Significance
Uniquely in B cells, deletion of the adaptor protein TRAF3 (TNF receptor-associated factor 3) causes enhanced survival; TRAF3 deficiency is observed in a substantial percentage of human B-cell malignancies. Here, we report that TRAF3 is a resident nuclear protein in B cells that regulates stability of the CREB (cAMP response element binding protein) transcription factor. This regulation is important for restraining B-cell survival by preventing transcription of prosurvival genes and may be a key mechanism by which loss of TRAF3 contributes to B-cell malignancies. Considering the high frequency of TRAF3 mutations in B-cell tumors, our findings of an important nuclear role for TRAF3 present a new paradigm in understanding both normal regulation of B-cell survival and pathogenesis of B-cell cancers.
Keywords: B cell, CREB, TRAF3
Abstract
The adaptor protein TNF receptor-associated factor 3 (TRAF3) regulates signaling through B-lymphocyte receptors, including CD40, BAFF receptor, and Toll-like receptors, and also plays a critical role inhibiting B-cell homoeostatic survival. Consistent with these findings, loss-of-function human TRAF3 mutations are common in B-cell cancers, particularly multiple myeloma and B-cell lymphoma. B cells of B-cell–specific TRAF3−/− mice (B-Traf3−/−) display remarkably enhanced survival compared with littermate control (WT) B cells. The mechanism for this abnormal homeostatic survival is poorly understood, a key knowledge gap in selecting optimal treatments for human B-cell cancers with TRAF3 deficiency. We show here for the first time to our knowledge that TRAF3 is a resident nuclear protein that associates with the transcriptional regulator cAMP response element binding protein (CREB) in both mouse and human B cells. The TRAF-C domain of TRAF3 was necessary and sufficient to localize TRAF3 to the nucleus via a functional nuclear localization signal. CREB protein was elevated in TRAF3−/− B cells, without change in mRNA, but with a decrease in CREB ubiquitination. CREB-mediated transcriptional activity was increased in TRAF3-deficient B cells. Consistent with these findings, Mcl-1, an antiapoptotic target of CREB-mediated transcription, was increased in the absence of TRAF3 and enhanced Mcl-1 was suppressed with CREB inhibition. TRAF3-deficient B cells were also preferentially sensitive to survival inhibition with pharmacologic CREB inhibitor. Our results identify a new mechanism by which nuclear TRAF3 regulates B-cell survival via inhibition of CREB stability, information highly relevant to the role of TRAF3 in B-cell malignancies.
TRAF3 (TNF receptor-associated factor 3) is an important member of the TRAF adaptor family with distinct cell and context-specific roles (1). Our group previously generated CD19CreTRAF3flox/flox mice (B-Traf3−/−) that lack TRAF3 specifically in B cells. This deletion results in remarkably enhanced B-cell survival in vitro and in vivo (2). Aged B-Traf3−/− mice have substantially higher incidence of spontaneous B-cell lymphoma, further supporting the tumor-suppressive role of TRAF3 in B cells (3). Studies of human tumors identified loss-of-function TRAF3 mutations in nearly 20% of multiple myeloma (MM); TRAF3 is now recognized as one of the top 11 genes mutated in two-thirds of MM tumors (4). Additionally, ≥15% of diffuse large B-cell lymphomas are now known to harbor TRAF3 mutations (5, 6). Lesions in human TRAF3 genes are also seen in Hodgkin’s lymphoma (7) and associated with particular chromosome 14 deletions in various B-cell lymphomas (8). Interestingly, Traf3 mutations are also common in canine B-cell lymphomas (5).
TRAF3 is a negative regulator of the noncanonical NF-κB (NF-κB2) pathway, and enhanced survival in TRAF3-deficient B cells is associated with constitutive activation of NF-κB2 (2, 9). BAFF binds to BAFF receptor (BAFFR) to activate a complex signaling cascade that includes TRAF3 degradation and NF-κB2 activation, ultimately promoting B-cell survival (10, 11). However, NF-κB2 activation is not sufficient to promote enhanced survival, because TRAF3-deficient T cells and macrophages lack the enhanced survival phenotype, although they display constitutive NF-κB2 activation (12, 13). TRAF3 degradation is neither necessary nor sufficient for B-cell NF-κB2 activation (14). These findings indicate that TRAF3 regulates additional important prosurvival pathways in B cells.
Nuclear localization of TRAF3 has been reported in several nonhematopoietic cell types (15, 16), but the function of TRAF3 in the nucleus is poorly understood. Pathway analysis of preliminary microarray gene expression data comparing B cells isolated from WT (littermate control) and B-Traf3−/− mice identified cAMP response element binding protein (CREB) as a transcription factor potentially dysregulated in the absence of TRAF3 specifically in B cells, as this pathway was not altered in TRAF3−/− T cells.
CREB is a ubiquitous transcription factor implicated in survival and proliferation of immune cells (17). CREB has also been suggested to promote BAFF-mediated B-cell survival, via its induction of Mcl-1, an antiapoptotic member of the Bcl-2 family, but the mechanism of this induction is unknown (10). In this study, we show that TRAF3 is a resident nuclear protein in B cells, and that nuclear, but not cytoplasmic TRAF3 interacts with CREB to inhibit CREB stability and CREB-mediated transcription. An important CREB-regulated transcriptional target, Mcl-1, is up-regulated in TRAF3-deficient B cells. These findings have important implications for both the regulation of homeostatic B-cell survival, as well as the pathogenesis of B-cell malignancies that harbor TRAF3 deficiency.
