Abstract
Objectives
We aimed to quantify the impact of fluoroquinolone resistance on the clinical outcome of paediatric shigellosis patients treated with fluoroquinolones in southern Vietnam. Such information is important to inform therapeutic management for infections caused by this increasingly drug-resistant pathogen, responsible for high morbidity and mortality in young children globally.
Methods
Clinical information and bacterial isolates were derived from a randomized controlled trial comparing gatifloxacin with ciprofloxacin for the treatment of paediatric shigellosis. Time–kill experiments were performed to evaluate the impact of MIC on the in vitro growth of Shigella and Cox regression modelling was used to compare clinical outcome between treatments and Shigella species.
Results
Shigella flexneri patients treated with gatifloxacin had significantly worse outcomes than those treated with ciprofloxacin. However, the MICs of fluoroquinolones were not significantly associated with poorer outcome. The presence of S83L and A87T mutations in the gyrA gene significantly increased MICs of fluoroquinolones. Finally, elevated MICs and the presence of the qnrS gene allowed Shigella to replicate efficiently in vitro in high concentrations of ciprofloxacin.
Conclusions
We found that below the CLSI breakpoint, there was no association between MIC and clinical outcome in paediatric shigellosis infections. However, S. flexneri patients had worse clinical outcomes when treated with gatifloxacin in this study regardless of MIC. Additionally, Shigella harbouring the qnrS gene are able to replicate efficiently in high concentrations of ciprofloxacin and we hypothesize that such strains possess a competitive advantage against fluoroquinolone-susceptible strains due to enhanced shedding and transmission.
Introduction
The Gram-negative bacterial genus Shigella are the most common cause of bacillary dysentery globally.1,2 Of the four species within the genus, Shigella flexneri and Shigella sonnei predominate, with S. sonnei currently replacing S. flexneri as the major species in industrializing regions.3 The WHO currently recommends the fluoroquinolone ciprofloxacin as the first-line therapy, with ceftriaxone and pivmecillinam as secondary alternatives.4 However, antimicrobial resistance (AMR) within the species is becoming more prevalent and may present a significant challenge for therapeutic management.
The primary target of the fluoroquinolones is the DNA gyrase, a type II topoisomerase essential for DNA replication and transcription.5 Mutations in the gyrA gene increase the MICs of fluoroquinolones for Shigella and other Enterobacteriaceae.6–9 Plasmid-mediated quinolone resistance (PMQR) genes can also be acquired, such as the qnr genes that encode pentapeptide repeat proteins that bind to and protect the DNA gyrase and topoisomerase from the action of fluoroquinolones.10 Complete ciprofloxacin resistance (MIC ≥4 mg/L11) has been recently reported in both domestic and imported S. sonnei isolates in the USA, Vietnam and elsewhere.12–14
The rapid evolution and global dissemination of fluoroquinolone resistance in the Enterobacteriaceae hampers effective treatment and is, therefore, a major threat to human health.15 The WHO has explicitly listed fluoroquinolone-resistant Shigella as one of its top concerns in the current international focus on AMR.15 AMR can lead to inappropriate choice of antimicrobial for initial therapy and may force clinicians to choose more toxic or more expensive antimicrobials.16 Furthermore, patients infected with fluoroquinolone-resistant Campylobacter and Salmonella infections in the USA have been shown to have a longer duration of diarrhoea compared with those infected with fluoroquinolone-susceptible strains.17,18 Although one study from Vietnam suggested a correlation between increasing AMR levels in S. sonnei and clinical severity,19 no rigorous evaluation of the impact of fluoroquinolone resistance or presence of gyrA mutations on clinical outcome of Shigella infections has been performed.
Here, we aimed to quantify the effect of fluoroquinolone choice, fluoroquinolone susceptibility and presence of gyrA mutations on fever clearance time (FCT) and total duration of illness in children with S. flexneri and S. sonnei infections in Vietnam. Additionally, we sought to compare the severity and AMR profiles between Shigella species as well as investigate the effect of elevated MIC and gyrA mutations on the in vitro activity of Shigella. Understanding the dynamics of increasing MICs of commonly used fluoroquinolones and clinical patient outcome in industrializing locations is important as it allows clinicians to be better informed when prescribing therapies for what can often be severe infections in young children.
Methods
Patient population
The source data for this study was a randomized controlled trial. The protocol (including justification for use of gatifloxacin) and results for this trial have been described previously in detail.20 Briefly, 500 children were enrolled into an open-label, randomized clinical trial comparing 3 day regimens of gatifloxacin (10 mg/kg/day orally in one dose) and ciprofloxacin (30 mg/kg/day orally in two doses) for the treatment of shigellosis in southern Vietnam. Children were enrolled between 2006 and 2009 at the Hospital for Tropical Diseases in Ho Chi Minh City and at Huu Nghi Hospital in Dong Thap province. Inclusion criteria included age <15 years and a history of bloody or mucoid stools in the 72 h prior to admission to hospital. Exclusion criteria included severe infection (shock, jaundice and extensive gastrointestinal bleeding), known treatment with a fluoroquinolone during the episode and concomitant infection requiring antimicrobial therapy.
Study procedures
Daily case report forms detailing clinical presentation were administered for each patient during the period of hospitalization. A case report form was also administered at a follow-up visit that occurred 7 days after discharge. Clinical failure was defined as fever (≥37.8°C) or the persistence of any signs or symptoms after 120 h of start of treatment (vomiting, abdominal pain or tenesmus with/without three or more loose stools with/without blood and/or mucus). Total duration of symptoms was defined as the time from admission until cessation of all listed symptoms. Microbiological failure was defined as a positive stool culture for the original infecting pathogen after day 3 of the antimicrobial therapy. FCT was defined as the time from admission until temperature was ≤37.8°C for ≥48 h.
