Abstract
Objectives
The objectives of this study were to study the presence of mutators in a set of Acinetobacter baumannii isolates and to explore whether there is a correlation between mutation rates and antibiotic resistance.
Methods
The variation in mutation rate was evaluated for 237 clinical A. baumannii isolates by determining the frequency of their mutation to rifampicin resistance. For each isolate, the antibiotic resistance profile was determined by disc diffusion and/or Etest. Isolates were divided into susceptible, resistant and MDR groups according to their resistance to five groups of different antibiotics. A comparison between differences in mutation frequency (f) and strain-specific factors was performed.
Results
Of the 237 isolates 32%, 18% and 50% were classified as susceptible, resistant and MDR, respectively. The f of rifampicin resistance varied between 2.2 × 10−10 and 1.2 × 10−6. Of the strains under investigation, 16% had an ≥2.5- to 166-fold higher f. The presence of mutators (definition ≥2.5-fold increase in f compared with ATCC 19606) in the MDR group (22%) was significantly higher (P < 0.05) than that in the susceptible and resistant groups (11% and 7%, respectively). Furthermore, f was significantly higher in the MDR group compared with that in the susceptible and resistant groups.
Conclusions
The facts that 26 of 37 mutator isolates (70%) in the population were MDR and that there was a significantly higher general f in isolates exhibiting an MDR profile suggest that hypermutability can be of advantage for the organism in a selective environment with extensive exposure to antimicrobials.
Introduction
Acinetobacter baumannii is increasingly involved in nosocomial infections, especially in the ICU setting. This pathogen is frequently MDR and has shown a propensity to adapt to the environment through up-regulation of intrinsic resistance and acquisition of antibiotic resistance determinants.1,2 Another parameter that modulates the progression of resistance is an increase in general mutation rate (hypermutability), which can influence resistance development through mutation,3 frequency of lateral gene transfer,4 up-regulated gene expression and compensatory evolution of a bacterial population.5
Studies of mutator distribution and level of hypermutability have shown that ≤50% of clinical isolates from a variety of bacterial species exhibit a mutator phenotype/genotype. Many of these mutators are found among infection-causing bacteria, which are isolated from an environment of high selective pressure due to intense antibiotic usage.6 No previous study has evaluated the existence of mutator isolates in an A. baumannii population, to our knowledge. The aims of this study were to determine the prevalence of mutators in A. baumannii and their hypermutability and to explore whether there is any correlation between changes in the mutation frequency (f) and antibiotic resistance.
Materials and methods
Bacterial isolates, antimicrobials and culture media
Two-hundred-and-thirty-seven A. baumannii isolates originating from Europe, the USA and Sweden between 1990 and 2007 were investigated.7,8 A. baumannii ATCC 19606 was chosen as a comparator strain for f assays due to its isolation in 1948 and exposure to less antibiotic selection pressure. The WT Escherichia coli MG1655, E. coli dam (damΔ 16::Kanr) and mutS (ΔmutS::FRT) mutants were used as quality-control strains.9 All experiments were performed in freshly prepared Mueller–Hinton (MH) broth and MH agar (BD, Sweden); rifampicin was purchased from Sigma-Aldrich, Sweden.
Susceptibility testing
Resistance profiles were determined using Etest (Biodisk/bioMérieux, Sweden) and/or disc diffusion (BD/Fisher Scientific, Sweden). The EUCAST clinical breakpoints were used for sorting isolates into antibiotic resistance profile groups (ARPGs) as follows; susceptible, susceptible to all antibiotic groups tested; resistant, resistant to one or two antibiotic groups; and MDR, resistant to three to five antibiotic groups.10 ARPGs are defined in Table 1.
Table 1.
