Abstract
Background
Schizophrenia is associated with lower pre-morbid intelligence (IQ) in addition to (pre-morbid) cognitive decline. Both schizophrenia and IQ are highly heritable traits. Therefore, we hypothesized that genetic variants associated with schizophrenia, including copy number variants (CNVs) and a polygenic schizophrenia (risk) score (PSS), may influence intelligence.
Method
IQ was estimated with the Wechsler Adult Intelligence Scale (WAIS). CNVs were determined from single nucleotide polymorphism (SNP) data using the QuantiSNP and PennCNV algorithms. For the PSS, odds ratios for genome-wide SNP data were calculated in a sample collected by the Psychiatric Genome-Wide Association Study (GWAS) Consortium (8690 schizophrenia patients and 11 831 controls). These were used to calculate individual PSSs in our independent sample of 350 schizophrenia patients and 322 healthy controls.
Results
Although significantly more genes were disrupted by deletions in schizophrenia patients compared to controls (p=0.009), there was no effect of CNV measures on IQ. The PSS was associated with disease status (R2=0.055, p=2.1×10−7) and with IQ in the entire sample (R2=0.018, p=0.0008) but the effect on IQ disappeared after correction for disease status.
Conclusions
Our data suggest that rare and common schizophrenia-associated variants do not explain the variation in IQ in healthy subjects or in schizophrenia patients. Thus, reductions in IQ in schizophrenia patients may be secondary to other processes related to schizophrenia risk.
Keywords: Cognition, deletion, duplication, endophenotype, IQ, polygenic, SNP
Introduction
Schizophrenia is a severe mental disorder with cognitive deficits as a core feature (Kraepelin, 1919; Elvevag & Goldberg, 2000). It has been associated with lower pre-morbid general intelligence (IQ) (Khandaker et al. 2011; Fowler et al. 2012), with cognitive decline prior to illness onset (Reichenberg et al. 2010) and with cognitive decline during the course of the illness (Hedman et al. 2012), although the latter is controversial. Both schizophrenia and IQ are largely determined by genetic factors, with estimated heritabilities of 80 % (Cardno et al. 1999) and 70 % (Fowler et al. 2012) respectively. Several studies have identified genetic variants that affect these respective phenotypes. For schizophrenia, the findings include an excess of de novo copy number variants (CNVs, genomic micro-deletions or microduplications from 1 kb to multiple Mb in size) (Stefansson et al. 2008; Walsh et al. 2008; Xu et al. 2008) and gene-containing CNV deletions (Buizer-Voskamp et al. 2011). Moreover, 10 individual single nucleotide polymorphisms (SNPs) have been associated with schizophrenia in genome-wide association studies (GWAS) (Ripke et al. 2011), and the combination of many (>900 000) SNPs captures 23 % of the variation in liability to develop schizophrenia (Lee et al. 2012). Similarly, rare CNVs are associated with general intelligence (Yeo et al. 2011) and mental retardation (Guilmatre et al. 2009). Although no large GWAS on intelligence in healthy subjects have been reported, the combined effects of multiple SNPs captures 48 % of the variation in intelligence in childhood (Deary et al. 2012). These data suggest that both common and rare alleles contribute to schizophrenia disease susceptibility in addition to general intelligence.
It is likely that subsets of schizophrenia-related SNPs are associated with distinct heritable disease-associated phenotypes. These are called endophenotypes when they are (1) associated with the illness, (2) heritable, (3) primarily state independent and (4) co-segregate with illness in families (Gottesman & Gould, 2003). Brain volume is an example of an endophenotype of schizophrenia, as it is robustly associated with disease (Wright et al. 2000), is highly heritable (van Haren et al. 2012) and cosegregates with illness in families (Boos et al. 2007). It is an objectively assessed quantitative measure. We revealed in a previous study that a combined score of common variants associated with schizophrenia (the polygenic schizophrenia score or PSS) significantly predicts total brain and white matter volume in schizophrenia patients and in healthy subjects (Terwisscha van Scheltinga et al. 2012b), suggesting that schizophrenia-associated SNPs may be involved in (normal) white matter development. Thus, novel candidate genes can be identified based on the overlap in high-ranking SNPs associated with schizophrenia and its endophenotypes. IQ is another candidate endophenotype for this approach because of its strong association with disease (Mesholam-Gately et al. 2009), high heritability (Fowler et al. 2012), normally distributed quantitative measures and relatively straightforward assessment.