Stool samples were collected on admission and standard microbiological techniques were employed to identify Shigella and Salmonella isolates.20 Antimicrobial susceptibility testing was performed by disc diffusion following methods prescribed by the CLSI.11 MICs were calculated by Etest as per the manufacturer's instructions (AB Biodisk, Sweden). Strains that were identified as resistant to ceftriaxone were subjected to further phenotypic tests to confirm ESBL production using discs containing only cefotaxime (30 μg) and both cefotaxime and ceftazidime combined with clavulanic acid (10 μg), according to current CLSI guidelines.11
Genomic DNA extraction, PCR and sequencing
Genomic DNA was purified using the Wizard genomic DNA extraction kit (Promega, USA) as recommended by the manufacturer. Extracted DNA was subjected to PCR targeting known mutation regions on gyrA and parC genes and the PMQR genes qnrA, qnrB, qnrC, qnrS, aac(6′)-Ib-cr and qepA. Primer sequences were GyrA_F: 5′-CGACCTTGCGAGAGAAAT-3′, GyrA_R: 5′-GTTCCATCAGCCCTTCAA-3′,21 ParC_F: 5′-AAACCTGTTCAGCGCCGCATT-3′ and ParC_R: 5′-GTGGTGCCGTTAAGCAAA-3′.22 The primers for the PMQR genes were as previously published.23–25 Taq DNA polymerase supplied by Bioline (UK) was used for the amplifications. Concentrations of reagents were as recommended by the manufacturers. PCR amplifications were performed under the following conditions: 1 cycle of 95°C for 5 min followed by 35 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 1 min. PCR amplicons were sequenced using an ABI 3700 system (ABI, USA) and sequencing reactions were prepared as recommended by the manufacturer. Resulting sequences were analysed using Bioedit software.
Time–kill analyses
One isolate from each mutation group was selected for in vitro time–kill experiments, all were S. flexneri isolates: (i) no gyrA mutation, ciprofloxacin MIC 0.023 mg/L and gatifloxacin MIC 0.023 mg/L; (ii) gyrA mutation A87T, ciprofloxacin MIC 0.094 mg/L and gatifloxacin MIC 0.125 mg/L; (iii) gyrA mutation S83L, ciprofloxacin MIC 0.19 mg/L and gatifloxacin MIC 0.19 mg/L; and (iv) gyrA mutation S83L, ciprofloxacin MIC 8 mg/L and gatifloxacin MIC 6 mg/L with qnrS gene. Strains were grown overnight in Mueller–Hinton (MH) broth. S. flexneri isolates were chosen due to the larger range of MICs of fluoroquinolones compared with those for S. sonnei. The bacterial cultures were diluted 1 : 1000 into 10 mL of fresh MH broth. The inoculation was incubated at 37°C with a circular agitation speed of 150 rpm for 1.5 h. The cultures were mixed with either ciprofloxacin or gatifloxacin, which had been prepared in MH broth to achieve the final volume of 20 mL of the desired concentration of drugs. Controls for each mutant were identical cultures but without supplementary antimicrobials. Bacterial cells were counted at time 0 and 30 min, 1 h, 2 h, 4 h, 6 h, 12 h and 24 h after incubation with antimicrobials. The experiment was performed in replicates of nine for each selected isolate. The limit of detection was 10 cfu/mL.26
Statistical analyses
All data were analysed in STATA v13 (TX, USA). Plots were made in R (R Foundation for Statistical Computing, Austria) using the ggplot2 package.27 Continuous data were compared between groups using the Kruskal–Wallis test. Categorical group data were compared using χ2 or Fisher's exact test. For MICs that were recorded as ‘greater than X’ or ‘less than X’, these values were converted into 2X and X/2, respectively. Logistic regression was used to evaluate the relationship between treatment arm, MICs and overall failure, with interaction between drug and MIC evaluated through the likelihood ratio test. We analysed the time to event endpoints of FCT and total duration of symptoms using Cox regression models. Interaction between species and treatment arm as well as MIC and treatment within species were evaluated using the likelihood ratio test. Age was included a priori as a covariate in all models.
Ethics approval
This study was approved by the institutional ethics review boards of the Hospital for Tropical Diseases and Huu Nghi Hospital and the Oxford Tropical Research Ethics Committee (OxTREC: 010-06). Written informed consent from the parent or guardian was required for enrolment into the trial.
Results
Baseline clinical and demographic characteristics
Of the 500 children enrolled, 6 withdrew after randomization, leaving 494 for analysis. A total of 107/494 (22%) enrolled trial patients were stool culture positive for Shigella spp. Of these, 72 (67%) were S. sonnei, 33 (31%) were S. flexneri and 2 (2%) were Shigella boydii. As shown in Table 1, S. sonnei patients were slightly younger (median: 30 months, IQR: 20–43) than S. flexneri patients (median: 36 months, IQR: 22–60) and were more likely to report a greater number of mucoid stools in the first 24 h after admission compared with S. flexneri patients. S. sonnei patients also had slightly elevated white cell counts (median 13.5 × 109/L, IQR: 10.7–16.9) compared with S. flexneri patients (median: 10.2 × 109/L, IQR: 7.3–16.4). S. flexneri patients, however, were more likely to report abdominal pain (91%) prior to admission than S. sonnei patients (72%).