Allocation of 237 isolates into ARPGs and the median and geometric mean f of isolates in ARPGs
| ARPG (n) | Percentage of total | Percentage of isolates in each group with a specific resistance phenotype (n) |
Median f | Geometric mean f (SD) | Median fold increase in f compared with that of ATCC 19606 | Geometric mean fold increase in f compared with that of ATCC 19606 (SD) | Percentage of mutators in ARPG | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TET | CEP | CAR | AMG | FQ | |||||||
| Susceptible (75) | 32 | 0 | 0 | 0 | 0 | 0 | 6.4E−09 | 9.6E−09 (1.2E−08) | 0.92 | 1.37 (1.67) | 10.7 |
| Resistant (43) | 18 | 16 (7) | 54 (23) | 0 | 42 (18) | 56 (24) | 6.2E−09 | 7.6E−09 (6.7E−09) | 0.90 | 1.09 (0.96) | 6.98 |
| MDR (119) | 50 | 55 (65) | 92 (109) | 35 (42) | 88 (105) | 70 (83) | 8.4E−09 | 2.3E−08 (1.1E−07) | 1.20 | 3.23 (15.3) | 21.8 |
| Total (237) | 100 | 30 (72) | 56 (132) | 18 (42) | 52 (123) | 45 (107) | 7.0E−09 | 1.6E−08 (7.6E−08) | 1.00 | 2.25 (10.8) | 15.6 |
TET, tetracycline-like group (tetracycline, tigecycline, doxycycline); CEP, cephalosporin group (cefepime, ceftazidime); CAR, carbapenem group (meropenem, imipenem); AMG, aminoglycoside group (gentamicin, tobramycin, amikacin); and FQ, fluoroquinolone group (ciprofloxacin, levofloxacin); SD, standard deviation.
Determination of f against rifampicin
To determine f, assays were performed as previously described by Baquero et al.11 Ten independent cultures for each isolate were grown overnight with shaking, and cells were plated onto MH plates with and without rifampicin (100 mg/L).
The distribution of f values in the population was categorized as previously described (Table 1).11,12 The f values were normalized to control strain ATCC 19606 (f = 7 × 10−9), which was set at 1. Isolates with an ≥2.5- to 10-fold higher f were defined as weak mutators and those with a >10-fold higher f were defined as strong mutators.
Data comparison and statistical methods
Isolates were grouped according to their ARPGs and further into hypo-normomutators (f < 2.5-fold) and mutators (f ≥ 2.5-fold) before analysis. The Mann–Whitney U-test and Kruskal–Wallis analysis of variance (ANOVA) statistical methods were used to compare resistance groups. For associations between ARPG and the number of mutator isolates in each group, the Pearson's χ2 test was used, and logistic regression was used to further investigate the relationship between these variables. P values <0.05 were considered significant.
Results
Antimicrobial susceptibility profiles
The division of isolates and the percentage of each isolate in each ARPG are shown in Table 1.
Distribution of f values in bacterial populations
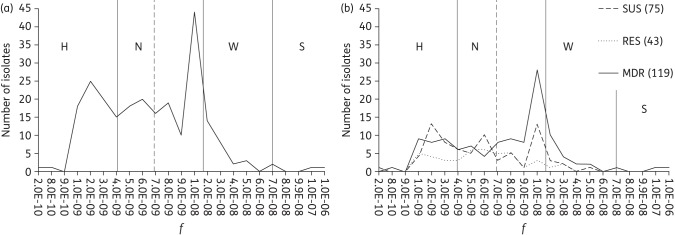
The frequency of rifampicin resistance was determined, and isolates were sorted according to their f, which varied between 2.2 × 10−10 and 1.2 × 10−6 (Figure 1). A sharp peak in frequency distribution was found at ∼10−8; however, 68% of the population had a lower frequency. To the right of the peak, 15% of the isolates had a moderately higher f, whereas only two isolates (<0.1%) had a frequency >10−7 (Figure 1a). No A. baumannii isolate displayed hypermutability as high as that of the E. coli mutS strain (f = 3 × 10−6), and only one isolate (f = 1.1 × 10−6) had a frequency higher than that of the E. coli dam-knockout strain (f = 3.9 × 10−7). When the population was sorted according to their ARPGs, the distribution of the susceptible group showed three peaks, at 2 × 10−9, 6 × 10−9 and 1 × 10−8, respectively, while the frequencies of the resistant group were spread over the whole interval, and for the MDR group, ∼45% of the isolates were clustered in a sharp peak, with f ranging between 8 × 10−9 and 2 × 10−8 (Figure 1b).