Although schizophrenia and IQ are phenotypically correlated and are both largely determined by genetic factors, these factors do not necessarily overlap. To what extent IQ-related genetic variants explain variation in schizophrenia is still unclear. A twin study in a sample of schizophrenia and healthy twins (n=263 subjects) found that IQ measured after disease onset explained 37 % of the variation in disease status (Toulopoulou et al. 2007). Shared genetic influences accounted for 92 % of this phenotypic correlation. However, a large Swedish population cohort study of twins and sib pairs (n=374 199 subjects) found that the broader phenotype of psychosis explained a much smaller amount (1 %) of variation in pre-morbid intelligence at age 18, although this phenotypic correlation was also largely determined by shared genetic influences (for 91 %). However, because the correlation was small, the genetic variance for pre-morbid IQ shared with psychosis was only 7 % (Fowler et al. 2012). This suggests that, of all genetic variants associated with pre-morbid IQ, few will influence psychosis, and vice versa. However, no studies have yet investigated the predictive value of measured genetic variants.
The pre-morbid cognitive deficits in schizophrenia patients were reported to be largest in the domains of problem solving, working memory and processing speed (Reichenberg et al. 2010). It is possible that schizophrenia-associated genetic factors influence these specific cognitive functions rather than general intelligence. However, previous studies indicate that IQ and working memory have a similar high heritability and a comparable genetic overlap with schizophrenia (Toulopoulou et al. 2007; Aukes et al. 2009). None of the cognitive subtests indicated a significantly better schizophrenia endophenotype than total IQ. Therefore, we used total IQ as our main outcome measure.
The aim of the current study was therefore to investigate the relationship between IQ and two distinct types of schizophrenia-related genetic variants : CNVs and common SNPs. There are several possible scenarios. The first option is that these genetic variants are associated with IQ in both patients and healthy controls. If so, this would imply that novel schizophrenia risk genes could be identified using population-based samples with IQ data. A second option is that these variants affect IQ in schizophrenia patients only, which would suggest that schizophrenia genes affect processes involved in cognition. Finally, if disease-associated variants do not predict IQ in schizophrenia patients or in healthy subjects, this could indicate that the cognitive deficits in patients are secondary to other processes related to the risk for schizophrenia, for example environmental factors such as the use of cannabis or stress. Alternatively, the cognitive deficits may be related to indirect genetic effects, for example by genes influencing early brain development, which in turn affects cognition.
Method
Subjects and IQ measurements
The study was approved by the Medical Ethical Committee of the University Medical Center Utrecht. The sample consisted of 350 patients (90 % with schizophrenia and 10 % with schizo-affective disorder) and 322 controls. Clinical diagnosis was determined using the Comprehensive Assessment of Symptoms and History (CASH; Andreasen et al. 1992). Unaffected controls had no history of psychiatric illness except for three subjects who had a history of anxiety disorder, obsessive–compulsive disorder and adjustment disorder respectively. None of the control subjects had first-degree family members with psychotic illness.
Total IQ was estimated with the Wechsler Adult Intelligence Scale (WAIS ; Stinissen et al. 1970). Four out of 11 subtests of WAIS versions III (n=97) and IIIR (n=575) were used (WAIS-III : block design, comprehension, vocabulary and picture arrangement ; WAIS-IIIR : block design, information, symbol search and arithmetic). Raw scores were multiplied by 11/4 to obtain an estimate of total IQ. Because of adjustment of the norm scores, mean IQ measures were significantly lower in the newer version compared to the older version. Therefore, IQ scores were corrected for version, by taking the unstandardized residual in a linear regression, with the newer version (IIIR) as reference. Estimated IQ scores were normally distributed both before and after correction for version.
Genotyping, CNV and PSS measurements
Subjects were genotyped at the University of California, Los Angeles (UCLA) Neurosciences Genomics Core (UNGC) using the Illumina HumanHap550 beadchip. CNVs were determined from SNP data using the QuantiSNP and PennCNV algorithms as described previously (Buizer-Voskamp et al. 2011; Terwisscha van Scheltinga et al. 2012a). In brief, deviations in log R ratios (a measure of the magnitude of the combined fluorescent signals from both sets of probes) and B allele frequencies (relative ratio of the fluorescent signals from one allelic probe to the other) for 10 consecutive SNPs were regarded as CNVs. Refseq genes within 50 kb of the CNV borders were counted using the March 2006 assembly on the University of California Santa Cruz (UCSC) browser (http://genome.ucsc.edu). The total numbers of CNVs and the total number of genes affected by CNVs were calculated for each individual.