Table 1.
Baseline demographic and clinical characteristics of all patients, S. sonnei patients and S. flexneri patients
| Characteristic | All patients, n = 494 | S. sonnei patients, n = 72 | S. flexneri patients, n = 33 | Pa |
|---|---|---|---|---|
| Site, n (%) | ||||
| Ho Chi Minh City | 194 (39.3) | 48 (66.7) | 20 (60.6) | 0.546 |
| Dong Thap | 300 (60.7) | 24 (33.3) | 13 (39.4) | |
| Study drug, n (%) | ||||
| ciprofloxacin | 245 (49.6) | 34 (47.2) | 12 (36.4) | 0.298 |
| gatifloxacin | 248 (50.2) | 38 (52.8) | 21 (63.6) | |
| Age (months), median (IQR) | 19 (10.5–32) | 30 (20–43) | 36 (22–60) | 0.062 |
| Male, n (%) | 291 (58.9) | 40 (55.6) | 14 (42.4) | 0.211 |
| Nutritional status, n (%) | ||||
| overweight | 4 (0.8) | 1 (1.4) | 0 (0) | 0.089 |
| normal | 363 (73.5) | 60 (83.3) | 26 (78.8) | |
| malnutrition I | 93 (18.8) | 10 (13.9) | 3 (9.1) | |
| malnutrition II | 29 (5.9) | 1 (1.4) | 4 (12.1) | |
| malnutrition III | 5 (1.0) | 0 (0) | 0 (0) | |
| Prior to admission | ||||
| illness duration (h), median (IQR) | 24 (16–48) | 20 (12–33) | 19 (12–24) | 0.785 |
| fever (≥37.8°C), n (%) | 429 (87.4) | 68 (94.4) | 33 (100) | 0.167 |
| history of febrile convulsions, n (%) | 40 (8.1) | 6 (8.3) | 2 (6.1) | 1.000 |
| history of diarrhoea with blood, n (%) | 210 (42.5) | 24 (33.3) | 11 (33.3) | 1.000 |
| history of mucoid diarrhoea without blood, n (%) | 284 (57.5) | 48 (66.7) | 22 (66.7) | 1.000 |
| vomiting, n (%) | 204 (41.3) | 34 (47.2) | 16 (48.5) | 0.904 |
| abdominal pain, n (%) | 365/492 (74.2) | 52 (72.2) | 30 (90.9) | 0.041 |
| tenesmus, n (%) | 339/490 (69.2) | 45/71 (63.4) | 22 (66.7) | 0.745 |
| Within 24 h of admission | ||||
| mucoid diarrhoea without blood, n (%) | 370 (74.9) | 59 (81.9) | 22 (66.7) | 0.083 |
| number of mucoid stools/24 h, median (IQR) | 3 (0–6) | 4 (2–10) | 2 (0–5) | 0.028 |
| diarrhoea with blood, n (%) | 445 (90.1) | 70 (97.2) | 33 (100) | 0.334 |
| number of bloody stools/24 h, median (IQR) | 1 (1–5) | 3 (1–6) | 3 (1–9) | 0.187 |
| maximum number of episodes/24 h, median (IQR) | 6 (3–10) | 8 (3–11) | 8 (4–10) | 0.806 |
| white blood cells in stool (cells/HPF), n (%) | ||||
| 0 | 214/479 (44.7) | 14/68 (20.6) | 8/32 (25.0) | 0.754 |
| 1–10 | 79/479 (16.5) | 11/68 (16.2) | 4/32 (12.5) | |
| 11–20 | 42/479 (8.8) | 5/68 (7.4) | 4/32 (12.5) | |
| 21–30 | 104/479 (21.7) | 26/68 (38.2) | 9/32 (28.1) | |
| >30 | 40/479 (8.4) | 12/68 (17.6) | 7/32 (21.9) | |
| white cell count ×109/L, median (IQR) | 11.3 (8–14.9) | 13.5 (10.7–16.9) | 10.2 (7.3–16.4) | 0.071 |
HPF, high power field.
aP value comparing S. sonnei and S. flexneri by Kruskal–Wallis test for continuous data or χ2/Fisher's exact test for categorical data.
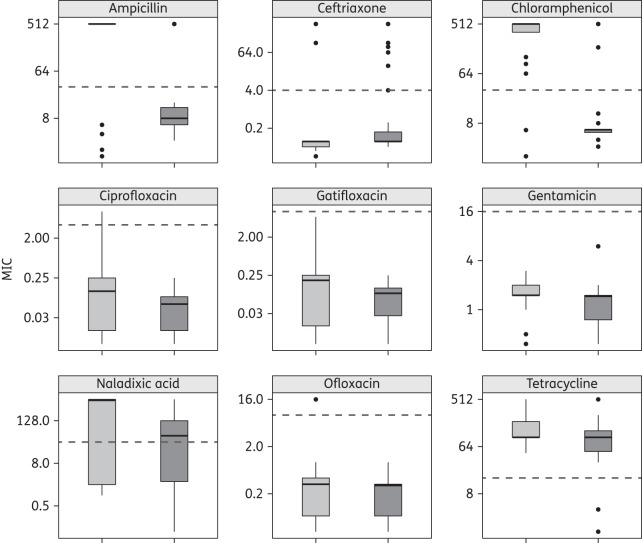
Antimicrobial susceptibility
As shown in Table 2, S. flexneri isolates were more likely to have higher MICs against the fluoroquinolones (with the exception of ofloxacin) compared to S. sonnei. In fact, S. flexneri had significantly higher MICs of all tested antimicrobials with the exception of ceftriaxone, for which S. sonnei had significantly elevated MICs over S. flexneri (P < 0.001, Kruskal–Wallis test) (Figure 1). Overall, S. flexneri isolates were more likely to be MDR (defined here as non-susceptible to more than three antimicrobial classes) (28/33, 85%) compared with S. sonnei (22/72, 31%) (P < 0.001, Fisher's exact test) while S. sonnei isolates were more likely to exhibit an ESBL phenotype (14/72, 19%) than S. flexneri (1/33, 3%) (P = 0.033, Fisher's exact test) (Figure S1, available as Supplementary data at JAC Online). Finally, 3/33 (9%) of the tested S. flexneri isolates and 1/72 (1%) of tested S. sonnei isolates were PCR amplification positive for the qnrS gene. No parC mutations, qnrA, qnrB, aac(6′)-Ib-cr or qepA genes were identified within any of the Shigella isolates.