Figure 1.
Distribution of f of 237 A. baumannii isolates. (a) Vertical continuous lines indicate the division of isolates into categories by their mutability status: H, hypomutators (f ≤4 × 10−9); N, normomutators (4 × 10−9 < f < 1.7 × 10−8); W, weak mutators (1.7 × 10−8 < f < 7 × 10−8); and S, strong mutators (f >7 × 10−8).11,12 The vertical broken line indicates f for the control strain A. baumannii ATCC 19606 (7 × 10−9). (b) Distribution of f when isolates are divided according to their ARPGs. Numbers in brackets indicate numbers of isolates in respective ARPGs. SUS, susceptible; RES, resistant.
Hypermutability and resistance profile in mutator isolates
Compared with the A. baumannii ATCC 19606 control strain, 200 isolates had an f ranging between 0.03-fold and <2.5-fold. Of these, 80 were classified as hypomutators (<1.0-fold), composed of 39% susceptible, 19% resistant and 43% MDR isolates, respectively. The overall occurrence of weak and strong mutators was ∼16%; when the population was divided into ARPGs, the prevalence of mutators in the MDR group (22%) was significantly higher (P < 0.05) than the prevalence in the susceptible and resistant groups (11% and 7%, respectively). The geometric mean and median of f for the population and the percentage of mutators in each ARPG are presented in Table 1.
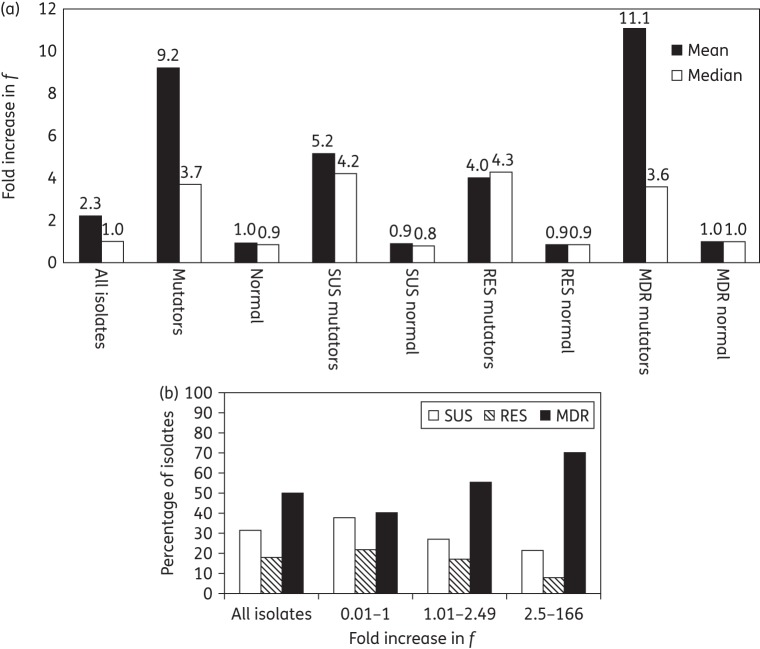
Thirty-seven isolates had an ≥2.5- to 166-fold increase in f (73% with ≥2.5- to 5-fold, 19% with >5- to 10-fold and 8% with >10-fold) (Table S1, available as Supplementary data at JAC Online). Of the mutators, 70% were MDR isolates with a median/mean increase f of 3.6/11.1-fold (Figure 2a). There was no significant difference in increased f between the mutators in the susceptible, resistant and MDR groups (P = 0.51–0.74) (Figure 2a). However, there was a significant correlation between MDR and increased f (P < 0.05) (Figure 2b).
Figure 2.
(a) Differences in geometric mean and median fold increase in f in the ARPGs. Each ARPG is subdivided into normal (f < 2.5-fold, hypo- and normomutators) and mutator (f ≥ 2.5-fold, weak and strong mutators) groups. (b) Distribution of isolates with increased f and increased multidrug resistance. The results of the Wilcoxon Mann–Whitney U-test showed that there was a significantly higher mean f among isolates with an MDR profile (P > 0.05). SUS, susceptible; RES, resistant.