We then calculated a PSS as described previously (Terwisscha van Scheltinga et al. 2012b). In short, data from the Schizophrenia Psychiatric GWAS Consortium (PGC) on 8690 schizophrenia patients and 11 831 controls were used as a discovery sample to identify the schizophrenia risk variants, their p values and odds ratios. For each individual in our independent sample of 350 patients and 322 controls, the PSS was calculated using PLINK (Purcell et al. 2007). For each SNP, the number of ‘ risk variants ’ an individual carried (0, 1 or 2) was multiplied by the logarithm of the odds ratio for that particular variant. ‘ Risk variants ’ are the alleles (nominally) associated with disease, including both true risk alleles and stochastic variation. Sets of SNPs with p values below different cut-offs for effect on schizophrenia (p value cut-offs for effect on schizophrenia or pcut-off) were defined. The following pcut-off were used : 0.01, 0.05, 0.1, 0.2, 0.3, 0.4 and 0.5. The score was summed over all SNPs in the pcut-off–SNP sets for each individual in the current sample to obtain the individual polygenic scores. PSSs were normally distributed.
Statistical analyses
First, the difference in the two CNV measures (the total number of CNVs and the total number of genes affected by CNVs) between patients and controls was calculated using a t test. Second, the two CNV measures were used as factors in ANOVAs together with disease status and a CNVrdisease status interaction as factors and total IQ as the dependent variable in the total sample. All analyses were repeated for deletions and duplications separately.
For the PSS, we performed linear regressions using the PSS as predictor and the total IQ estimates as the outcome measure. Ten population stratification dimensions, an inbreeding coeficient, the number of SNPs used for scoring and the number of CNVs were used as covariates (for details see Terwisscha van Scheltinga et al. 2012b). Similar to the previous analysis, we performed secondary analyses while including effects for disease status and PSS × disease status interactions. The association of the PSSs with disease status was analyzed using logistic regression. The variance explained, adjusted for the number of predictors in the model (R2), was used as the main outcome parameter. p values were Bonferroni corrected for multiple comparisons with a=0.008 [0.05/6 (because there are three ‘ independent ’ genetic measures (two CNV and one PSS)r2 tests : main effect and interaction effect)]. Post-hoc we analyzed the association between the PSS and the four subtests of the WAIS-IIIR separately.
Results
As shown in Table 1, there were significantly more males in the patient group than in the control group (78 % v. 49 %, p<0.0001). The groups did not differ in age and parental education. The years of education and estimated IQ were significantly lower in the schizophrenia patients.
Table 1.
Demographic information
| Schizophrenia patients |
Healthy controls |
Significance, p |
|
|---|---|---|---|
| n | 350 | 322 | n.a. |
| Gender (M/F) | 272/78 | 158/164 | <0.0001 |
| Age (years) | 30.2 (9.0) | 31.6 (11.9) | n.s. |
| Age of first psychotic symptoms (years) | 22.5 (5.4) | n.a. | |
| Duration of illness (years) | 7.5 (7.9) | n.a. | |
| Level of education (years) | 12.6 (2.4) | 13.9 (2.3) | <0.0001 |
| Level of parental education (years) | 13.4 (3.1) | 13.3 (3.1) | n.s. |
| Estimated IQ | 95.8 (16.6) | 111.2 (14.1) | <0.0001 |
M, Male; F, female; estimated IQ, a measure of intelligence quotient as calculated from the scores on four Wechsler Adult Intelligence Scale (WAIS) subtests; n.a., not applicable, n.s., not significant.
Means are displayed with the standard deviation in parentheses.
Table 2 shows that there was a trend towards more deletions in schizophrenia patients compared to healthy controls (p=0.09), whereas significantly more genes were affected by deletions in patients (p=0.009). Neither of the CNV measures showed a significant association with IQ.
Table 2.
The association between CNV measures and IQ
| p sz | p IQ | p interaction | |
|---|---|---|---|
| CNVs | 0.67 | 0.99 | 0.53 |
| Genes in CNVs | 0.22 | 0.97 | 0.24 |
| Deletions | 0.09 | 0.82 | 0.95 |
| Genes in deletions | 0.009 | 0.43 | 0.21 |
| Duplications | 0.38 | 0.60 | 0.73 |
| Genes in duplications | 0.63 | 0.74 | 0.97 |
CNV, Copy number variant; p sz, the p value of the main effect of the CNVs on schizophrenia disease status tested with a t test; p IQ, the p value of the main effect of the CNV measures on IQ, investigated with ANOVAs in which the CNV measures, disease status and the CNV × disease status interactions were included as factors in the analyses; p interaction, the p value of the interaction effect between the CNV measure and disease status on IQ.