Table 2.
Comparison of MICs of fluoroquinolones for S. sonnei and S. flexneri isolates
| Antimicrobial | S. sonnei MIC (mg/L), n = 72 | S. flexneri MIC (mg/L), n = 33 | Pa |
|---|---|---|---|
| Nalidixic acid | |||
| median (range) | 48 (0.09–512) | 512 (1–512) | 0.046 |
| geometric mean | 35.3 | 68.4 | |
| Ciprofloxacin | |||
| median (range) | 0.064 (0.01–0.25) | 0.125 (0.01–8) | 0.011 |
| geometric mean | 0.05 | 0.08 | |
| Gatifloxacin | |||
| median (range) | 0.094 (0.01–0.25) | 0.19 (0.01–6) | 0.008 |
| geometric mean | 0.06 | 0.09 | |
| Ofloxacin | |||
| median (range) | 0.38 (0.05–1) | 0.38 (0.05–16) | 0.135 |
| geometric mean | 0.25 | 0.3 | |
aP value comparing MIC between species by Kruskal–Wallis test.
Figure 1.
MICs (mg/L) of a range of antimicrobials for the Shigella isolates in this study (log2 scale). Box plots show the median (black line across each box) and the 5th and 95th percentiles. MICs for S. sonnei are shown in dark grey and MICs for S. flexneri are in light grey. The broken line in each plot represents the current CLSI breakpoint for resistance.11
The MICs of fluoroquinolones were highly correlated for both S. sonnei and S. flexneri (P < 0.001 for all correlations). When normalized to the mean of the current CLSI resistance breakpoint (ciprofloxacin: ≥4 mg/L, gatifloxacin: ≥8 mg/L),11 both S. flexneri and S. sonnei had higher relative median log2 MICs of ciprofloxacin than of gatifloxacin (P < 0.001 in both cases, Kruskal–Wallis test of z-score) (Figure S2). Furthermore, the presence of a single gyrA mutation and/or the qnrS gene dramatically increased the MICs of fluoroquinolones for both S. sonnei and S. flexneri (Table 3 and Figure S3).
Table 3.
MICs of fluoroquinolones for S. sonnei and S. flexneri by gyrA mutation and qnrS gene, median (range)
| Antimicrobial |
S. sonnei MIC (mg/L) |
S. flexneri MIC (mg/L) |
||||||
|---|---|---|---|---|---|---|---|---|
| no mutation, n = 19 | A87T, n = 40 | S83L, n = 9 | S83L/qnrS, n = 1 | no mutation, n = 11 | S83L, n = 16 | qnrS, n = 1 | S83L/qnrS, n = 2 | |
| Nalidixic acid | 1.50 (0.09–3.0) | 64 (32–512) | 96 (64–512) | 512 | 2.0 (1.0–4.0) | 512 (512–512) | 1.50 | 512 (512–512) |
| Ciprofloxacin | 0.01 (0.01–0.02) | 0.06 (0.05–0.13) | 0.19 (0.13–0.25) | 0.25 | 0.02 (0.01–0.05) | 0.19 (0.09–0.25) | 0.01 | 4.13 (0.25–8.0) |
| Gatifloxacin | 0.01 (0.01–0.13) | 0.09 (0.06–0.13) | 0.19 (0.13–0.25) | 0.19 | 0.01 (0.01–0.06) | 0.19 (0.13–0.38) | 0.01 | 3.13 (0.25–6.0) |
| Ofloxacin | 0.06 (0.05–0.09) | 0.38 (0.25–0.50) | 0.75 (0.5–1.0) | 0.75 | 0.09 (0.05–0.19) | 0.5 (0.38–1.0) | 0.05 | 8.25 (0.5–16) |
All pairwise comparisons within species across antimicrobials were statistically significantly different by Kruskal–Wallis tests (all P < 0.001) and all pairwise comparisons of A87T and S83L (regardless of qnrS) in S. sonnei were statistically significantly different (P < 0.001), except for A87T versus S83L for nalidixic acid (P = 0.03), by Kruskal–Wallis test.