Of the 34 weak mutators, 7, 3 and 24 of the isolates had a susceptible, resistant and MDR phenotype, respectively. All the weak resistant mutators (f 3.19–4.29-fold) were susceptible to cephalosporins and carbapenems, with a varied range of susceptibility to the other ARPGs. Four weak MDR mutators (f 2.51–3.25-fold), which displayed antibiotic resistance to three of five antibiotic groups, were all susceptible to the carbapenem group in combination with susceptibility to either the tetracycline-like, aminoglycoside or fluoroquinolone group of antibiotics. Of the remaining 20 weak MDR isolates, nine were resistant to all antibiotic groups (f 3.14–8.29-fold), 5 were resistant to all except tetracycline-like antibiotics (f 2.57–5.89-fold) and 6 were resistant to all except carbapenem-group antibiotics (f 2.71–10.0-fold) (Table S1).
The remaining three strong mutators, one susceptible isolate and two MDR isolates, displayed 11.1-fold (AB95), 19.5-fold (AB279) and 166-fold (AB190) increased f (Table S1). In comparison with the control mutator E. coli strains (dam and mutS mutants, 55.7- and 429-fold increased f), the AB190 isolate can be regarded as a strong mutator.
Discussion
This study is the first one, to our knowledge, on the occurrence of mutators in an A. baumannii population, and we have shown that 16% of isolates from our collection displayed an ≥2.5- to 166-fold increase in f. The prevalence of mutators and their f values were significantly higher in the MDR group compared with those of the susceptible and resistant groups. We also found 0.42% and 15.2% of isolates to be strong and weak mutators, respectively, which are around the same range as findings for E. coli urinary tract isolates, where the proportions were ∼0.5%–1% and 25%–40% for strong and weak mutators, respectively.11,13
Variation in mutation rates and its effect on resistance development and bacterial adaptability have been studied extensively over recent years. It was shown that even a small (2–4-fold) increase in mutation rate can drive the evolution of fluoroquinolone resistance, and, furthermore, a slight increase in f favours the evolution of MDR.9,14 In E. coli, selection with rifampicin and ciprofloxacin showed that mutator strains generated both higher resistance levels and resistance mutations with ≤1000-fold higher f.15
Studies of clinical isolates have revealed that weak mutators can be present at earlier stages of infection, and a modestly elevated mutation rate can give them an adaptive advantage.16
Another aspect of mutators is the notion that some genotypes can exhibit increased frequency of recombination, interspecies recombination and transformation.17,18 These events might explain in part the ability of A. baumannii to adapt, whereby a mutator can integrate DNA that carries resistance markers and/or increase the chance of gaining mutations that promote the survival of the organism in clinical settings.
Carbapenems are still the drugs of choice to treat A. baumannii infections, even though resistance rates are increasing.1 To understand the progression of carbapenem resistance, Zander et al.19 investigated three isolates recovered from patients during an outbreak in a hospital in Krakow, Poland. Sequencing of blaOXA 51-like genes showed that carbapenem resistance was caused by a conversion of OXA-66 into OXA-82 and that blaOXA-82 was also associated with the IS element ISAba1. A second study looked at related isolates possessing similar plasmids that encode the carbapenemase OXA-58 and exhibit varying levels of carbapenem resistance. Sequencing revealed genetic variability composed of multiple copies of the blaOXA 58 gene and that extra copies were due to IS-element transposition or recombination events.20 These adaptive modifications could be elevated in a population composed of mutators, where hypermutability can drive the progression of survival and evolution of genetic elements such as β-lactamase genes. However, whether any of the genetic changes seen in these studies was due to an altered mutation rate is not known.
In conclusion, we have shown that 16% of the A. baumannii strains were weak-to-strong mutators and that there was a strong correlation with an increased f with an MDR phenotype. The fact that a high percentage of MDR A. baumannii isolates show an increased mutability in clinical settings calls for further studies that could form the basis of novel treatment strategies.
Funding
This work was supported by grants from the Scandinavian Society for Antimicrobial Chemotherapy Foundation.
