Abstract
Composite retrotransposons are widely distributed in the plant and animal kingdoms. Some of the most complex of these are found in hominoid primates. SVA, LAVA, PVA and FVA combine simple repeats, Alu fragments, a VNTR (Variable Number of Tandem Repeats) and variable 3′ domains, which are, except for PVA, derived from other retrotransposons. Although a likely precursor of SVA–a “tailed VNTR” named SVA2–had been identified in the Rhesus genome, the exact sequence and mechanism of the assembly of this type of composite retrotransposon had been elusive. The discovery of LAVA, PVA and FVA in gibbons provided the opportunity to delineate the order of assembly of the components of VNTR-containing retrotransposons. Our recent analysis suggests that an extinct “Alu-SVA2” acquired variant 3′ ends by splicing. In this commentary I will discuss the mode of assembly of VNTR composites in the context of their capacity to engage in alternative splicing to co-mobilize host RNA sequences and to become exonized. The second part will focus on structural determinants of VNTR composite retrotransposon mobilization in the context of lineage-specific expansion of particular families/subfamilies of these elements.
Keywords: retrotransposon, SVA, VNTR, hominoids, splicing
Abbreviations
- SVA
SINE-R-VNTR-Alu
- LAVA
L1-Alu-VNTR-Alu
- PVA
PTGR2-VNTR-Alu
- FVA
FRAM-VNTR-Alu
- VNTR
Variable Number of Tandem Repeats
- FRAM
Free Right Alu Monomer
- SINE
Short INterspersed Element
- L1
LINE-1.
VNTR Composites–Generation of New Families and New Exons through Splicing to Host Sequences
VNTR (Variable Number of Tandem Repeats) containing non-LTR retrotransposons are specific to hominoid primates. They are characterized by a shared 5′ hexameric (CCTCCC) repeat and Alu-like region, followed by a VNTR central core. The 3′ domain is family-specific and consists of endogenous retrovirus derived sequence (SINE-R in SVA1), an Alu and an L1 fragment separated by simple repeats (LA in LAVA2), exon 4 and part of intron 4 of the PTGR2 gene (in PVA3) or a partial FRAM element surrounded by non-repetitive sequence (in FVA4). SVA expanded in great apes. LAVA, PVA and FVA, by contrast, are found in gibbons only. SVA comprises a total of 2700 elements in humans, divided in 7 subfamilies (SVA_A to SVA_F15-8). Around 1800 and 2500 SVA elements are found in the orangutan9 and chimpanzee10 genomes, respectively. The genome of the Northern white cheeked gibbon, Nomascus leucogenys, harbours ca. 1800 LAVA copies,11 150 PVAs, 9 FVA elements and 29 SVAs.4 Through sequence analysis we could show that the variant 3′ ends of all 4 families of VNTR composites have been acquired by splicing–either to the VNTR or to the SVA2 unique 3′ sequence4 (Fig. 1A). The latter finding provides final proof that SVA2 is the precursor of this superfamily of non-LTR retrotransposons. In addition, it complements the analysis of Hancks and Kazazian who suggested splicing as the mechanism of assembly of the SVA 5′ Alu-like region12 and results independently obtained by 3 different groups providing evidence that the MAST2 derived sequence present in the SVA_F1 family members has been fused to the Alu-like region by splicing.6–8
Figure 1.