Time–kill curves
Mutations in gyrA, the presence of qnrS and elevated MICs of ciprofloxacin substantially increased the ability of S. flexneri to replicate in the presence of ciprofloxacin (Figure 2). The S. flexneri strain lacking a gyrA mutation with a low MIC of ciprofloxacin was rapidly killed by the antimicrobial (Figure 2). While the isolate with the gyrA mutation A87T (Figure 2) was still effectively killed at higher concentrations of ciprofloxacin, bacterial growth remained elevated at lower concentrations compared with the S. flexneri without a gyrA mutation. The S83L mutation (Figure 2) reduced the bactericidal activity of ciprofloxacin even further, with cfu/mL reduced by >5 logs at high concentrations. Finally, the S. flexneri isolate with an S83L gyrA mutation, qnrS gene and MIC of ciprofloxacin of 8 mg/L (Figure 2) was able to grow in the presence of all tested concentrations of ciprofloxacin during the first hour post-inoculation, with growth falling by 2 logs only within the second hour post-inoculation. Further, the concentration-dependent killing effect was lost with the qnrS mutant, as the 2× to 16× MIC curves exhibited almost identical profiles. The same pattern was observed with the corresponding gatifloxacin time–kill curve (data not shown).
Figure 2.
Time–kill curves of S. flexneri gyrA (qnrS) genotypes on exposure to increasing concentrations of ciprofloxacin. Plots showing the mean time–kills of S. flexneri isolates grown with increasing concentrations of ciprofloxacin based on the MIC for the original isolate at different timepoints post-inoculation (log2 scale). The S. flexneri isolates are: (a) no gyrA mutation, ciprofloxacin MIC = 0.023 mg/L; (b) gyrA mutation A87T, ciprofloxacin MIC = 0.094 mg/L; (c) gyrA mutation S83L, ciprofloxacin MIC = 0.19 mg/L; and (d) gyrA mutation S83L, ciprofloxacin MIC = 8 mg/L with the qnrS gene. The broken line represents the limit of detection for the assay (10 cfu/mL).
Clinical outcome in shigellosis with fluoroquinolone therapy
Overall, there were 8 (7.6%) failures among the 105 Shigella cases, all of which were S. sonnei (8/72, 11%). Two of these cases were microbiological failures and six were classified as clinical failures. Overall failure amongst the S. sonnei isolates was not significantly associated with either treatment drug (OR: 0.5, 95% CI: 0.1–2.3, P = 0.37) or MIC of nalidixic acid, ciprofloxacin, gatifloxacin or ofloxacin. Additionally, although 5/8 (63%) failures had the A87T gyrA mutation and 1/8 (13%) had the S83L gyrA mutation, there was no significant effect of a gyrA mutation on failure (OR: 1.03, 95% CI: 0.3–3.3, P = 0.96).
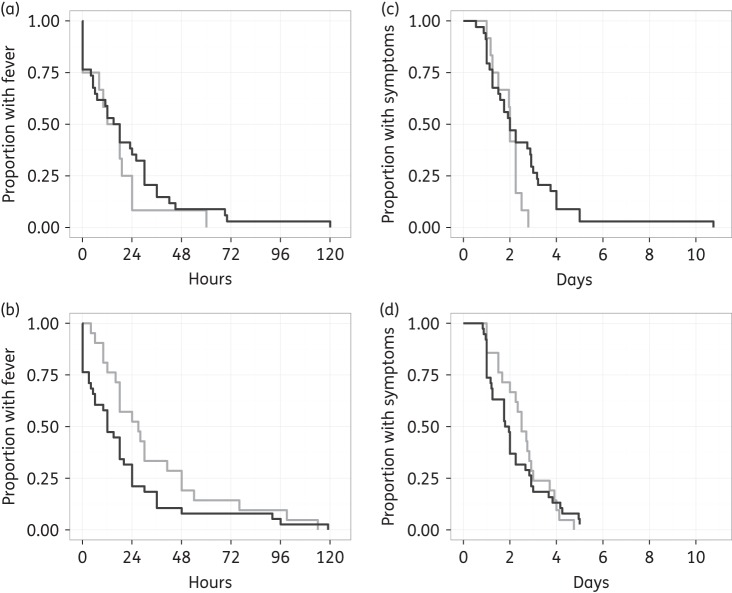
Overall, the FCTs of S. sonnei and S. flexneri were not significantly different (HR: 1.2, 95% CI: 0.8–1.8, P = 0.47). There was no difference in FCT between the two species in the ciprofloxacin arm (HR: 0.73, 95% CI: 0.4–1.4, P = 0.355) and although S. flexneri patients had moderately longer FCTs in the gatifloxacin arm (median: 27 h, IQR: 16–48) compared with S. sonnei (median: 12 h, IQR: 3–24), the difference was not statistically significant after controlling for age (HR: 1.52, 95% CI: 0.9–2.7, P = 0.13) (Figure 3a and b). Notably, the FCTs of S. flexneri treated with gatifloxacin (median: 27 h, IQR: 16–48) were significantly longer than for S. flexneri patients treated with ciprofloxacin (median: 15, IQR: 4–22) (HR: 0.40, 95% CI: 0.2–0.9, P = 0.02). Similar patterns were observed for total duration of symptoms (Figure 3c and d), as S. flexneri patients treated with gatifloxacin (median: 60 h, IQR: 4–72) had significantly longer duration of symptoms compared with S. flexneri patients treated with ciprofloxacin (median: 48 h, IQR: 33–54) (HR: 0.39, 95% CI: 0.2–0.9, P = 0.02). No difference in total duration of symptoms was detected between the treatment arms for S. sonnei patients. The presence of gyrA mutations did not have a significant effect on either FCT or total duration of symptoms (P > 0.05 for both comparisons).
Figure 3.