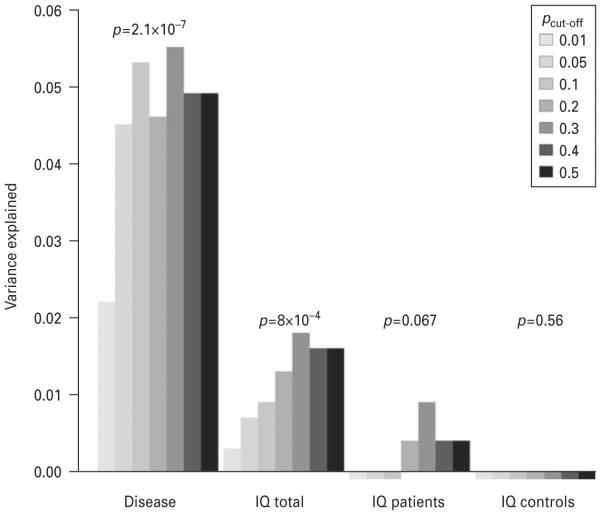
The PSS significantly predicted disease status in the current sample (R2=0.055, p=2.1×10−7, for pcut-off 0.3). A significant association was found between the PSS and IQ (R2=0.018, p=0.0008, for pcut-off 0.3) as shown in Fig. 1. The results for the other pcut-off are also shown in Fig. 1. However, after correction for disease status the association was no longer significant (pPSS=0.97, pPSS×disease status=0.17). Furthermore, no association of the PSS with IQ was found within the patient and control groups [patients : R2=0.009, p=0.067 and controls : R2=×0.003 (negative value because of the adjustment for the number of predictors in the model), p=0.82]. Excluding subjects tested with the older version of the WAIS did not change the results.
Fig. 1.
The variance explained by the polygenic schizophrenia score (PSS) in schizophrenia disease status and IQ for different pcut-off single nucleotide polymorphism (SNP) sets. For ‘ schizophrenia ’ the association between the PSS and schizophrenia disease status was tested. In ‘ IQ total ’ the association between PSS and IQ was tested in the total sample whereas in ‘ IQ patients ’ and ‘ IQ controls ’ this association was tested respectively in the groups of patients and controls only. Variance explained (R2) is adjusted for the number of predictors in the model. The p values shown are the p values for the pcut-off set with the lowest p value within that phenotype.
Discussion
We have investigated, in 350 schizophrenia patients and 322 healthy subjects, the association of schizophrenia-related genetic variants with IQ, a highly heritable phenotype associated with schizophrenia. Although deletions disrupted significantly more genes in schizophrenia patients, these deletions and the other CNV measures did not exert major effects on IQ in schizophrenia patients or in healthy controls.
The combined score of schizophrenia-associated SNPs was associated with disease status, indicating that a PSS can predict disease in an independent sample. The PSS was also significantly associated with IQ in the entire sample of patients and controls. However, after correcting for disease status, this association disappeared. The PSS does not predict IQ within the patient and control groups. This result may seem puzzling at first sight, but can be explained by assuming that both IQ and the PSS are independently associated with schizophrenia.
Another possibility is that schizophrenia-associated genetic factors influence specific cognitive functions rather than general intelligence. Therefore, we repeated the analyses post-hoc for each of the four subtests of the WAIS-IIIR separately. Similar results were found as for the total IQ scores : the PSS explained some of the variation in the subtest in the whole sample but the association disappeared after correction for disease status. We estimated that we had 80 % power to detect a significant effect with α=0.008 when the PSS explained 1.8 % or more of the variance in IQ. Thus, if there is any effect of schizophrenia-related SNPs on total IQ or one of the cognitive subdomains, this effect is likely to be small.
The findings are in agreement with the small (7 %) genetic variance for psychosis shared with pre-morbid IQ (Fowler et al. 2012). Thus, although there is a robust association between schizophrenia and intelligence, this may not be due mainly to shared genetic factors. Instead, the cognitive deficits in schizophrenia patients may be secondary to other processes. We showed previously, in a partly overlapping sample, that the variance in total brain and white matter volume explained by the PSS is much larger (4.8 and 5.1 %) (Terwisscha van Scheltinga et al. 2012b). This effect was found in schizophrenia patients and also in healthy controls. Hence, the cognitive deficits in schizophrenia patients could be a result of a generalized effect of schizophrenia genes on early brain growth and maturation, perhaps in interaction with the effects of disease-related environmental factors.
There are several other factors that may have influenced the results and that should be acknowledged as potential limitations of this study. First, IQ measurements in patients could be influenced by the presence of psychotic symptoms, or the use of medication or illicit drugs, which could have partly masked the genetic effects. Second, total IQ was estimated by four out of 11 WAIS subtests, which is less precise than an estimate based on all 11 subtests, although the shortened version is more widely used (Fowler et al. 2012). Furthermore, the lack of effect of global CNV measures on IQ does not exclude an effect of individual CNVs at specific chromosomal locations on schizophrenia susceptibility and IQ. However, because CNVs at specific locations are usually rare, these are likely to go unnoticed in the current sample.