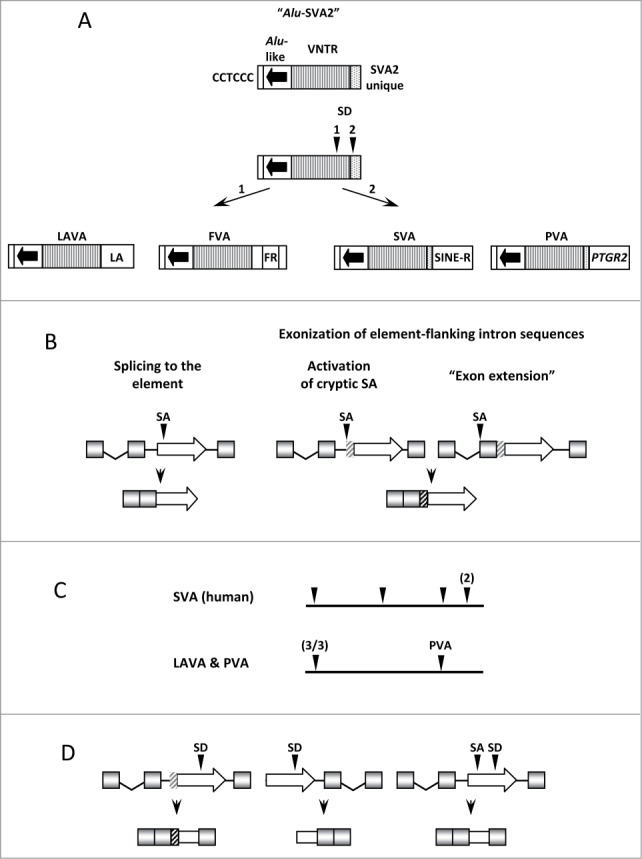
VNTR-containing retrotransposons and splicing. (A) The variant 3′ ends of VNTR composites have been acquired by splicing to either the VNTR (1 - in LAVA and FVA) or the SVA2 unique 3′ sequence (2 - in SVA and PVA) of an extinct “Alu-SVA2” SD–splice donor. (B) VNTR composites function as exon traps and generate new exons through splicing. Elements integrated in introns in sense use acceptor sites in the element, in introns and acceptor sites of the upstream exons. (C) Positions of splice acceptor sites (arrowheads) in the Alu-like regions of human SVA elements7 and gibbon LAVA11 and PVA4 elements. (D) SVAs can be exonized through splice donor sites located in the elements. Three different scenarios are shown: the splice donor is complemented by an intronic acceptor upstream of the element (left panel; the SVA constitutes the first exon (middle panel) and the element provides both splice acceptor and donor–parts of the element are exonized (right panel, all based on Kwon et al.13). Intronic sequences that become exonized are shown as gray and black hatched boxes, respectively.
The extraordinary propensity of VNTR composite RNAs to engage in alternative splicing became first obvious when Hancks and colleagues identified RNAs comprising SVA sequences as well as exons of cellular genes which had been spliced to them.8 In parallel, we identified SVAs containing spliced 5′ transductions resulting from the activation of SVA upstream cryptic splice acceptors in introns.7 In both cases, splicing to the SVA sequence and activation of upstream cryptic acceptor sites, new exons are generated (Fig. 1B, left and middle). The analysis of spliced RNAs transduced by LAVA11 and PVA4 elements uncovers a third variant of VNTR composite induced alternative splicing: extension of an existing exon mediated by elements residing either in an exon or in close proximity downstream of an exon (Fig. 1B, right). Interestingly, in cases where RNAs are spliced to the elements, LAVA and PVA differ from human SVA. Whereas in human SVA splice acceptor sites used are spread over the entire Alu-like region, LAVA and PVA preferentially use an acceptor at the 5′ end of the Alu-like region (Fig. 1C).
Exon trapping and 5′ transduction of spliced RNAs involves splice acceptors provided, activated or used by VNTR composites. A recent survey of intragenic SVA elements13 shows that they can also provide splice donor sites. In all cases reported the donor localizes to the VNTR region of the elements (Fig. 1D).
Taken together, there is “splicing all around” VNTR composites - leading to the emergence of new families as well as to the generation of new exons of cellular genes and shuffling of host sequences through transduction.
