Ca2+ signals provide an evolutionarily conserved and crucial means of communication and coordination within animal cells. Unlike other critical messengers in cells, Ca2+ cannot be synthesized or degraded. Instead, it must be sequestered within and exported from cells by powerful Ca2+ pumps, and then delivered with extreme temporal and spatial precision through accurately controlled Ca2+ channels. Prominent Ca2+ signals in excitable cells trigger rapid contractile or excitatory responses through the opening of Ca2+ channels in the endoplasmic reticulum (ER) and/or plasma membranes (PM) in response to voltage or neurotransmitters. More subtle but equally vital Ca2+ signals are generated in all cell types in the form of oscillating Ca2+ waves or spikes that emanate from Ca2+ stores and are mediated and maintained by a complex array of sensing and signaling proteins (Berridge et al., 2003). In this issue of Molecular Cell, Kar and Parekh (2015) reveal the remarkable intricacy and accuracy of these Ca2+ signals in differentially controlling the function of closely related transcription factors.
Ca2+ signals are unique also in being able to impose “frequency-modulated” responses within cells (Berridge et al., 2003). Periodic release of Ca2+ from ER stores in response to receptor-induced production of the powerful second messenger, inositol 1,4,5-trisphosphate (InsP3), results in pulses or spikes of Ca2+ which rapidly traverse cells (Fig. 1). The frequency of these Ca2+ oscillations reflects receptor occupancy, providing an accurate “digital” response in cells, and one that can penetrate and coordinate events across the cell with remarkable fidelity (Brandman and Meyer, 2008). Ca2+ spike generation, depletes ER stored Ca2+, and triggers a second axis of Ca2+ signal transduction, mediated by the ubiquitous ER STIM proteins (Soboloff et al., 2012). STIMs are accurate ER luminal Ca2+ sensors which, triggered by lowered ER luminal Ca2+, undergo activation and migrate into ER-PM junctions where they tether and gate PM Orai channels (Amcheslavsky et al., 2015). The Orais are exquisitely Ca2+-selective channels mediating “store-operated Ca2+ entry” (SOCE) that triggers critical cellular events and also replenishes ER Ca2+ to sustain the generation of Ca2+ spikes (Fig. 1A–C). Despite the large fluxes of Ca2+ from stores that culminate in global Ca2+ spikes, the small PM-localized trickle of Ca2+ ions through Orai channels is responsible for activation of the ubiquitous NFAT family of transcription factors (Baine et al., 2009). Thus, calmodulin and calcineurin lurking close to the PM, are specifically triggered by this spatially restricted Ca2+ signal alone leading to dephosphorylation of NFAT molecules and exposure of nuclear localization signals (Kar et al., 2014).
Figure 1.
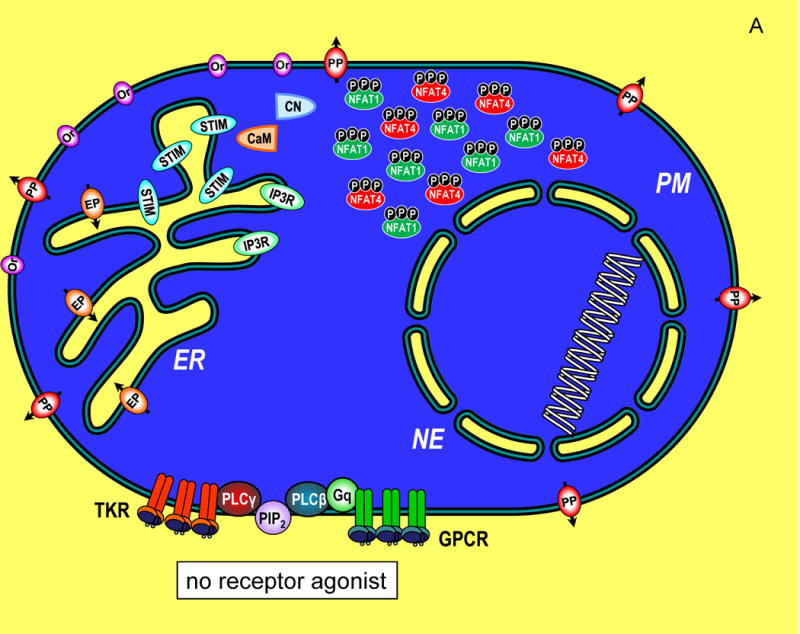
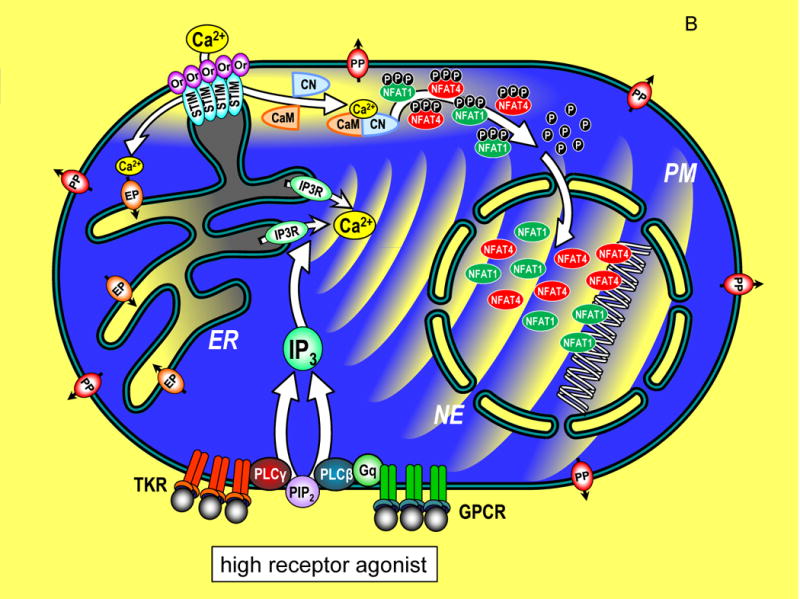
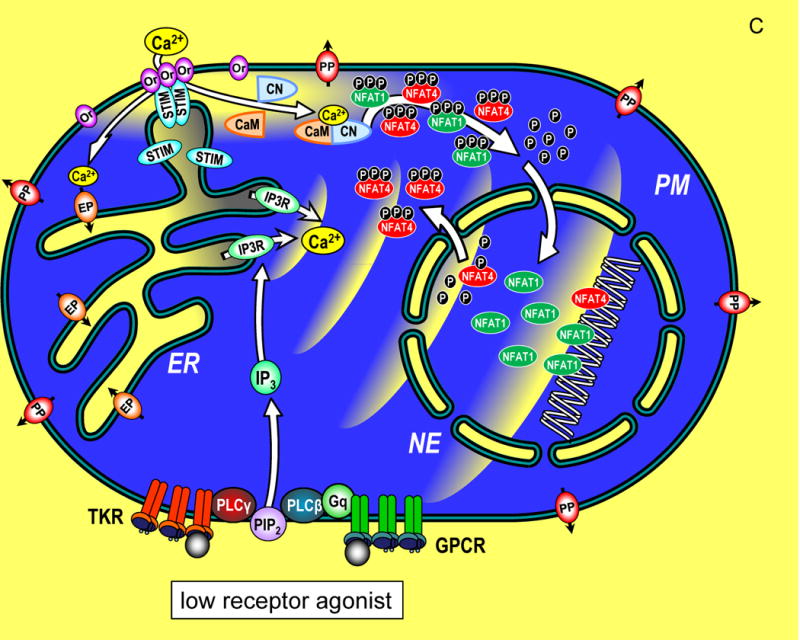
