics in the pathogenesis of Alzheimer’s disease. J. Alzheimers Dis., 33(Suppl. 1), S253–S262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonda D.J., Wang X., Perry G., Nunomura A., Tabaton M., Zhu X., Smith M.A. (2010) Oxidative stress in Alzheimer disease: a possibility for prevention. Neuropharmacology, 59, 290–294. [DOI] [PubMed] [Google Scholar]
- 5.Reddy P.H., Tripathi R., Troung Q., Tirumala K., Reddy T.P., Anekonda V., Shirendeb U.P., Calkins M.J., Reddy A.P., Mao P. et al. (2012) Abnormal mitochondrial dynamics and synaptic degeneration as early events in Alzheimer's disease: implications to mitochondria-targeted antioxidant therapeutics. Biochim. Biophys. Acta, 1822, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X., Su B., Lee H.G., Li X., Perry G., Smith M.A., Zhu X. (2009) Impaired balance of mitochondrial fission and fusion in Alzheimer's disease. J. Neurosci., 29, 9090–9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Escudero V., Martin-Maestro P., Perry G., Avila J. (2013) Deconstructing mitochondrial dysfunction in Alzheimer disease. Oxid. Med. Cell. Longev., 2013, 162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leuner K., Schutt T., Kurz C., Eckert S.H., Schiller C., Occhipinti A., Mai S., Jendrach M., Eckert G.P., Kruse S.E. et al. (2012) Mitochondrion-derived reactive oxygen species lead to enhanced amyloid beta formation. Antioxid. Redox Signal., 16, 1421–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santa-Maria I., Hernandez F., Martin C.P., Avila J., Moreno F.J. (2004) Quinones facilitate the self-assembly of the phosphorylated tubulin binding region of tau into fibrillar polymers. Biochemistry, 43, 2888–2897. [DOI] [PubMed] [Google Scholar]
- 10.Taga M., Mouton-Liger F., Paquet C., Hugon J. (2011) Modulation of oxidative stress and tau phosphorylation by the mTOR activator phosphatidic acid in SH-SY5Y cells. FEBS Lett., 585, 1801–1806. [DOI] [PubMed] [Google Scholar]
- 11.Bonda D.J., Castellani R.J., Zhu X., Nunomura A., Lee H.G., Perry G., Smith M.A. (2011) A novel perspective on tau in Alzheimer's disease. Curr. Alzheimer Res., 8, 639–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belkacemi A., Ramassamy C. (2012) Time sequence of oxidative stress in the brain from transgenic mouse models of Alzheimer's disease related to the amyloid-beta cascade. Free Radic. Biol. Med., 52, 593–600. [DOI] [PubMed] [Google Scholar]
- 13.Kopeikina K.J., Carlson G.A., Pitstick R., Ludvigson A.E., Peters A., Luebke J.I., Koffie R.M., Frosch M.P., Hyman B.T., Spires-Jones T.L. (2011) Tau accumulation causes mitochondrial distribution deficits in neurons in a mouse model of tauopathy and in human Alzheimer's disease brain. Am. J. Pathol., 179, 2071–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature, 392, 605–608. [DOI] [PubMed] [Google Scholar]
- 15.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G. et al. (2004) Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science, 304, 1158–1160. [DOI] [PubMed] [Google Scholar]
- 16.Lin W., Kang U.J. (2008) Characterization of PINK1 processing, stability, and subcellular localization. J. Neurochem., 106, 464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol., 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjorkoy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol., 171, 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiba-Fukushima K., Imai Y., Yoshida S., Ishihama Y., Kanao T., Sato S., Hattori N. (2012) PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci. Rep., 2, 1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okatsu K., Oka T., Iguchi M., Imamura K., Kosako H., Tani N., Kimura M., Go E., Koyano F., Funayama M. et al. (2012) PINK1 autophosphorylation upon membrane potential dissipation is essential for Parkin recruitment to damaged mitochondria. Nat. Commun., 3, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. (2010) PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol., 12, 119–131. [DOI] [PubMed] [Google Scholar]