Results
Association of Nuclear TRAF3 with CREB in B Cells.
In search of novel targets of TRAF3 that contribute to its B-cell–unique function in regulating homeostatic survival, we identified CREB as a transcription factor pathway, dysregulated specifically in B cells in the absence of TRAF3. In B cells, CREB is activated in response to B-cell antigen receptor (BCR) (18) or BAFFR stimulation, possibly downstream of the kinase Akt (10). Enhanced B-cell survival resulting from absence of TRAF3 is BAFF independent (2), and it was recently reported that TRAF3 is an important regulator of transcription factor degradation (19). We thus hypothesized that TRAF3 regulates CREB in B cells to restrain their homeostatic survival.
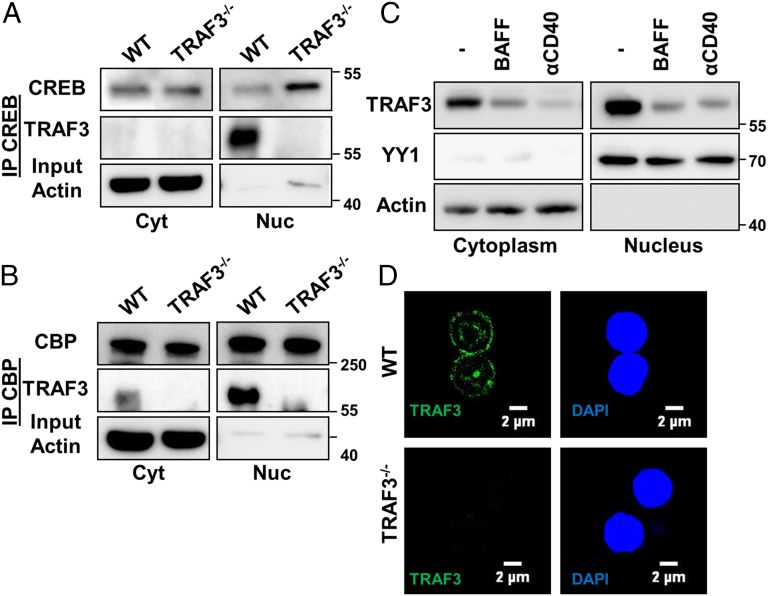
To investigate a potential association between TRAF3 and CREB, we immunoprecipitated CREB and its coactivator CREB-binding protein (CBP) from cytoplasmic and nuclear lysates of parental (TRAF3+/+) and TRAF3−/− subclones of the CH12.LX mouse B-cell lymphoma line (20). Both CREB and CBP specifically associated with TRAF3 in parental cells, but the association was seen almost exclusively in the nucleus and not the cytoplasm (Fig. 1 A and B).
Fig. 1.
Association of nuclear TRAF3 with CREB in B cells. (A and B) CH12.LX or CH12.TRAF3−/− B cells were fractionated into nuclear and cytoplasmic lysates. CREB (A) and CBP (B) were immunoprecipitated from the cytoplasmic and nuclear fractions. Western blots were probed for indicated proteins. Data from one of three independent experiments are shown. (C and D) Primary B cells were isolated from spleens of littermate WT and B-Traf3−/− mice. (C) The 5 × 106 WT B cells were stimulated with 200 ng/mL of recombinant BAFF or 5 μg/mL of anti-mouse CD40 mAb for 20 h at 37 °C. Nuclear and cytoplasmic cell lysates were immunoblotted for TRAF3, with actin and YY1 serving as loading controls. Representative blots from one of three independent experiments are shown. (D) Splenic B cells were stained with anti-TRAF3 Ab and assayed with immunofluorescence confocal microscopy. Representative images from one of three independent experiments are shown.
We confirmed nuclear localization of TRAF3 in mouse primary B cells via both cellular fractionation and in intact cells, using confocal microscopy (Fig. 1 C and D). CD40 and BAFFR are TNFR superfamily receptors that when stimulated promote TRAF3 degradation (21). Consistent with published studies, TRAF3 was susceptible to CD40 and BAFFR-mediated degradation both in the cytoplasm and the nucleus of primary B cells (Fig. 1C). These results show that TRAF3 is a resident nuclear protein that associates with CREB in B cells.
Requirement for the TRAF-C Domain for Nuclear Localization of TRAF3.