Transcriptional control through integrated Ca2+ signal transduction pathways. (A) In a resting cell, cytoplasmic Ca2+ is maintained at approximately 100 nM (blue) by endoplasmic reticulum (ER) Ca2+ pumps (EP) and plasma membrane (PM) Ca2+ pumps (PP). Ca2+ levels outside the cell and within the ER lumen and nuclear envelope (NE) are much higher, approximately 1 mM (yellow). Components of the Ca2+ signaling pathway include tyrosine kinase receptors (TKR), phospholipase C-γ (PLC-γ), G protein-coupled receptors (GPCR), G protein q (Gq), phospholipase C-β (PLC-β), phosphatidylinositol 4,5-bisphosphate (PIP2), inositol 1,4,5-trisphosphate (IP3), InsP3 receptor (IP3R), STIM proteins (STIM), Orai channels (Or), calmodulin (CaM), calcineurin (CN), nuclear factor of activated T cells (NFAT1 and NFAT4) shown in their cytoplasmic, phosphorylated state. (B) A cell in which Ca2+ signals are induced by maximal levels of TKR or GPCR agonists, inducing PIP2 breakdown to release high levels of InsP3 which activates large pulsatile ER Ca2+ release through InsP3Rs, manifested as global Ca2+ oscillations that traverse the cytoplasm to activate cellular responses. The cytosolic Ca2+ pulses penetrate within the nucleoplasmic space through permeable nuclear envelope pores. Ca2+ release from ER, depletes luminal stored Ca2+ (grey shading). STIM proteins, widely distributed across ER in the resting state, become activated and translocate into ER-PM junctions where they contact the PM and expose an Orai channel binding domain that tethers and gates Orai channels to allow Ca2+ to enter the cytosol. The Orai-mediated Ca2+ entry provides highly localized Ca2+ signals (within a few nm of the PM; yellow shading). The entering Ca2+ binds to CaM and triggers CN association and activates CN phosphatase activity which dephosphorylates NFAT1 and NFAT4 exposing nuclear import signal sequences. The dephosphorylated NFAT1 and NFAT4 molecules enter the nucleus wherein they each target distinct arrays of genes. The continuation of Ca2+ signals induces retention of NFAT molecules in the nucleus and prolonged gene expression. (C) A cell in which submaximal levels of TKR or GPCR agonists induces modest InsP3 levels and smaller, less frequent pulses of Ca2+ in the cytoplasm. STIM and Orai proteins are still activated, albeit to a lower extent, and Ca2+ entry triggers NFAT1 and NFAT4 phosphorylation and nuclear import. Within the nucleus, the smaller pulsatile rises in Ca2+ are insufficient to retain NFAT4, but NFAT1 remains activated in the nucleus despite the lower nucleoplasmic Ca2+. The lower nuclear Ca2+ may specifically trigger an increased rate of phosphorylation and faster export of NFAT4 from the nucleus compared to NFAT1. Since NFAT1 and NFAT4 target different genes, the pattern of transcription is tuned by both the Orai-mediated Ca2+ entry, and the frequency of Ca2+ pulses released from ER.