Transparency declarations
P. G. H. was the recipient of a Pfizer Investigator Initiated Research grant. All other authors: none to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
Part of these data were presented at the Nineteenth European Congress of Clinical Microbiology and Infectious Diseases, Helsinki, Finland, 2009 (O443).
We thank Professor Diarmaid Hughes for sharing the mutator control strains with us and Oscar Klockars for his help with part of the f experiments.
References
- 1.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 2008; 21: 538–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poirel L, Bonnin RA, Nordmann P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life 7 2011; 63: 1061–7. [DOI] [PubMed] [Google Scholar]
- 3.Woodford N, Ellington MJ. The emergence of antibiotic resistance by mutation. Clin Microbiol Infect 2007; 13: 5–18. [DOI] [PubMed] [Google Scholar]
- 4.Townsend JP, Nielsen KM, Fisher DS et al. . Horizontal acquisition of divergent chromosomal DNA in bacteria: effects of mutator phenotypes. Genetics 2003; 164: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maisnier-Patin S, Andersson DI. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res Microbiol 2004; 155: 360–9. [DOI] [PubMed] [Google Scholar]
- 6.Hall LMC, Henderson-Begg SK. Hypermutable bacteria isolated from humans—a critical analysis. Microbiology 2006; 152: 2505–14. [DOI] [PubMed] [Google Scholar]
- 7.Higgins PG, Janssen K, Fresen MM et al. . Molecular epidemiology of Acinetobacter baumannii bloodstream isolates obtained in the United States from 1995 to 2004 using rep-PCR and multilocus sequence typing. J Clin Microbiol 2012; 50: 3493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wisplinghoff H, Hippler C, Bartual SG et al. . Molecular epidemiology of clinical Acinetobacter baumannii and Acinetobacter genomic species 13TU isolates using a multilocus sequencing typing scheme. Clin Microbiol Infect 2008; 14: 708–15. [DOI] [PubMed] [Google Scholar]
- 9.Orlen H, Hughes D. Weak mutators can drive the evolution of fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother 2006; 50: 3454–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 4.0. EUCAST. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_4.0.pdf.
- 11.Baquero MR, Nilsson AI, Turrientes MC et al. . Polymorphic mutation frequencies in Escherichia coli: emergence of weak mutators in clinical isolates. J Bacteriol 2004; 186: 5538–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turrientes MC, Baquero MR, Sánchez MB et al. . Polymorphic mutation frequencies of clinical and environmental Stenotrophomonas maltophilia populations. Appl Environ Microbiol 2010; 76: 1746–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komp Lindgren P, Karlsson A, Hughes D. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother 2003; 47: 3222–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denamur E, Tenaillon O, Deschamps C et al. . Intermediate mutation frequencies favor evolution of multidrug resistance in Escherichia coli. Genetics 2005; 171: 825–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller K, O'Neill AJ, Chopra I. Response of Escherichia coli hypermutators to selection pressure with antimicrobial agents from different classes. J Antimicrob Chemother 2002; 49: 925–34. [DOI] [PubMed] [Google Scholar]
- 16.Feliziani S, Marvig RL, Luján AM et al. . Coexistence and within-host evolution of diversified lineages of hypermutable Pseudomonas aeruginosa in long-term cystic fibrosis infections. PLoS Genet 2014; 10: e1004651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matic I, Rayssiguier C, Radman M. Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell 1995; 80: 507–15. [DOI] [PubMed] [Google Scholar]
- 18.Denamur E, Matic I. Evolution of mutation rates in bacteria. Mol Microbiol 2006; 60: 820–7. [DOI] [PubMed] [Google Scholar]
- 19.Zander E, Chmielarczyk A, Heczko P et al. . Conversion of OXA-66 into OXA-82 in clinical Acinetobacter baumannii isolates and association with altered carbapenem susceptibility. J Antimicrob Chemother 2013; 68: 308–11. [DOI] [PubMed] [Google Scholar]
- 20.Bertini A, Poirel L, Bernabeu S et al. . Multicopy blaOXA-58 gene as a source of high-level resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother 2007; 51: 2324–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.