Clinical outcome comparison between S. sonnei and S. flexneri infections treated with fluoroquinolones. Unadjusted Kaplan–Meier plots showing FCT in hours in patients treated with (a) ciprofloxacin (n = 46) and (b) gatifloxacin (n = 59). Total duration of symptoms in days is shown in patients treated with (c) ciprofloxacin and (d) gatifloxacin. S. sonnei are shown in dark grey and S. flexneri are shown in light grey.
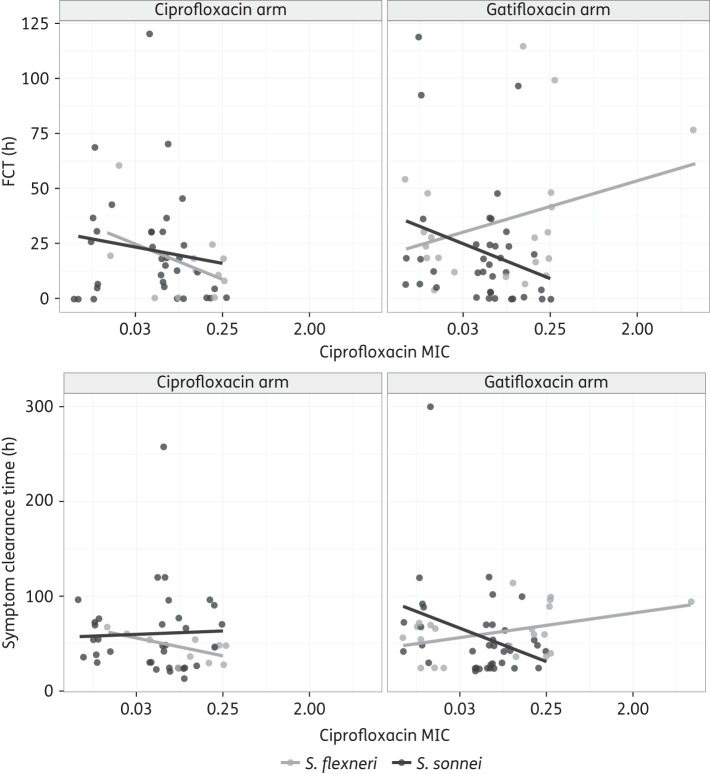
When evaluating the effect of MICs on FCT and duration of symptoms, we found a significant interaction between treatment arm and gatifloxacin and ciprofloxacin MICs for S. flexneri patients (P < 0.03 in all comparisons). Although increasing ciprofloxacin MICs appeared to be associated with increased FCT and symptom duration in S. flexneri patients treated with gatifloxacin (Figure 4), these trends were not statistically significant (effect of log2 ciprofloxacin MIC on (i) FCT: HR: 0.92 (95% CI: 0.8–1.1, P = 0.33); and (ii) symptom duration: HR: 0.86 (95% CI: 0.7–1.1, P = 0.15). This pattern was comparable for the gatifloxacin MIC (data not shown). Finally, there was no significant effect of MIC of either antimicrobial in the ciprofloxacin treatment arm or amongst S. sonnei patients in either treatment arm.
Figure 4.
Effect of MIC of ciprofloxacin (mg/L) on clinical outcome for S. sonnei and S. flexneri infections. Associations between ciprofloxacin and FCT (top two plots) and symptom clearance time (bottom two plots) are shown. Patients treated with ciprofloxacin are shown on the left and patients treated with gatifloxacin are on the right. Patients infected with S. sonnei are shown in dark grey and patients infected with S. flexneri are in light grey. The lines represent the best-fit linear model for each set of patients.
Discussion
The bacterial genus Shigella are a considerable public health problem, responsible for >7 million disability-adjusted life years and >100 000 deaths annually, which are mainly concentrated in children living in developing countries.1,2,28,29 The current drug of choice for treating Shigella infections is ciprofloxacin. However, fluoroquinolone resistance is threatening to make the management of this diarrhoeal pathogen even more challenging.15 Here, we aimed to quantify the impact of treatment choice, fluoroquinolone MIC and presence of gyrA mutations on clinical outcome of paediatric S. flexneri and S. sonnei patients treated with fluoroquinolones. We additionally evaluated the ability of Shigella strains of varying resistance profiles to grow in the presence of ciprofloxacin through time–kill experiments rather than single timepoint MIC testing.
While we hypothesized that poorer clinical outcome was associated with higher fluoroquinolone MIC and observed this trend in our data, we were unable to conclude that such relationships were statistically significant. This is likely due to small numbers of patients and relatively low MICs for isolates collected during the time period (2006–09). Mutations in gyrA were not found to be associated with an inferior clinical outcome in either species, although S83L and A87T mutations did result in isolates with higher MICs of fluoroquinolones. We hypothesize that fluoroquinolone MICs may play a greater role in clinical outcome above the CLSI breakpoint as we have shown conclusively that below the current breakpoints, MIC does not have a significant impact on patient outcome. Therefore, we surmise that the current CLSI breakpoints for fluoroquinolones are appropriate for Shigella therapy.
We identified differences in clinical response between both the Shigella species and treatment arms. S. flexneri patients had significantly longer FCTs and duration of symptoms when treated with gatifloxacin compared with ciprofloxacin. Gatifloxacin is presumed to be a more efficacious drug than ciprofloxacin,30 yet S. flexneri isolates were more likely to have higher MICs of ciprofloxacin (relative to the CLSI breakpoint) compared with gatifloxacin. In the absence of additional data assessing the penetration of gatifloxacin into the epithelial cells lining the gastrointestinal tract, we hypothesize this difference between antimicrobials in S. flexneri patients may be due to more effective killing of the local commensal gut microbiota by gatifloxacin, which may allow Shigella to more efficiently invade the gut tissue.31–33 Further investigation into the effects of fluoroquinolones on the gut microbiota are required.