- 22.Gegg M.E., Cooper J.M., Chau K.Y., Rojo M., Schapira A.H., Taanman J.W. (2010) Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet., 19, 4861–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fedorowicz M.A., de Vries-Schneider R.L., Rub C., Becker D., Huang Y., Zhou C., Alessi Wolken D.M., Voos W., Liu Y., Przedborski S. (2014) Cytosolic cleaved PINK1 represses Parkin translocation to mitochondria and mitophagy. EMBO Rep., 15, 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nixon R.A., Wegiel J., Kumar A., Yu W.H., Peterhoff C., Cataldo A., Cuervo A.M. (2005) Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J. Neuropathol. Exp. Neurol., 64, 113–122. [DOI] [PubMed] [Google Scholar]
- 25.Scheper W., Nijholt D.A., Hoozemans J.J. (2011) The unfolded protein response and proteostasis in Alzheimer disease: preferential activation of autophagy by endoplasmic reticulum stress. Autophagy, 7, 910–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuervo A.M. (2004) Autophagy: in sickness and in health. Trends Cell Biol., 14, 70–77. [DOI] [PubMed] [Google Scholar]
- 27.Nixon R.A. (2005) Endosome function and dysfunction in Alzheimer's disease and other neurodegenerative diseases. Neurobiol. Aging, 26, 373–382. [DOI] [PubMed] [Google Scholar]
- 28.Lee J.H., Yu W.H., Kumar A., Lee S., Mohan P.S., Peterhoff C.M., Wolfe D.M., Martinez-Vicente M., Massey A.C., Sovak G. et al. (2010) Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell, 141, 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreira P.I., Siedlak S.L., Wang X., Santos M.S., Oliveira C.R., Tabaton M., Nunomura A., Szweda L.I., Aliev G., Smith M.A. et al. (2007) Autophagocytosis of mitochondria is prominent in Alzheimer disease. J. Neuropathol. Exp. Neurol., 66, 525–532. [DOI] [PubMed] [Google Scholar]
- 30.Moreira P.I., Siedlak S.L., Wang X., Santos M.S., Oliveira C.R., Tabaton M., Nunomura A., Szweda L.I., Aliev G., Smith M.A. et al. (2007) Increased autophagic degradation of mitochondria in Alzheimer disease. Autophagy, 3, 614–615. [DOI] [PubMed] [Google Scholar]
- 31.Yu W.H., Kumar A., Peterhoff C., Shapiro Kulnane L., Uchiyama Y., Lamb B.T., Cuervo A.M., Nixon R.A. (2004) Autophagic vacuoles are enriched in amyloid precursor protein-secretase activities: implications for beta-amyloid peptide over-production and localization in Alzheimer's disease. Int. J. Biochem. Cell Biol., 36, 2531–2540. [DOI] [PubMed] [Google Scholar]
- 32.Cecchi C., Fiorillo C., Sorbi S., Latorraca S., Nacmias B., Bagnoli S., Nassi P., Liguri G. (2002) Oxidative stress and reduced antioxidant defenses in peripheral cells from familial Alzheimer's patients. Free. Radic. Biol. Med., 33, 1372–1379. [DOI] [PubMed] [Google Scholar]
- 33.Citron M., Vigo-Pelfrey C., Teplow D.B., Miller C., Schenk D., Johnston J., Winblad B., Venizelos N., Lannfelt L., Selkoe D.J. (1994) Excessive production of amyloid beta-protein by peripheral cells of symptomatic and presymptomatic patients carrying the Swedish familial Alzheimer disease mutation. Proc. Natl Acad. Sci. USA, 91, 11993–11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Vicente M., Talloczy Z., Wong E., Tang G., Koga H., Kaushik S., de Vries R., Arias E., Harris S., Sulzer D. et al. (2010) Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci., 13, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo S.H., Tang G., Ma K., Babij R., Cortes E., Vonsattel J.P., Faust P.L., Sulzer D., Louis