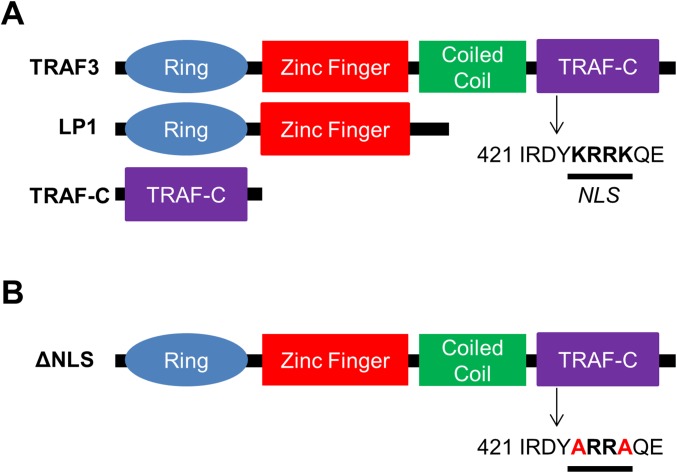
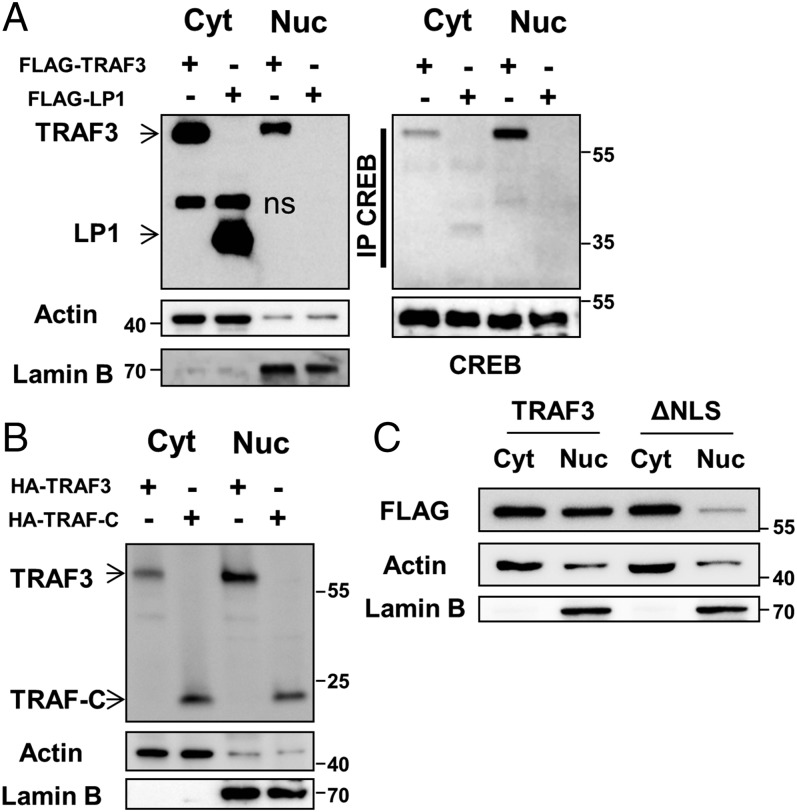
The nuclear localization signal (NLS) amino acid sequence targets proteins to the nucleus; abrogation of this signal inhibits nuclear import (22). We analyzed the TRAF3 amino acid sequence with the cNLS mapper program (23), and identified a putative monopartite NLS in the TRAF-C domain (Fig. S1A). Screening of human MM samples revealed a rather high frequency of TRAF3 mutations (4). One such mutant, a truncated form of TRAF3 lacking the TRAF-C domain, was identified in the LP1 MM cell line (LP1 mutant) (24) (Fig. S1A). When stably and inducibly expressed by transfection in TRAF3−/− CH12.LX B cells, LP1 promotes elevated NF-κB2 activation (14). Interestingly, LP1 failed to localize to the nucleus or associate with CREB (Fig. 2A). Furthermore, the TRAF-C domain alone was sufficient for nuclear localization (Fig. 2B). To confirm the role of the NLS in targeting TRAF3 to the nucleus, we generated a point mutant (ΔNLS) that has two lysines replaced with alanines in the NLS sequence (Fig. S1B). The ΔNLS mutant showed markedly inhibited capacity for nuclear localization (Fig. 2C). These findings demonstrate that the NLS in the TRAF-C domain of TRAF3 is critical for its trafficking to the nucleus. The presence of truncated TRAF3 mutants lacking that domain in human B-cell malignancies (24) strongly suggests that loss of nuclear TRAF3 may specifically contribute to disease pathogenesis.
Fig. S1.
Schematic representation of TRAF3 mutants LP1, TRAF-C (A), and ΔNLS (B). Location of the NLS sequence in the TRAF-C domain is indicated.
Fig. 2.
Requirement for the TRAF-C domain for nuclear localization of TRAF3. (A) CH12.TRAF3−/− B cells stably transfected for inducible expression of FLAG-tagged TRAF3 or the TRAF3 LP1 mutant were fractionated into nuclear and cytoplasmic lysates postinduction with IPTG for 24 h. CREB was immunoprecipitated from cytoplasmic and nuclear fractions and blots were probed for CREB and TRAF3. Blots of input lysates were probed for TRAF3 and LP1 expression with anti-FLAG Ab, with actin and lamin B serving as loading controls. Data are representative of three independent experiments. (B) Nuclear and cytoplasmic lysates from CH12.LX B cells, stably transfected for inducible expression of the HA-tagged TRAF-C mutant, were analyzed for TRAF-C expression with anti-HA Ab. Representative data from one of two independent experiments are shown. (C) CH12.TRAF3−/− B cells stably transfected for inducible expression of FLAG-tagged TRAF3 or the ΔNLS mutant were fractionated into nuclear and cytoplasmic lysates postinduction with IPTG for 24 h. Fractionated cell lysates were probed for TRAF3 and ΔNLS expression with anti-FLAG Ab, with actin and lamin B serving as loading controls. Data are representative of two independent experiments.
TRAF3-Mediated Negative Regulation of CREB.