The time–kill experiments clearly demonstrate that an elevated MIC, and most notably presence of the combination of a qnrS gene and a gyrA mutation, allows Shigella to replicate efficiently in high concentrations of fluoroquinolone. We hypothesize that such enhanced persistence in the gastrointestinal tract during therapy endows fluoroquinolone-resistant Shigella with a competitive advantage over fluoroquinolone-susceptible strains. AMR Shigella are not only more likely to survive therapy but also to be shed and further transmitted in the community, over time replacing the fluoroquinolone-susceptible strains. For example, the acquisition of chloramphenicol resistance is understood to have afforded certain clones of Salmonella enterica serovar Typhimurium the ability to replace susceptible strains in sub-Saharan Africa over several years.34 Furthermore, there is limited evidence that suggests that an elevated MIC of nalidixic acid lengthens the duration of excretion of Shigella.35
The time–kill results also suggest that acquisition of qnrS in addition to an S83L mutation in gyrA may lead to a loss of concentration-dependent killing by fluoroquinolones. This has been observed previously in Escherichia coli with the qnrS gene36 and may relate to the mechanism of action of qnrS whereby peptides bind to the DNA gyrase and block fluoroquinolone activity. Although higher concentrations of fluoroquinolones are generally preferred in therapy to reduce the likelihood of development of resistant strains,37,38 qnrS mutants may not respond to high doses given our time–kill observations. Therefore, combination therapy in patients with qnrS-positive Shigella infections could be warranted. Further pharmacodynamic work on qnrS mutants in the Enterobacteriaceae is clearly required.
This study has some limitations. First, FCT and duration of symptoms may not be the most appropriate measures of clinical outcome. We therefore may have under- or overestimated the effect of fluoroquinolone resistance on patient outcome. Second, we had a limited range of fluoroquinolone MICs and a limited number of treatment failures, which made it difficult to better evaluate trends in the isolates with elevated MICs or in those that failed treatment. It will be important to repeat this work with contemporary strains as higher levels of fluoroquinolone resistance will permit such an investigation.39 Furthermore, collection of longitudinal stool samples after treatment is warranted to understand the effect of resistance on excretion duration. Nonetheless, the work presented here offers the first rigorous analysis of fluoroquinolone resistance on patient outcome to our knowledge and fills an important gap in the knowledge of this increasingly antimicrobial-resistant pathogen.
We conclude that below the CLSI breakpoint, MICs of fluoroquinolones do not strongly impact patient outcome in shigellosis. Therefore, the current CLSI breakpoints are warranted for Shigella infections. However, our data suggest that the choice of fluoroquinolone is important in the management of Shigella infections, as S. flexneri patients treated with gatifloxacin had poorer outcomes compared with those treated with ciprofloxacin. Further, we demonstrate that qnrS-harbouring Shigella are able to grow effectively in vitro at high concentrations of ciprofloxacin and hypothesize that fluoroquinolone-resistant strains outcompete susceptible strains, as they are maintained during therapy, shed and therefore more likely to be transmitted in the community. Continued evaluation of the impact of fluoroquinolone resistance on the clinical outcome of Shigella patients over time is critical to help inform clinical treatment decisions for diarrhoeal infections.
Funding
This work was supported by the Wellcome Trust of the UK, through core funding (089276/2/09/2). S. B. is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and the Royal Society (100087/Z/12/Z).
Transparency declarations
None to declare.
Supplementary data
Figures S1 to S3 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
We wish to thank all the staff of ward B at the Hospital for Tropical Diseases in Ho Chi Minh City and at Huu Nghi Hospital in Dong Thap province for assisting in sample collection, data collection and patient care, and Ms Song Chau for her ongoing efforts.
References
- 1.Kotloff KL, Nataro JP, Blackwelder WC et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case–control study. Lancet 2013; 382: 209–22. [DOI] [PubMed] [Google Scholar]
- 2.Platts-Mills JA, Babji S, Bodhidatta L et al. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health 2015; 3: e564–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thompson CN, Thanh DP, Baker S. The rising dominance of Shigella sonnei: an intercontinental shift in the etiology of bacillary dysentery. PLoS Negl Trop Dis 2015; 9: e0003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. The Treatment of Diarrhoea: A Manual for Physicians and other Senior Health Workers. Geneva, 2005. whqlibdoc.who.int/publications/2005/9241593180.pdf. [Google Scholar]
- 5.Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother 2005; 56: 463–9. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother 2003; 51: 1109–17. [DOI] [PubMed] [Google Scholar]
- 7.Morgan-Linnell SK, Zechiedrich L. Contributions of the combined effects of topoisomerase mutations toward fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother 2007; 51: 4205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heisig P, Tschorny R. Characterization of fluoroquinolone-resistant mutants of Escherichia coli selected in vitro. Antimicrob Agents Chemother 1994; 38: 1284–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hooper DC, Jacoby GA. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci 2015; 1354: 12–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodríguez-Martínez JM, Cano ME, Velasco C et al. Plasmid-mediated quinolone resistance: an update. J Infect Chemother 2011; 17: 149–82. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-fourth Informational Supplement M100-S24. CLSI, Wayne, PA, USA, 2014. [Google Scholar]