In their paper, Kar and Parekh (2015), address the interesting question of how Ca2+ signals in a single cell mediate quite disparate temporal patterns of activation of two major transcription factors, NFAT1 and NFAT4 (Yissachar et al., 2013). NFAT1 and NFAT4 have similar structures and both are activated by the same Orai channel-mediated cytoplasmic Ca2+ entry signals restricted to just a few nm below the Orai channel mouth (Fig. 1B). Both NFATs undergo similar calcineurin-induced dephosphorylation, exposure of nuclear localization sequence, and translocation into the nucleus. Perplexing, however, was why NFAT1 lingered within the nucleus whereas NFAT4 appeared to undergo rapid export out the nucleus. Clever use of a genetically-encoded nuclear-directed Ca2+ buffer, revealed that the Ca2+ level within the nucleus was the determining factor for nuclear retention of NFAT4. Export of NFAT4 from the nucleus was much faster when nuclear Ca2+ remained low whereas high nuclear Ca2+ lowered this export rate. Thus, only when Ca2+ within the nucleus was high did NFAT4 remain within the nucleus. In contrast, NFAT1 was relatively less sensitive to the levels of nuclear Ca2+ and hence could remain to activate transcription within the nucleus for some period after termination of the agonist-induced signal.
The picture emerging from these studies is that both the highly localized Orai-mediated Ca2+ entry and the global InsP3R-mediated Ca2+ spikes, contribute dually to specific patterns of NFAT1 or NFAT4 nuclear localization and hence patterns of gene expression. Under conditions of strong agonist-induced receptor activation, higher levels of InsP3 induce rapid Ca2+ spikes, deeper depletion of stores, resulting in greater STIM aggregation and translocation into ER-PM junctions and hence greater Ca2+ entry through Orai channels (Fig. 1B). Both NFAT1 and NFAT4 are activated and move into the nucleus under this condition. The prominent global Ca2+ spikes penetrating the nucleus are required for maintaining NFAT4 within the nucleus. Under conditions of lower agonist intensity, whereas NFAT1 and NFAT4 are still both activated (albeit NFAT4 at a reduced level), the decreased intensity of Ca2+ spikes results in lower nuclear Ca2+ and a greatly increased rate of NFAT4 export (Fig. 1C). Conversely, NFAT1 is indifferent to lower nuclear Ca2+ and is not exported. Kar and Parekh (2015) noted some structural differences between NFAT1 and NFAT4 that correlated with their distinct nuclear Ca2+-sensitivity. Mechanistically, we did not learn whether enhanced nuclear export of NFAT4 is mediated by increased kinase or decreased phosphatase activity. Perhaps low nuclear Ca2+ may preferentially prevent dephosphorylation of NFAT4 through calcineurin which also moves into nuclei during receptor activation. Alternatively, although nuclear export kinases are not Ca2+ sensitive, the rate of rephosphorylation of NFAT4 may be faster than NFAT1 with lower nuclear Ca2+.