To conclude, rare and common schizophrenia-related genetic variants do not predict variation in IQ in healthy subjects or within the schizophrenia patient group. This suggests that cognitive deficits and the risk for schizophrenia cannot be attributed to shared genes. The cognitive dysfunction in schizophrenia patients is therefore more likely to be secondary to other processes related to schizophrenia risk. Thus, although we did not investigate this directly, our study suggests that population-based studies aiming to identify schizophrenia candidate genes by investigating the association of genetic variants with IQ may not be fruitful.
Acknowledgements
This work was supported by a Veni grant to S.C.B. from Zorg Onderzoek Nederland, Medische Weten-schappen (ZON-MW, the Netherlands Organization for Health Research and Development, project no. 91686137), by Top Institute Pharma (TI-Pharma, project T5-203) and by the National Institute of Mental Health grant RO1 MG 078075 to R.A.O.
Appendix. The Schizophrenia Psychiatric GWAS Consortium
S. Ripke, A. R. Sanders, K. S. Kendler, D. F. Levinson, P. Sklar, P. A. Holmans, D.-Y. Lin, J. Duan, R. A. Ophoff, O. A. Andreassen, E. Scolnick, S. Cichon, D. St Clair, A. Corvin, H. Gurling, T. Werge, D. Rujescu, D. H. R. Blackwood, C. N. Pato, A. K. Malhotra, S. Purcell, F. Dudbridge, B. M. Neale, L. Rossin, P. M. Visscher, D. Posthuma, D. M. Ruderfer, A. Fanous, H. Stefansson, S. Steinberg, B. J. Mowry, V. Golimbet, M. De Hert, E. G. Jönsson, I. Bitter, O. P. H. Pietiläinen, D. A. Collier, S. Tosato, I. Agartz, M. Albus, M. Alexander, R. L. Amdur, F. Amin, N. Bass, S. E. Bergen, D. W. Black, A. D. Børglum, M. A. Brown, R. Bruggeman, N. G. Buccola, W. F. Byerley, W. Cahn, R. M. Cantor, V. J. Carr, S. V. Catts, K. Choudhury, C. R. Cloninger, P. Cormican, N. Craddock, P. A. Danoy, S. Datta, L. de Haan, D. Demontis, D. Dikeos, S. Djurovic, P. Donnelly, G. Donohoe, L. Duong, S. Dwyer, A. Fink-Jensen, R. Freedman, N. B. Freimer, M. Friedl, L. Georgieva, I. Giegling, M. Gill, B. Glenthøj, S. Godard, M. Hamshere, M. Hansen, T. Hansen, A. M. Hartmann, F. A. Henskens, D. M. Hougaard, C. M. Hultman, A. Ingason, A. V. Jablensky, K. D. Jakobsen, M. Jay, G. Jürgens, R. S. Kahn, M. C. Keller, G. Kenis, E. Kenny, Y. Kim, G. K. Kirov, H. Konnerth, B. Konte, L. Krabbendam, R. Krasucki, V. K. Lasseter, C. Laurent, J. Lawrence, T. Lencz, F. B. Lerer, K.-Y. Liang, P. Lichtenstein, J. A. Lieberman, D. H. Linszen, J. Lönnqvist, C. M. Loughland, A. W. Maclean, B. S. Maher, W. Maier, J. Mallet, P. Malloy, M. Mattheisen, M. Mattingsdal, K. A. McGhee, J. J. McGrath, A. McIntosh, D. E. McLean, A. McQuillin, I. Melle, P. T. Michie, V. Milanova, D. W. Morris, O. Mors, P. B. Mortensen, V. Moskvina, P. Muglia, I. Myin-Germeys, D. A. Nertney, G. Nestadt, J. Nielsen, I. Nikolov, M. Nordentoft, N. Norton, M. M. Nöthen, C. T. O’Dushlaine, A. Olincy, L. Olsen, F. A. O’Neill, T. F. Ørntoft, M. J. Owen, C. Pantelis, G. Papadimitriou, M. T. Pato, L. Peltonen, H. Petursson, B. Pickard, J. Pimm, A. E. Pulver, V. Puri, D. Quested, E. M. Quinn, H. B. Rasmussen, J. M. Réthelyi, R. Ribble, M. Rietschel, B. P. Riley, M. Ruggeri, U. Schall, T. G. Schulze, S. G. Schwab, R. J. Scott, J. Shi, E. Sigurdsson, J. M. Silverman, C. C. A. Spencer, K. Stefansson, A. Strange, E. Strengman, T. S. Stroup, J. Suvisaari, L. Terenius, S. Thirumalai, J. H. Thygesen, S. Timm, D. Toncheva, E. van den Oord, J. van Os, R. van Winkel, J. Veldink, D. Walsh, A. G. Wang, D. Wiersma, D. B. Wildenauer, H. J. Williams, N. M. Williams, B. Wormley, S. Zammit, P. F. Sullivan, M. C. O’Donovan, M. J. Daly and P. V. Gejman.