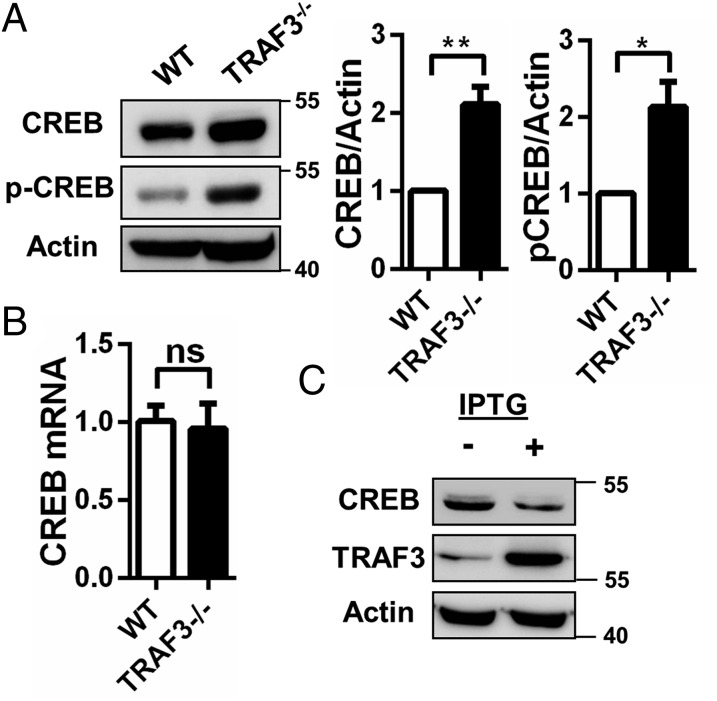
TRAF3 negatively regulates stability of certain transcription factors in macrophages (19). We thus hypothesized that TRAF3 interaction with CREB regulates CREB nuclear stability in B cells. B cells isolated from B-Traf3−/− (TRAF3−/−) mice had increased levels of both total and phosphorylated cellular CREB protein (Fig. 3A). Levels of mRNA were unchanged (Fig. 3B), indicating a posttranslational mechanism for TRAF3-mediated CREB regulation. Induction of TRAF3 expression in TRAF3−/− CH12.LX cells resulted in a decrease in CREB abundance, confirming that TRAF3 is specifically affecting CREB (Fig. 3C).
Fig. 3.
TRAF3-mediated negative regulation of CREB. (A–C) B cells were isolated from littermate WT and B-Traf3−/− mice. (A) Whole cell lysates of WT and TRAF3−/− B cells were analyzed with Western blotting for CREB and p-CREB (Ser133) expression. CREB and p-CREB band intensities were normalized to actin. Graphs depict mean values ± SEM from four independent experiments. (B) CREB mRNA levels in WT and TRAF3−/− B cells were assayed with RT-PCR and analyzed as described in Materials and Methods. Data were normalized to GAPDH and fold change was determined using the comparative Ct method. n = 3 mice with mean values ± SEM are shown. Student’s t test was used to evaluate differences for statistical significance in A and B (ns, not significant; *P < 0.05, **P < 0.01). (C) FLAG-tagged TRAF3 expression was induced in TRAF3−/− CH12.LX cells with IPTG for 24 h. Lysates of samples with and without IPTG treatment were analyzed for TRAF3 (with anti-FLAG antibody) and CREB expression with Western blotting.
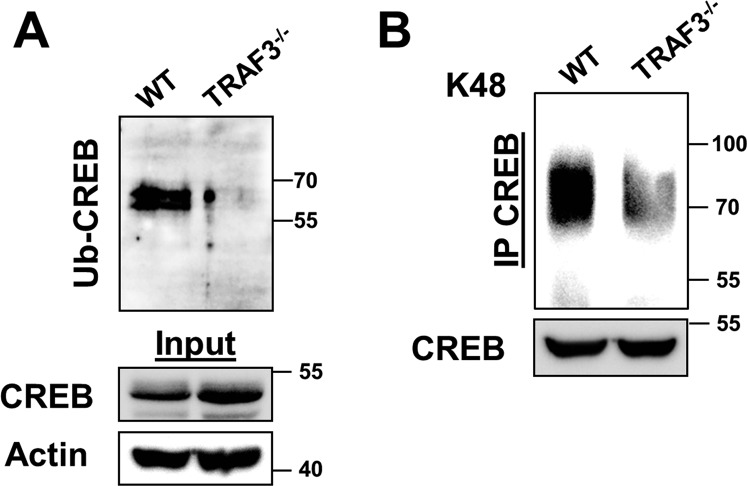
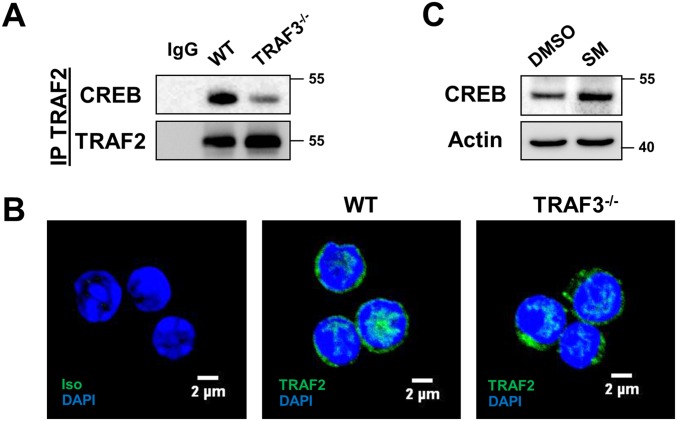
TRAF3 is known to regulate protein stability by forming an ubiquitin ligase complex with TRAF2 and cIAP1/2 (19). Loss of TRAF3 resulted in a decrease in total and K48 ubiquitination of CREB (Fig. S2 A and B). TRAF2 associated with CREB, and this association was substantially reduced in the absence of TRAF3 (Fig. S3A), although TRAF2 localized to the nucleus of B cells independently of TRAF3 (Fig. S3B). Smac mimetics (SMs) are small molecule inhibitors of IAP proteins (25), and CREB accumulated in B cells with SM treatment (Fig. S3C). Taken together, these data suggest that a TRAF3/TRAF2/cIAP1/2 complex is recruited to CREB in B cells and support the conclusion that TRAF3 regulates CREB stability by promoting its ubiquitination and degradation.