So far VNTR composites are the only known compound non-LTR retrotransposons that have been assembled by splicing. SINEs (Short INterspersed Elements), which show a modular organization, are transcribed by RNA polymerase III. Their transcripts are not targeted to the spliceosomal pathway. Mechanisms discussed for their assembly and the exchange of modules between different families include template switching of the reverse transcriptase and non-allelic SINE-SINE recombination/gene conversion. In squid, for example, the emergence of the chimeric Sepioth-SINE2A has been explained by template switching from Sepioth-SINE1 to Sepioth-SINE2B.14 Although non-allelic recombination has been extensively studied in the context of e.g. Alu non-traditional sequence evolution,15 there is, to my knowledge, no report providing evidence for the emergence of a new modular SINE family through this mechanism.
Lineage-Specific Expansion of VNTR Composite Families: More Questions than Answers - Yet
SVA elements are found in both gibbons and great apes. LAVA elements are present in gibbons and 5′ truncated copies were identified in the genomes of great apes2 (A. Damert, unpublished data). The “founder” elements of both families–SVA and LAVA–must therefore have been present in the common ancestor of gibbons and great apes. Intriguingly, SVA did not amplify to appreciable copy numbers in gibbons. LAVA, by contrast, did not proliferate in great apes. The molecular basis of these lineage-specific differences is not yet understood.
Proliferation of non-autonomous retrotransposons in a particular genome is dependent on their expression in the germline and/or early embryo and on their efficient interaction with the proteins synthesized from their autonomous partner. Mechanisms governing SVA/LAVA expression in germ cells are still elusive. The postulated element-internal promoter could not be identified to date. To what extent transcriptional repression by KRAB zinc-finger proteins and KAP1/TRIM28–recently demonstrated for SVA in human embryonic stem cells16,17–plays a role in regulation of VNTR composite expression in the germline and/or the early embryo remains to be seen. Efficient interaction of the RNA of non-autonomous elements with the proteins encoded by the autonomous partner (LINE-1 in case of VNTR composites) requires (i) co-localization in the same cellular compartment and (ii) specific sequence and/or structural features of the RNA that facilitate the contact. The only data available on sub-cellular localization of SVA RNA show its presence in nuclear aggregates and distributed throughout the cytoplasm.18 It is unclear, where the RNA interacts with the LINE-1 (L1) encoded proteins. Considerable progress has been made in the last years with regard to the determination of sequence/structural features enabling VNTR composites to be efficiently mobilized by the L1-encoded proteins. Hancks and colleagues identified the 5′ hexameric repeat/Alu-like domain as the minimal active human SVA.19 Our recent results suggest that a particular sequence-based structure of this domain is required for efficient mobilization of SVA. Surprisingly, this same structure was found to be non-functional when combined with LAVA-derived central VNTRs and 3′ regions. Based on this finding we concluded that structural determinants of mobilization differ for elements of the 2 families.4 Subsequent experiments using domain swaps between active and inactive LAVA elements revealed that key determinants of their mobilization potential reside in the central VNTR domain.20 Thus, although the modular organization of LAVA and SVA is similar, the function of the individual modules differs between the families.
Differences in their RNA structure combined with the activity of specific L1 subfamilies could provide a starting point to explain the lineage-specific expansion of particular VNTR composite families (Fig. 2). The prototype SVA and LAVA elements already assembled in the common ancestor of gibbons and great apes are characterized by an ancestral Alu-like region. This type of the Alu-like domain persists in great ape SVA_A and its gibbon counterpart SVANLE. It is also found in the gibbon-specific PVA and FVA families and appears to confer only moderate mobilization potential–as judged by copy numbers and retrotransposition rates obtained in vitro.4 In gibbons there is no further evolution of this domain in SVAs. In the lineage leading to the great apes major changes in the SVA structure occur only after the split of orangutans: a 5 bp duplication at its 5′ end and an 11 bp deletion give rise to the structure of the Alu-like region as it is found in all evolutionary younger chimpanzee and human SVAs.5 Concurrently to the changes in the Alu-like region, longer repeat units and repeat unit arrays emerge in the VNTR20 and the SVA2-derived sequence upstream of the SINE-R is lost.5 Taken together, these complex changes eventually give rise to the highly organized SVA structure found in currently active human and chimpanzee subfamilies. In gibbons, by contrast, SVA remains largely silent–possibly due to the persistence of the ancestral-type Alu-like region.