Footnotes
Declaration of Interest
This Declaration of Interest statement concerns members of the Schizophrenia Psychiatric GWAS Consortium, not any of the main authors of this paper. Eli Lilly funded portions of the genotyping for CATIE and TOP. P.F.S. received research funding from Eli Lilly in connection with CATIE. T.S.S. received research funding from Eli Lilly and consulting fees from Janssen Pharmaceutica, GlaxoSmithKline and Bristol-Myers Squibb. J.A.L. received research funding from AstraZeneca Pharmaceuticals, Bristol-Myers Squibb, GlaxoSmithKline, Janssen Pharmaceutica and Pfizer and consulting and educational fees from AstraZeneca Pharmaceuticals, Bristol-Myers Squibb, Eli Lilly, Forest Pharmaceuticals, GlaxoSmithKline, Janssen Pharmaceutica, Novartis, Pfizer and Solvay. D.St.C. received research funding from GlaxoSmithKline and Generation Scotland, Genetics Health Initiative. F.A. received funds from Pfizer, Organon and the Foundation for the National Institutes of Health. D.W.B. has received research support from Shire and Forest, has been on the Speakers ’ Bureau for Pfizer and has received consulting honoraria from Forest and Jazz. T.W. has received consulting and lecture fees from H. Lundbeck A/S. O.A.A. has received a Speaker’s honorarium from AstraZeneca, Janssen, Bristol–Myers Squibb and GlaxoSmithKline. I.M. has received a Speaker’s honorarium from Janssen and AstraZeneca. A.K.M. has received consulting fees or honoraria from Eli Lilly & Company, Janssen Pharmaceutica, Merck, Bristol–Meyers Squibb, Pfizer, PGxHealth (a division of Clinical Data, Inc.), Roche Diagnostics and Vanda Pharmaceuticals and has received research support from Eli Lilly & Company. T.L. has received consulting fees or honoraria from Merck, Eli Lilly & Company, Golden Helix, Inc., InforMed Insights and PGxHealth (a division of Clinical Data, Inc.). I.B. has been an advisory board member, consultant and lecturer for AstraZeneca, Bristol-Myers Squibb, Eli Lilly, EGIS, Janssen, H. Lundbeck A/S, Novartis, Pfizer, Richter and Schering-Plough and received a grant for an investigator-initiated study from H. Lundbeck A/S. J.J.M. has received consulting and speaker’s fees from Johnson & Johnson, Schering-Plough and Eli Lilly. C.P. has received grant support from Janssen-Cilag, Eli Lilly, Hospira (Mayne) and AstraZeneca, provided consultancy to Janssen-Cilag, Eli Lilly, Hospira (Mayne), AstraZeneca, Pfizer and Schering-Plough and has undertaken investigator-initiated studies supported by Eli Lilly, Hospira, Janssen Cilag and AstraZeneca. The Denmark-Aarhus group (The GEMS Stud with principal investigators A.D.B., O.M. and P.B.M.) received research funding from H. Lundbeck A/S. E.G.J. has served as an unpaid consultant for Eli Lilly. All other authors declare no confiict of interest in relation to the subject of this study.
References
- Andreasen NC, Flaum M, Arndt S. The Comprehensive Assessment of Symptoms and History (CASH). An instrument for assessing diagnosis and psychopathology. Archives of General Psychiatry. 1992;49:615–623. doi: 10.1001/archpsyc.1992.01820080023004. [DOI] [PubMed] [Google Scholar]
- Aukes MF, Alizadeh BZ, Sitskoorn MM, Kemner C, Ophoff RA, Kahn RS. Genetic overlap among intelligence and other candidate endophenotypes for schizophrenia. Biological Psychiatry. 2009;65:527–534. doi: 10.1016/j.biopsych.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Boos HB, Aleman A, Cahn W, Hulshoff Pol HE, Kahn RS. Brain volumes in relatives of patients with schizophrenia : a meta-analysis. Archives of General Psychiatry. 2007;64:297–304. doi: 10.1001/archpsyc.64.3.297. [DOI] [PubMed] [Google Scholar]
- Buizer-Voskamp JE, Muntjewerff JW, Strengman E, Sabatti C, Stefansson H, Vorstman JA, Ophoff RA. Genome-wide analysis shows increased frequency of copy number variation deletions in Dutch schizophrenia patients. Biological Psychiatry. 2011;70:655–662. doi: 10.1016/j.biopsych.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardno AG, Marshall EJ, Coid B, Macdonald AM, Ribchester TR, Davies NJ, Venturi P, Jones LA, Lewis SW, Sham PC, Gottesman II, Farmer AE, McGuffin P, Reveley AM, Murray RM. Heritability estimates for psychotic disorders : the Maudsley twin psychosis series. Archives of General Psychiatry. 1999;56:162–168. doi: 10.1001/archpsyc.56.2.162. [DOI] [PubMed] [Google Scholar]