E.D. (2012) Macroautophagy abnormality in essential tremor. PLoS ONE, 7, e53040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozawa K., Komatsubara A.T., Nishimura Y., Sawada T., Kawafune H., Tsumoto H., Tsuji Y., Zhao J., Kyotani Y., Tanaka T. et al. (2013) S-nitrosylation regulates mitochondrial quality control via activation of parkin. Sci. Rep., 3, 2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowman E.J., Siebers A., Altendorf K. (1988) Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl Acad. Sci. USA, 85, 7972–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshimori T., Yamamoto A., Moriyama Y., Futai M., Tashiro Y. (1991) Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem., 266, 17707–17712. [PubMed] [Google Scholar]
- 39.Katsuragi Y., Ichimura Y., Komatsu M. (2015) p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J., 282, 4672–4678. [DOI] [PubMed] [Google Scholar]
- 40.Braak H., Braak E. (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol., 82, 239–259. [DOI] [PubMed] [Google Scholar]
- 41.Hokama M., Oka S., Leon J., Ninomiya T., Honda H., Sasaki K., Iwaki T., Ohara T., Sasaki T., LaFerla F.M. et al. (2014) Altered expression of diabetes-related genes in Alzheimer's disease brains: the Hisayama study. Cereb. Cortex, 24, 2476–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X., Wang W., Li L., Perry G., Lee H.G., Zhu X. (2014) Oxidative stress and mitochondrial dysfunction in Alzheimer's disease. Biochim. Biophys. Acta, 1842, 1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chu C.T., Zhu J., Dagda R. (2007) Beclin 1-independent pathway of damage-induced mitophagy and autophagic stress: implications for neurodegeneration and cell death. Autophagy, 3, 663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu J.H., Horbinski C., Guo F., Watkins S., Uchiyama Y., Chu C.T. (2007) Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am. J. Pathol., 170, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polgar L., Csoma C. (1987) Dissociation of ionizing groups in the binding cleft inversely controls the endo- and exopeptidase activities of cathepsin B. J. Biol. Chem., 262, 14448–14453. [PubMed] [Google Scholar]
- 46.Lonskaya I., Shekoyan A.R., Hebron M.L., Desforges N., Algarzae N.K., Moussa C.E. (2013) Diminished parkin solubility and co-localization with intraneuronal amyloid-beta are associated with autophagic defects in Alzheimer's disease. J. Alzheimers Dis., 33, 231–247. [DOI] [PubMed] [Google Scholar]
- 47.Witte M.E., Bol J.G., Gerritsen W.H., van der Valk P., Drukarch B., van Horssen J., Wilhelmus M.M. (2009) Parkinson's disease-associated parkin colocalizes with Alzheimer's disease and multiple sclerosis brain lesions. Neurobiol. Dis., 36, 445–452. [DOI] [PubMed] [Google Scholar]
- 48.Khandelwal P.J., Herman A.M., Hoe H.S., Rebeck G.W., Moussa C.E. (2011) Parkin mediates beclin-dependent autophagic clearance of defective mitochondria and ubiquitinated Abeta in AD models. Hum. Mol. Genet., 20, 2091–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.del Arco A., Satrustegui J. (2004) Identification of a novel human subfamily of mitochondrial carriers with calcium-binding domains. J. Biol. Chem., 279, 24701–24713. [DOI] [PubMed] [Google Scholar]
- 50.Mosmann T. (1983) Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods, 65, 55–63. [DOI] [PubMed] [Google Scholar]
- 51.da Costa C.A., Sunyach C., Giaime E., West A., Corti O., Brice A., Safe S., Abou-Sleiman P.M., Wood N.W., Takahashi H. et al. (2009) Transcriptional repression of p53 by parkin and impairment by mutations associated with autosomal recessive juvenile Parkinson's disease. Nat. Cell Biol., 11, 1370–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.