Figure 2.
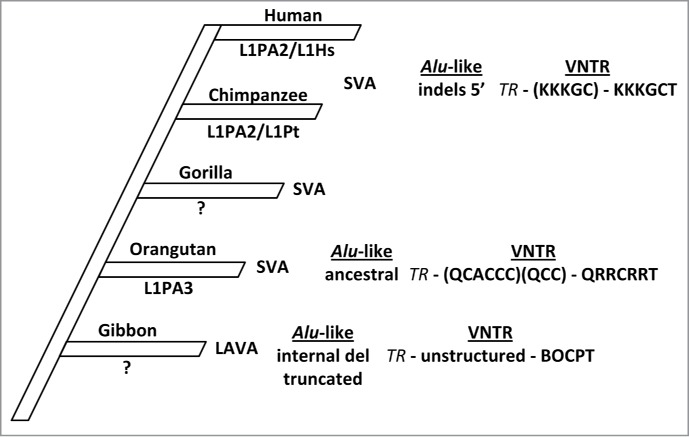
Lineage-specific evolution of VNTR composite retrotransposons in hominoid primates. Specific characteristics of the Alu-like domain and the central VNTR are shown for the evolutionary youngest and presumably currently active VNTR-containing retrotransposon subfamilies in each of the lineages. TR indicates the invariable 5′ part of the central domain that is conserved within subfamilies. Upper case letters designate VNTR repeat unit types as identified in Lupan et al.20 Internal arrays of repeat units are shown in parentheses. The currently active L1 subfamilies are indicated below the lineage name. There are no data available for gorilla. del–deletions.
Against the background that the ancestral type Alu-like region supports only moderate mobilization rates in vitro, the lineage-specific amplification of SVAs carrying this type of 5′ domain in orangutans is the currently most puzzling observation. If lineage-specific insertions per million years are taken as proxy, then SVA has been about 3 quarter as active in orangutans as in humans.9 It could be possible that the specific structure of the VNTR that developed in orangutans20 contributed to the expansion of SVA in this lineage. Alternatively, host factors involved in mobilization might have evolved in orangutan in a way as to facilitate efficient retrotransposition of the elements.
In LAVA a first 10bp deletion in the Alu-like domain occurs early on in evolution. This is followed by a larger internal deletion of 32bp in LAVA_C. The final step in evolution to LAVA_F1 reduces the Alu-like region to only about half of its original length.4,11 This rudimentary domain has only a modulatory function in retrotransposition. Overall, the structural requirements of LAVA mobilization appear to be less stringent than those of human/chimpanzee SVAs: a particular structure of the 5′ domain does not appear to be necessary. The central VNTR, much less organized than that found in SVAs, appears to play a critical role. Whether a specific arrangement of internal repeat units or simply a minimum length make up an “active” VNTR cannot be decided based on the data available. However, all data on mobilization determinants of LAVA have been obtained using a human L1 element in a human cell line.4,20 Considering the evolutionary distance between gibbons and humans and the fact that most likely a different L1 subfamily is active in gibbons than in humans, the results obtained so far might not reflect the “real” requirements for retrotransposition of LAVA by its–as yet uncharacterized–gibbon LINE-1 partner.