Fig. S2.
Changes in CREB ubiquitination in the absence of TRAF3. (A) Ubiquitinated fractions recovered from cell lysates of WT and TRAF3−/− B cells as described in Materials and Methods were blotted for CREB. Actin and CREB levels in input lysates are also shown. Data are representative of two independent experiments. (B) CREB was immunoprecipitated from lysates of WT and TRAF3−/− mouse primary B cells. Samples were analyzed with Western blotting for K48 linkage-specific ubiquitination. Data are representative of two independent experiments.
Fig. S3.
TRAF2 and cIAP1/2 involvement in TRAF3-mediated CREB regulation. (A) TRAF2 was immunoprecipitated from whole cell lysates of CH12.LX and CH12.TRAF3−/− cells. Western blots were probed for CREB and TRAF2. Lanes labeled IgG indicate samples were precipitated with isotype control Ab. Data are representative of two independent experiments. (B) WT and TRAF3−/− B cells were stained with anti-TRAF2 Ab and TRAF2 localization was assayed with immunofluorescence confocal microscopy. DAPI staining was used to identify nuclei. IgG control is also included. Representative images from one of three independent experiments are shown. (C) WT B cells were treated with 2.5 μM Smac mimetic (SM) for 24 h and CREB expression was assayed with Western blotting.
CREB-Mediated Induction of Mcl-1 in the Absence of TRAF3.
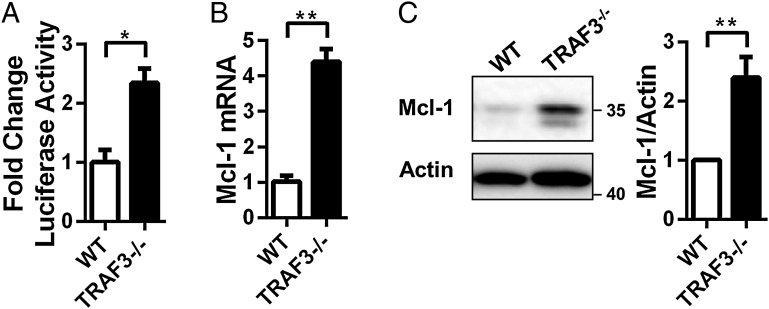
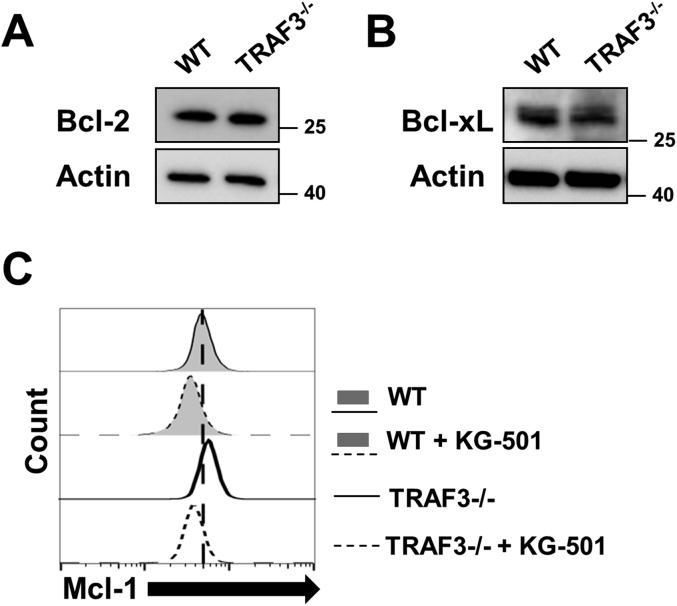
CREB regulates expression of many genes. Mcl-1, an antiapoptotic member of the Bcl-2 family of proteins, is induced by CREB (26) and is implicated in pathogenesis of a variety of malignancies (27). BAFFR signals induce Mcl-1 expression in an NF-κB–independent manner critical for promoting BAFF-mediated B-cell survival (11). Because loss of TRAF3 is also sufficient to promote BAFF-independent enhanced B-cell survival, we hypothesized that B-cell TRAF3 deficiency leads to CREB-mediated induction of Mcl-1. Loss of TRAF3 resulted in a significant increase in CREB-dependent reporter gene activation (Fig. 4A), indicating that CREB accumulation led to an increase in its transcriptional activity. Both mRNA and protein expression of Mcl-1 were increased in the absence of TRAF3 in mouse primary B cells (Fig. 4 B and C). Expression of other members of the Bcl-2 family, Bcl-2 and Bcl-xL, did not change with loss of TRAF3 (Fig. S4 A and B). From these results, we conclude that in the absence of TRAF3, protein accumulation leads to increased CREB activity and induction of Mcl-1.