- 12.Bowen A, Hurd J, Hoover C et al. Importation and domestic transmission of Shigella sonnei resistant to ciprofloxacin—United States, May 2014–February 2015. Morb Mortal Wkly Rep 2015; 64: 318–20. [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JS, Kim JJ, Kim SJ et al. Outbreak of ciprofloxacin-resistant Shigella sonnei associated with travel to Vietnam, Republic of Korea. Emerg Infect Dis 2015; 21: 1247–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Lappe N, O'Connor J, Garvey P et al. Ciprofloxacin-resistant Shigella sonnei associated with travel to India. Emerg Infect Dis 2015; 21: 894–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO. Antimicrobial Resistance: Global Report on Surveillance. Geneva, 2014. http://www.who.int/drugresistance/documents/surveillancereport/en/. [Google Scholar]
- 16.Travers K, Barza M. Morbidity of infections caused by antimicrobial-resistant bacteria. Clin Infect Dis 2002; 34 Suppl 3: S131–4. [DOI] [PubMed] [Google Scholar]
- 17.Nelson JM, Smith KE, Vugia DJ et al. Prolonged diarrhea due to ciprofloxacin-resistant Campylobacter infection. J Infect Dis 2004; 190: 1150–7. [DOI] [PubMed] [Google Scholar]
- 18.Smith KE, Besser JM, Hedberg CW et al. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998. N Engl J Med 1999; 340: 825–31. [DOI] [PubMed] [Google Scholar]
- 19.Vinh H, Nhu NTK, Nga TVT et al. A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect Dis 2009; 9: 204–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vinh H, Anh VTC, Anh ND et al. A multi-center randomized trial to assess the efficacy of gatifloxacin versus ciprofloxacin for the treatment of shigellosis in Vietnamese children. PLoS Negl Trop Dis 2011; 5: e1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weigel LM, Steward CD, Tenover FC. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob Agents Chemother 1998; 42: 2661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talukder KA, Khajanchi BK, Islam MA et al. Fluoroquinolone resistance linked to both gyrA and parC mutations in the quinolone resistance-determining region of Shigella dysenteriae type 1. Curr Microbiol 2006; 52: 108–11. [DOI] [PubMed] [Google Scholar]
- 23.Vien LTM, Minh NNQ, Thuong TC et al. The co-selection of fluoroquinolone resistance genes in the gut flora of Vietnamese children. PLoS One 2012; 7: e42919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eun SK, Jeong JY, Jun JB et al. Prevalence of aac(6′)-Ib-cr encoding a ciprofloxacin-modifying enzyme among Enterobacteriaceae blood isolates in Korea. Antimicrob Agents Chemother 2009; 53: 2643–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cattoir V, Poirel L, Nordmann P. Plasmid-mediated quinolone resistance pump QepA2 in an Escherichia coli isolate from France. Antimicrob Agents Chemother 2008; 52: 3801–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chau TT, Campbell JI, Galindo CM et al. Antimicrobial drug resistance of Salmonella enterica serovar Typhi in Asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob Agents Chemother 2007; 51: 4315–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer, 2009. [Google Scholar]
- 28.Lozano R, Naghavi M, Foreman K et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380: 2095–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray CJL, Vos T, Lozano R et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380: 2197–223. [DOI] [PubMed] [Google Scholar]
- 30.Lu T, Zhao X, Drlica K. Gatifloxacin activity against quinolone-resistant gyrase: allele-specific enhancement of bacteriostatic and bactericidal activities by the C-8-methoxy group. Antimicrob Agents Chemother 1999; 43: 2969–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grasela DM. Clinical pharmacology of gatifloxacin, a new fluoroquinolone. Clin Infect Dis 2000; 31 Suppl 2: S51–8. [DOI] [PubMed] [Google Scholar]
- 32.Pédron T, Sansonetti P. Commensals, bacterial pathogens and intestinal inflammation: an intriguing ménage à trois. Cell Host Microbe 2008; 3: 344–7. [DOI] [PubMed] [Google Scholar]
- 33.Lupp C, Robertson ML, Wickham ME et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007; 2: 119–29. [DOI] [PubMed] [Google Scholar]
- 34.Okoro CK, Kingsley RA, Connor TR et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet 2012; 44: 1215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vinh H, Wain J, Chinh MT et al. Treatment of bacillary dysentery in Vietnamese children: two doses of ofloxacin versus 5-days nalidixic acid. Trans R Soc Trop Med Hyg 2000; 94: 323–6. [DOI] [PubMed] [Google Scholar]
- 36.Cengiz M, Sahinturk P, Sonal S et al. In vitro bactericidal activity of enrofloxacin against gyrA mutant and qnr-containing Escherichia coli isolates from animals. Vet Rec 2013; 172: 474. [DOI] [PubMed] [Google Scholar]
- 37.Linde HJ, Lehn N. Mutant prevention concentration of nalidixic acid, ciprofloxacin, clinafloxacin, levofloxacin, norfloxacin, ofloxacin, sparfloxacin or trovafloxacin for Escherichia coli under different growth conditions. J Antimicrob Chemother 2004; 53: 252–7. [DOI] [PubMed] [Google Scholar]
- 38.Randall LP, Cooles SW, Piddock LJV et al. Mutant prevention concentrations of ciprofloxacin and enrofloxacin for Salmonella enterica. J Antimicrob Chemother 2004; 54: 688–91. [DOI] [PubMed] [Google Scholar]
- 39.Thompson CN, Phan Vu Tra M, Nguyen Van Minh H et al. A prospective multi-center observational study of children hospitalized with diarrhea in Ho Chi Minh City, Vietnam. Am J Trop Med Hyg 2015; 95: 1045–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.