In orangutans, chimpanzees and humans different subfamilies of L1 are assumed to be currently active based on the presence of species-specific insertions (or have been active in the not so far past). L1Pt/L1PA2 and L1Hs/L1PA2 copies are found to be still polymorphic in chimpanzees and humans, respectively.21 L1PA3 elements dominate the landscape of mobile element insertions in orangutans.22 There are no published data on gibbon-specific L1 subfamilies. It is possible that L1 subfamilies display differential affinity for the different structures found in human/chimpanzee SVA, orangutan SVA and gibbon LAVA. In 2012 Wagstaff and colleagues addressed the question of a preferential association of Alu–the other non-autonomous, non-LTR retrotransposon present in hominoid genomes–and L1 subfamilies. Using reconstructed L1 elements from extinct subfamilies in a present-day human cellular background, they found only limited evidence for co-evolution with Alu and considered it likely that host factors influence the interaction between the autonomous and the non-autonomous elements.23 It will be interesting to see how efficiently L1 elements derived from different hominoids can mobilize different VNTR composites in heterologous as well as homologous cell systems. The prerequisite for such experiments is now given with the establishment of a protocol for the derivation of retrotransposition-competent induced pluripotent stem cells from non-human primate fibroblasts.24
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon-intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem 1994; 269(11):8466-76; PMID:8132574 [PubMed] [Google Scholar]
- 2.Carbone L, Harris RA, Mootnick AR, Milosavljevic A, Martin DI, Rocchi M, Capozzi O, Archidiacono N, Konkel MK, Walker JA et al.. Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome Biol Evol 2012; 4(7):648-58; PMID:22593550; http://dx.doi.org/ 10.1093/gbe/evs048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hara T, Hirai Y, Baicharoen S, Hayakawa T, Hirai H, Koga A. A novel composite retrotransposon derived from or generated independently of the SVA (SINE/VNTR/Alu) transposon has undergone proliferation in gibbon genomes. Genes Genet Syst 2012; 87(3):181-90; http://dx.doi.org/ 10.1266/ggs.87.181 [DOI] [PubMed] [Google Scholar]
- 4.Ianc B, Ochis C, Persch R, Popescu O, Damert A. Hominoid composite non-LTR retrotransposons-variety, assembly, evolution, and structural determinants of mobilization. Mol Biol Evol 2014; 31(11):2847-64; PMID:25216663; http://dx.doi.org/ 10.1093/molbev/mst256 [DOI] [PubMed] [Google Scholar]
- 5.Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: a hominid-specific retroposon family. J Mol Biol 2005; 354(4):994-1007; PMID:16288912; http://dx.doi.org/ 10.1016/j.jmb.2005.09.085 [DOI] [PubMed] [Google Scholar]
- 6.Bantysh OB, Buzdin AA. Novel family of human transposable elements formed due to fusion of the first exon of gene MAST2 with retrotransposon SVA. Biochemistry (Mosc) 2009; 74(12):1393-9; PMID:19961423; http://dx.doi.org/ 10.1134/S0006297909120153 [DOI] [PubMed] [Google Scholar]
- 7.Damert A, Raiz J, Horn AV, Lower J, Wang H, Xing J, Batzer MA, Lower R, Schumann GG. 5′-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res 2009; 19(11):1992-2008; PMID:19652014; http://dx.doi.org/ 10.1101/gr.093435.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hancks DC, Ewing AD, Chen JE, Tokunaga K, Kazazian HH Jr. Exon-trapping mediated by the human retrotransposon SVA. Genome Res 2009; 19(11):1983-91; PMID:19635844; http://dx.doi.org/ 10.1101/gr.093153.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P et al.. Comparative and demographic analysis of orang-utan genomes. Nature 2011; 469(7331):529-33; PMID:21270892; http://dx.doi.org/ 10.1038/nature09687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CSAC . Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 2005; 437(7055):69-87; PMID:16136131; http://dx.doi.org/ 10.1038/nature04072 [DOI] [PubMed] [Google Scholar]