Fig. 4.
CREB-mediated induction of Mcl-1 in the absence of TRAF3. (A) WT and TRAF3−/− B cells were transfected with a vector expressing luciferase under the control of the CRE promoter as described in Materials and Methods. Firefly luminescence was measured and normalized to Renilla luminescence. Mean values ± SEM of three independent experiments are shown, with values in WT B cells set as one. (B) Mcl-1 mRNA levels in WT and TRAF3−/− B cells were assayed with RT-PCR and analyzed as described in Materials and Methods. Data were normalized to GAPDH with fold change shown. n = 3 mice. Graphs depict mean values ± SEM (C) Whole cell lysates of splenic B cells were analyzed with Western blotting for Mcl-1. Mcl-1 band intensities were normalized to actin. Graphs depict mean values ± SEM from five independent experiments. Student’s t test was used for statistical significance in A–C (*P < 0.05, **P < 0.01).
Fig. S4.
B-cell expression of Bcl-2 family members. (A and B) B cells were isolated from littermate WT and B-Traf3−/− mice. Whole cell lysates of WT and TRAF3−/− B cells were analyzed with Western blotting for Bcl-2 (A) and Bcl-xL (B) expression. Representative blots of three independent experiments are shown. (C) WT and TRAF3−/− B cells were treated with either DMSO or 5 μM KG-501 for 24 h. Mcl-1 expression in intact cells was assayed with flow cytometry. Representative data from three independent experiments are shown.
TRAF3-Mediated Regulation of CREB in Human B Cells.
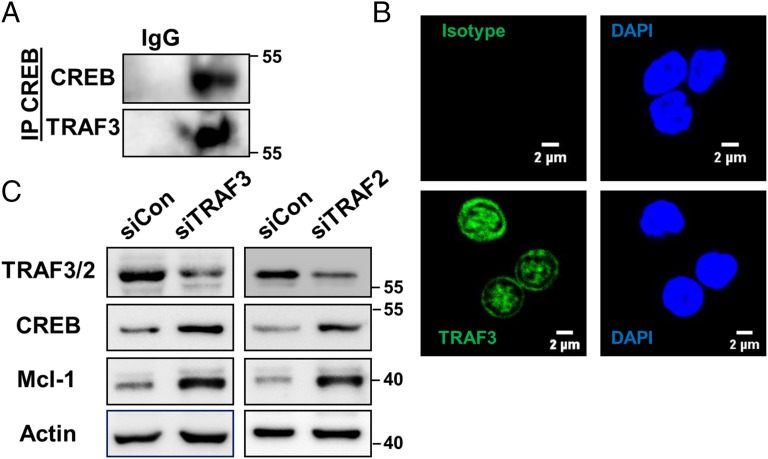
To further validate our findings in mouse models, we confirmed TRAF3 association with CREB and its nuclear localization in human B cells isolated from the peripheral blood mononuclear cells (PBMCs) of healthy donors (Fig. 5 A and B). Inhibition of TRAF3 or TRAF2 with specific siRNA resulted in increases in both CREB and Mcl-1 (Fig. 5C), indicating that TRAF3 regulates CREB in both mouse and human B cells.
Fig. 5.
TRAF3-mediated regulation of CREB in human B cells. (A) CREB was immunoprecipitated from lysates of human B cells and Western blots were probed for CREB and TRAF3. IgG Ab isotype control Ab is also shown. Data are representative of two independent experiments. (B) Human B cells were stained with anti-TRAF3 Ab and TRAF3 expression was assayed with immunofluorescence confocal microscopy. Isotype control is also shown. Representative images of one of three independent experiments are shown. (C) Human B cells were transfected with siRNA targeting TRAF3 (Left) or TRAF2 (Right) and control siRNA for 48 h. Whole cell lysates were immunoblotted for indicated proteins. Data are representative of three independent experiments.
Role of CREB in Survival of TRAF3−/− B Cells.
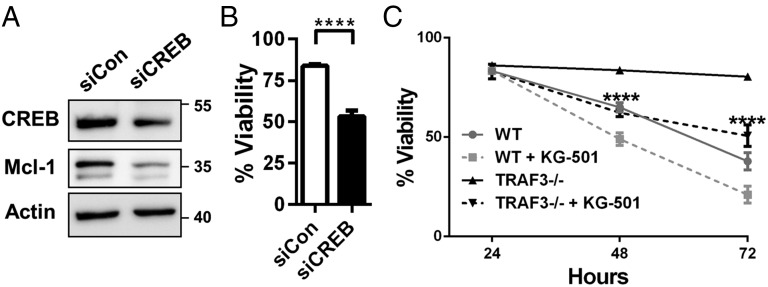
B-cell–specific deletion of TRAF3 results in enhanced B-cell survival in vitro and in vivo (2), and CREB promotes survival in B cells (28). We thus hypothesized that CREB-mediated induction of Mcl-1 plays a key role in the extended survival of TRAF3-deficient B cells. Inhibition of CREB expression with siRNA in TRAF3−/− mouse B cells resulted in decreases in both Mcl-1 levels and viability (Fig. 6 A and B). KG-501 is a chemical inhibitor that inhibits transcription factor complex assembly and expression of CREB target genes (29). Treatment of TRAF3−/− B cells with this inhibitor significantly attenuated cell survival (Fig. 6 C and D) with a greater overall impact than the effect of the inhibitor on WT B-cell viability. Mcl-1 expression was also reduced with inhibitor treatment (Fig. S4C). These findings implicate CREB activation as an important factor in the enhanced survival seen with loss of TRAF3 in B cells.