The studies of Kar and Parekh (2015) provide a glimpse of how global Ca2+ spikes and local Ca2+ influx can dually impinge upon the activation Ca2+-dependent nuclear regulators. Together, these distinct Ca2+ signals tune the fidelity of transcriptional responses mediated by two closely related transcription factors. For immune cells such as T cells, Ca2+ is therefore not just a triggering signal that sets things rolling – Ca2+ signals continue to spatially and temporally coordinate and refine the ultimate response to graded receptor activation. Despite intense investigation, the nature of nuclear Ca2+ signaling remains a contentious issue. The nuclear envelope sequesters Ca2+ and the outer membrane has the same Ca2+ signaling machinery as normal ER. Although, the inner membrane is entirely separated from the outer membrane by nuclear pores and contains very different protein machinery, Ca2+ release channels and other signaling proteins do appear to be present in the inner membrane (Resende et al., 2013). However, given the complete permeability of nuclear pores to Ca2+ the case for considering any autonomy in nucleoplasmic Ca2+ signaling is a difficult one (Bading, 2013). Indeed, Kar and Parekh (2015) nicely reveal the coincidence of nuclear and cytoplasmic Ca2+ signals, and their results militate against local nuclear Ca2+ changes as mediating NFAT activation. Nevertheless, just as the authors show that the subtle local Ca2+ signals at the PM have such a profound role in initiating transcription factor responses, it would be interesting to investigate whether local nuclear Ca2+ signals may play an important role in refining and coordinating other transcriptional and chromosomal regulatory events within the nucleus.
References
- Amcheslavsky A, Wood ML, Yeromin AV, Parker I, Freites JA, Tobias DJ, Cahalan MD. Molecular Biophysics of Orai Store-Operated Ca Channels. Biophys J. 2015;108:237–246. doi: 10.1016/j.bpj.2014.11.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bading H. Nuclear calcium signalling in the regulation of brain function. Nature Rev Neuroscience. 2013;14:593–608. doi: 10.1038/nrn3531. [DOI] [PubMed] [Google Scholar]
- Baine I, Abe BT, Macian F. Regulation of T-cell tolerance by calcium/NFAT signaling. Immunol Rev. 2009;231:225–240. doi: 10.1111/j.1600-065X.2009.00817.x. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- Brandman O, Meyer T. Feedback loops shape cellular signals in space and time. Science. 2008;322:390–395. doi: 10.1126/science.1160617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar P, Parekh AB. Distinct spatial Ca2+ signatures selectively activate different NFAT transcription factor isoforms. Mol Cell. 2015 doi: 10.1016/j.molcel.2015.02.027. (In Press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar P, Samanta K, Kramer H, Morris O, Bakowski D, Parekh AB. Dynamic assembly of a membrane signaling complex enables selective activation of NFAT by Orai1. Curr Biol. 2014;24:1361–1368. doi: 10.1016/j.cub.2014.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende RR, Andrade LM, Oliveira AG, Guimaraes ES, Guatimosim S, Leite MF. Nucleoplasmic calcium signaling and cell proliferation: calcium signaling in the nucleus. Cell Commun & and Signaling. 2013;11:1–7. doi: 10.1186/1478-811X-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soboloff J, Rothberg BS, Madesh M, Gill DL. STIM proteins: dynamic calcium signal transducers. Nat Rev Mol Cell Biol. 2012;13:549–565. doi: 10.1038/nrm3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yissachar N, Sharar Fischler T, Cohen AA, Reich-Zeliger S, Russ D, Shifrut E, Porat Z, Friedman N. Dynamic response diversity of NFAT isoforms in individual living cells. Mol Cell. 2013;49:322–330. doi: 10.1016/j.molcel.2012.11.003. [DOI] [PubMed] [Google Scholar]