- 11.Carbone L, Harris RA, Gnerre S, Veeramah KR, Lorente-Galdos B, Huddleston J, Meyer TJ, Herrero J, Roos C, Aken B et al.. Gibbon genome and the fast karyotype evolution of small apes. Nature 2014; 513(7517):195-201; PMID:25209798; http://dx.doi.org/ 10.1038/nature13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hancks DC, Kazazian HH Jr. SVA retrotransposons: Evolution and genetic instability. Semin Cancer Biol 2010; 20(4):234-45; PMID:20416380; http://dx.doi.org/ 10.1016/j.semcancer.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon YJ, Choi Y, Eo J, Noh YN, Gim JA, Jung YD, Lee JR, Kim HS. Structure and Expression Analyses of SVA Elements in Relation to Functional Genes. Genomics Inform 2013; 11(3):142-8; PMID:24124410; http://dx.doi.org/ 10.5808/GI.2013.11.3.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akasaki T, Nikaido M, Nishihara H, Tsuchiya K, Segawa S, Okada N. Characterization of a novel SINE superfamily from invertebrates: “Ceph-SINEs” from the genomes of squids and cuttlefish. Gene 2010; 454(1–2):8-19; PMID:19914361; http://dx.doi.org/ 10.1016/j.gene.2009.11.005 [DOI] [PubMed] [Google Scholar]
- 15.Roy-Engel AM, Carroll ML, El-Sawy M, Salem AH, Garber RK, Nguyen SV, Deininger PL, Batzer MA. Non-traditional Alu evolution and primate genomic diversity. J Mol Biol 2002; 316(5):1033-40; PMID:11884141; http://dx.doi.org/ 10.1006/jmbi.2001.5380 [DOI] [PubMed] [Google Scholar]
- 16.Turelli P, Castro-Diaz N, Marzetta F, Kapopoulou A, Raclot C, Duc J, Tieng V, Quenneville S, Trono D. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res 2014; 24(8):1260-70; PMID:24879559; http://dx.doi.org/ 10.1101/gr.172833.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 2014; 516(7530): 242-5; http://dx.doi.org/ 10.1038/nature13760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodier JL, Mandal PK, Zhang L, Kazazian HH Jr. Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum Mol Genet 2010; 19(9):1712-25; PMID:20147320; http://dx.doi.org/ 10.1093/hmg/ddq048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancks DC, Mandal PK, Cheung LE, Kazazian HH Jr. The minimal active human SVA retrotransposon requires only the 5'-hexamer and Alu-like domains. Mol Cell Biol 2012; 32(22):4718-26; PMID:23007156; http://dx.doi.org/ 10.1128/MCB.00860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupan I, Bulzu P, Popescu O, Damert A. Lineage specific evolution of the VNTR composite retrotransposon central domain and its role in retrotransposition of gibbon LAVA elements. BMC Genomics 2015; 16:389; PMID:25981446; http://dx.doi.org/ 10.1186/s12864-015-1543-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J, Cordaux R, Han K, Wang J, Hedges DJ, Liang P, Batzer MA. Different evolutionary fates of recently integrated human and chimpanzee LINE-1 retrotransposons. Gene 2007; 390(1–2):18-27; PMID:17055192; http://dx.doi.org/ 10.1016/j.gene.2006.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gokcumen O, Tischler V, Tica J, Zhu Q, Iskow RC, Lee E, Fritz MH, Langdon A, Stutz AM, Pavlidis P et al.. Primate genome architecture influences structural variation mechanisms and functional consequences. Proc Natl Acad Sci U S A 2013; 110(39):15764-9; PMID:24014587; http://dx.doi.org/ 10.1073/pnas.1305904110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagstaff BJ, Kroutter EN, Derbes RS, Belancio VP, Roy-Engel AM. Molecular reconstruction of extinct LINE-1 elements and their interaction with nonautonomous elements. Mol Biol Evol 2012; 30(1):88-99; PMID:22918960; http://dx.doi.org/ 10.1093/molbev/mss202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchetto MC, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola AC, Desai KN, Herai RH, Weitzman MD et al.. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 2013; 503(7477):525-9; PMID:24153179; http://dx.doi.org/ 10.1038/nature12686 [DOI] [PMC free article] [PubMed] [Google Scholar]