Fig. 6.
Role of CREB in survival of TRAF3−/− B cells. (A and B) TRAF3−/− mouse primary B cells were transfected with siRNA targeting CREB and control siRNA for 24 h. Samples were analyzed for protein expression with Western blotting (A). Cell viability was determined using propidium iodide staining and normalized to survival after electroporation. Graph depicts mean values ± SEM. n = 6 transfections from two independent experiments. Student’s t test was used to evaluate differences for statistical significance (****P < 0.0001). (C) WT and TRAF3−/− B cells were treated with DMSO or 3 μM KG-501. Percentage of viable cells identified with the Trypan Blue exclusion assay ± SEM is shown at indicated time points. Two-way ANOVA was used to analyze results for statistical significance and adjusted P value was calculated using the Bonferroni method (n = 3 or more from three independent experiments; **P < 0.01, ****P < 0.0001).
Discussion
Results presented here revealing the important nuclear role of B-cell TRAF3 in regulating B-cell homeostatic survival uncover a new paradigm of TRAF3-mediated regulation of cellular processes and help to explain the striking B-cell–specific effect of enhanced cellular survival that results from TRAF3 deficiency. As a resident nuclear protein, B-cell TRAF3 associates with the transcription factor CREB, regulating its availability and thus transcriptional impact. The importance of this regulation for inhibiting B-cell survival suggests that reduced nuclear TRAF3 promotes aberrant B-cell viability. Supporting this conclusion, the MM-derived TRAF3 mutant LP1 failed to localize to the nucleus or associate with CREB. This is consistent with previous findings that deliberate expression of LP1 in B cells promotes increased basal as well as CD40- and BAFFR-induced signaling (14). Determining the subcellular localization of truncated forms of TRAF3 in human tumors may thus provide a valuable insight into the possible contribution that loss of nuclear TRAF3 makes to pathogenesis of TRAF3-deficient B-cell malignancies. We identified a functional NLS sequence in the TRAF-C terminal of TRAF3; deletion of the NLS inhibited TRAF3 nuclear localization. Future studies are needed to determine the effect of NLS ablation on TRAF3 function in vivo and in vitro.
CD40 and BAFFR stimulation led to degradation of both cytoplasmic and nuclear TRAF3, indicating that receptor-mediated regulation of TRAF3 occurs in the nucleus as well as the cytoplasm. Mechanisms that regulate trafficking of TRAF3 in and out of the nucleus are an intriguing avenue of future research. One possibility is that TRAF3 can be SUMOylated (30), as SUMO modification is known to regulate nuclear import of proteins (31). Identification of signals that regulate shuttling of TRAF3 will help delineate nuclear and cytoplasmic roles of this versatile protein.
Our work shows that TRAF3 in the nucleus regulates degradation of proteins, similarly to its cytoplasmic role. TRAF3 association with nuclear CREB and CBP suggests that it also may regulate gene expression directly as part of a transcription factor complex or by direct interaction with DNA possibly through its Zn finger motifs, further expanding the regulatory potential of nuclear TRAF3.
Loss of TRAF3 led to increased Mcl-1 expression in a CREB-dependent manner but did not affect the expression of Bcl-2 or Bcl-xL. Interestingly, BAFF does not induce expression of Bcl-2 and Bcl-xL (32), suggesting that similarly to loss of TRAF3, BAFF-mediated B-cell survival is not due to induction of these members of the Bcl-2 family. CREB has been shown to induce Bcl-2 expression in response to BCR stimulation (18, 28). Cross-talk between BAFF and the BCR in regulating B-cell homeostasis is complex (33). Further studies delineating mechanistic distinctions in CREB activation and expression of target genes between these receptors are needed.
TRAF3 deficiency is associated with increased tumor susceptibility to proteasome inhibitors in MM (24). Identifying other agents with similar properties but potentially greater specificity is clinically desirable. Our findings show that TRAF3-deficient B cells undergo cell death when treated with CREB inhibitors, and CREB has been suggested as a potential target for cancer therapy (34). This suggests that B-cell malignancies characterized by loss of TRAF3 expression may show greater susceptibility to treatment strategies targeting CREB.
The current paradigm of TRAF3 function has been focused on its role as a cytoplasmic adaptor protein that regulates NF-κB2. The characterization of nuclear TRAF3 as a CREB regulator in B cells advances our understanding of its functions and highlights this organelle-specific role of TRAF3, a role that is likely very important to the observed frequency of TRAF3 deficiency associated with human B-cell cancers.
Materials and Methods
Mice.
TRAF3flox/flox mice bred with CD19Cre mice and extensively backcrossed onto the C57BL/6 background were previously described (2). Mice of 4–6 mo of age were used for all experiments. All mice were maintained under specific pathogen-free conditions and were used in accordance with National Institutes of Health guidelines under an animal protocol approved by the Animal Care and Use Committee at the University of Iowa.
Cell Lines and Mouse Primary B-Cell Isolation.