- Deary IJ, Yang J, Davies G, Harris SE, Tenesa A, Liewald D, Luciano M, Lopez LM, Gow AJ, Corley J, Redmond P, Fox HC, Rowe SJ, Haggarty P, McNeill G, Goddard ME, Porteous DJ, Whalley LJ, Starr JM, Visscher PM. Genetic contributions to stability and change in intelligence from childhood to old age. Nature. 2012;482:212–215. doi: 10.1038/nature10781. [DOI] [PubMed] [Google Scholar]
- Elvevag B, Goldberg TE. Cognitive impairment in schizophrenia is the core of the disorder. Critical Reviews in Neurobiology. 2000;14:1–21. [PubMed] [Google Scholar]
- Fowler T, Zammit S, Owen MJ, Rasmussen F. A population-based study of shared genetic variation between premorbid IQ and psychosis among male twin pairs and sibling pairs from Sweden. Archives of General Psychiatry. 2012;69:460–466. doi: 10.1001/archgenpsychiatry.2011.1370. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry : etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, Laumonnier F, Odent S, Le Vacon G, Joly-Helas G, David V, Bendavid C, Pinoit JM, Henry C, Impallomeni C, Germano E, Tortorella G, Di Rosa G, Barthelemy C, Andres C, Faivre L, Frebourg T, Saugier VP, Campion D. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Archives of General Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman AM, van Haren NE, van Baal GC, Brans RG, Hijman R, Kahn RS, Hulshoff Pol HE. Is there change in intelligence quotient in chronically ill schizophrenia patients? A longitudinal study in twins discordant for schizophrenia. Psychological Medicine. 2012;42:2535–2541. doi: 10.1017/S0033291712000694. [DOI] [PubMed] [Google Scholar]
- Khandaker GM, Barnett JH, White IR, Jones PB. A quantitative meta-analysis of population-based studies of premorbid intelligence and schizophrenia. Schizophrenia Research. 2011;132:220–227. doi: 10.1016/j.schres.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraepelin E. Dementia Praecox and Paraphrenia. E & S Livingstone; Edinburgh: 1919. [Google Scholar]
- Lee SH, Decandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, Keller MC, Visscher PM, Wray NR. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nature Genetics. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesholam-Gately RI, Giuliano AJ, Goff KP, Faraone SV, Seidman LJ. Neurocognition in first-episode schizophrenia : a meta-analytic review. Neuropsychology. 2009;23:315–336. doi: 10.1037/a0014708. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK : a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenberg A, Caspi A, Harrington H, Houts R, Keefe RS, Murray RM, Poulton R, Moffitt TE. Static and dynamic cognitive deficits in childhood preceding adult schizophrenia : a 30-year study. American Journal of Psychiatry. 2010;167:160–169. doi: 10.1176/appi.ajp.2009.09040574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin DY, Duan J, Ophoff RA, Andreassen OA, Scolnick E, Cichon S, St Clair D, Corvin A, Gurling H, Werge T, Rujescu D, Blackwood DH, Pato CN, Malhotra AK, Purcell S, Dudbridge F, Neale BM, Rossin L, Visscher PM, Posthuma D, Ruderfer DM, Fanous A, Stefansson H, Steinberg S, Mowry BJ, Golimbet V, de Haan M, Jonsson EG, Bitter I, Pietilainen OP, Collier DA, Tosato S, Agartz I, Albus M, Alexander M, Amdur RL, Amin F, Bass N, Bergen SE, Black DW, Borglum AD, Brown MA, Bruggeman R, Buccola NG, Byerley WF, Cahn W, Cantor RM, Carr VJ, Catts SV, Choudhury K, Cloninger CR, Cormican P, Craddock N, Danoy PA, Datta S, de Hann L, Demontis D, Dikeos D, Djurovic S, Donnelly P, Donohoe G, Duong L, Dwyer S, Fink-Jensen A, Freedman R, Freimer NB, Friedl M, Georgieva L, Giegling I, Gill M, Glenthoj B, Godard S, Hamshere M, Hansen M, Hansen T, Hartmann AM, Henskens FA, Hougaard DM, Hultman CM, Ingason A, Jablensky AV, Jakobsen KD, Jay M, Jurgens G, Kahn RS, Keller MC, Kenis G, Kenny E, Kim Y, Kirov GK, Konnerth H, Konte B, Krabbendam L, Krasucki R, Lasseter VK, Laurent C, Lawrence J, Lencz T, Lerer FB, Liang KY, Lichtenstein P, Lieberman JA, Linszen DH, Lonnqvist J, Loughland CM, MacLean AW, Maher BS, Maier W, Mallet J, Malloy P, Mattheisen M, Mattingsdal M, McGhee KA, McGrath JJ, McIntosh A, McLean DE, McQuillin A, Melle I, Michie PT, Milanova V, Morris DW, Mors O, Mortensen PB, Moskvina V, Muglia P, Myin-Germeys I, Nertney DA, Nestadt G, Nielsen J, Nikolov I, Nordentoft M, Norton N, Nothen MM, O’Dushlaine CT, Olincy A, Olsen L, O’Neill FA, Orntoft TF, Owen MJ, Pantelis C, Papadimitriou G, Pato MT, Peltonen L, Petursson H, Pickard B, Pimm J, Pulver AE, Puri V, Quested D, Quinn EM, Rasmussen HB, Rethelyi JM, Ribble R, Rietschel M, Riley BP, Ruggeri M, Schall U, Schulze TG, Schwab SG, Scott RJ, Shi J, Sigurdsson E, Silverman JM, Spencer CC, Stefansson K, Strange A, Strengman E, Stroup TS, Suvisaari J, Terenius L, Thirumalai S, Thygesen JH, Timm S, Toncheva D, van den Oord E, van Os J, van Winkel R, Veldink J, Walsh D, Wang AG, Wiersma D, Wildenauer DB, Williams HJ, Williams NM, Wormley B, Zammit S, Sullivan PF, O’Donovan MC, Daly MJ, Gejman PV. Genome-wide association study identifies five new schizophrenia loci. Nature Genetics. 2011;10:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Kahn RS, Linszen D, van Os J, Wiersma D, Bruggeman R, Cahn W, Germeys I, de Haan L, Krabbendam L, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinissen J, Willems PJ, Coetsier P, Hulsman WWL. Manual of the Dutch Translation of the Wechsler Adult Intelligence Scale. Swets : Amsterdam, The Netherlands. 1970 [Google Scholar]
- Terwisscha van Scheltinga A, Bakker S, van Haren N, Buizer-Voskamp J, Boos H, Vorstman J, Cahn W, Hulshoff Poll H, Ophoff R, Kahn R. Association study of copy number variants with brain volume in schizophrenia patients and healthy controls. Psychiatry Research. 2012a;30:1011–1013. doi: 10.1016/j.psychres.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Terwisscha van Scheltinga AF, Bakker SC, van Haren NEM, Derks EM, Buizer-Voskamp JE, Boos HB, Cahn W, Hulshoff Pol HE, Ripke S, Ophoff RA, Kahn RS, Psychiatric GWAS Consortium Genetic schizophrenia risk variants jointly modulate total brain and white matter volume. Biological Psychiatry. 2012b doi: 10.1016/j.biopsych.2012.08.017. Published online : 3 October 2012. doi :10.1016/j.biopsych.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulopoulou T, Picchioni M, Rijsdijk F, Hua-Hall M, Ettinger U, Sham P, Murray R. Substantial genetic overlap between neurocognition and schizophrenia : genetic modeling in twin samples. Archives of General Psychiatry. 2007;64:1348–1355. doi: 10.1001/archpsyc.64.12.1348. [DOI] [PubMed] [Google Scholar]
- van Haren NEM, Rijsdijk F, Schnack HG, Picchioni MM, Toulopoulou T, Weisbrod M, Sauer H, van Erp TG, Cannon TD, Huttunen MO, Boomsma DI, Hulshoff Pol HE, Murray RM, Kahn RS. The genetic and environmental determinants of the association between brain abnormalities and schizophrenia : the Schizophrenia Twins and Relatives Consortium. Biological Psychiatry. 2012;71:915–921. doi: 10.1016/j.biopsych.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, Stray SM, Rippey CF, Roccanova P, Makarov V, Lakshmi B, Findling RL, Sikich L, Stromberg T, Merriman B, Gogtay N, Butler P, Eckstrand K, Noory L, Gochman P, Long R, Chen Z, Davis S, Baker C, Eichler EE, Meltzer PS, Nelson SF, Singleton AB, Lee MK, Rapoport JL, King MC, Sebat J. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Wright IC, Rabe-Hesketh S, Woodruff PW, David AS, Murray RM, Bullmore ET. Meta-analysis of regional brain volumes in schizophrenia. American Journal of Psychiatry. 2000;157:16–25. doi: 10.1176/ajp.157.1.16. [DOI] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nature Genetics. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Yeo RA, Gangestad SW, Liu J, Calhoun VD, Hutchison KE. Rare copy number deletions predict individual variation in intelligence. PloS One. 2011;6:e16339. doi: 10.1371/journal.pone.0016339. [DOI] [PMC free article] [PubMed] [Google Scholar]