The mouse B-cell line CH12.LX and its TRAF3-deficient subclones have been described (20). CH12.TRAF3−/− cells lacking endogenous TRAF3, and subclones of these cells inducibly expressing FLAG-tagged human TRAF3 or its truncated LP1 mutant were described in detail in ref. 14. CH12.LX cells inducibly expressing HA-tagged mouse TRAF3 or the TRAF-C mutant were also previously reported (35). In these cell lines, expression is under control of the Lac repressor and can be induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich) treatment for 24 h. Resting splenic B cells were obtained as described (2). Briefly, high-density splenic B cells were isolated by anti-CD43 Ab-mediated negative selection, using a magnetic bead kit (Miltenyi Biotec), according to the manufacturer’s protocol. Cells were maintained in RPMI medium 1640 (Life Technologies) containing 10 μM 2-β-mercaptoethanol (Sigma), 10% heat-inactivated FCS (Atlanta Biologicals), 2 mM L-glutamine (Life Technologies), and 100 units/mL of penicillin–streptomycin antibiotics (Life Technologies). Medium additionally contained 400 μg/mL of G418 disulfate and 200 μg/mL of hygromycin (Life Technologies) for subclones expressing FLAG-tagged WT or LP1 TRAF3, and HA-tagged WT or TRAF-C mutant TRAF3.
Antibodies and Reagents.
IPTG, mouse anti-actin antibody (Ab), and anti-FLAG M2-HRP Ab were purchased from Sigma. Rabbit IgG isotype control, anti-CREB, anti–p-CREB anti-CBP, anti–Mcl-1, anti–Bcl-2 and anti–Bcl-xL Abs were purchased from Cell Signaling Technology. Rabbit anti-TRAF3 Ab for Western blotting was purchased from Santa Cruz Biotechnology and for immunofluorescence, from Abcam. Rabbit anti-TRAF2 Ab was from MBL. Rabbit K-48 linkage-specific Ab against ubiquitin was also purchased from Abcam. Stimulatory hamster anti-CD40 Ab was from eBioscience. Recombinant BAFF was from Peprotech. siRNA for TRAF2, TRAF3, and CREB was purchased from Santa Cruz and control siRNA from Origene. KG-501 was purchased from EMD Millipore. Smac mimetic was purchased from Tetralogic Pharmaceuticals.
Cellular Fractionation.
Cells were incubated in hypotonic buffer (10 mM Hepes pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 5 mM MgCl2) supplemented by 0.2 mM sodium vanadate, 1 mM DTT, and a protease inhibitor tablet (Roche) for 30 min. Swollen cells were lysed with 0.1% Nonidet P-40 and a 20-s vortex. Cytoplasmic lysate was removed and the pellet was incubated with hypotonic buffer + 0.1% Triton X-100 for 30 min to dissolve the membrane fraction. The nuclear pellet was resuspended in hypertonic buffer (20 mM Hepes pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA) supplemented by 0.2 mM sodium vanadate, 1 mM DTT, and a protease inhibitor tablet (Roche) and incubated for 30 min. The nuclear fraction was retrieved and the pellet was discarded.
Immunoprecipitation.
The 10 × 106 cells were lysed following the fractionation protocol. Nuclear and cytoplasmic lysates were incubated with an Ab against the protein of interest or isotype control Ab (Cell Signaling) overnight at 4 °C with constant agitation. The immune complex was precipitated with Dyna Protein G beads (Life Technologies), washed and resuspended in SDS/PAGE loading buffer, and heated to 95 °C for 10 min. For whole cell lysates, IP Lite lysis buffer (0.5% Triton X, 40 mM Tris, 100 mM NaCl, 1 mM CaCl2) containing a protease inhibitor tablet (Roche) and 100 μg/mL DNase I (Roche) was used. When assaying K-48 ubiquitination, lysis buffer was supplemented with 20 mM N-ethylmaleimide (Sigma).
Immunoprecipitation from Human B-Cell Lysates.
PBMCs were obtained from human whole blood leukocyte reduction cones obtained from the DeGowin Blood Center at the University of Iowa. Healthy donors 18–55 y of age provided written consent for their blood to be used in research projects, in compliance with the University of Iowa’s Institutional Review Board. Mononuclear cells were isolated with lymphocyte separation media (Mediatech). CD19+ B cells were purified with Miltenyi positive selection beads according to the manufacturer’s protocol. B cells were stimulated for 48 h with 5 μg/mL F(ab)′2 fragments of goat anti-IgM Ab (Jackson Immunoresearch), 5 μM CpG2006 (class B) purified oligonucleotides (Integrated DNA Technologies), and 2.5 μg/mL anti-human CD40 mAb (G28.5; hybridoma obtained from the American Type Culture Collection). Cells were rested for 6 h, washed with medium, and rested for 2 more hours. Cells were lysed with IP Lite and the immunoprecipitation protocol outlined above was followed.
Immunofluorescence Analysis of Cellular Protein Localization.
Cells were fixed with ice cold 100% methanol for 5 min. Rat serum and anti-CD16/32 Ab (eBioscience) blockade was performed for 30 min. Cells were stained overnight at 4 °C with primary Ab. Anti-TRAF3 Ab was conjugated to Alexa Fluor 488. For anti-TRAF2 Ab staining, cells were incubated in the dark at room temperature for 2 h with anti-rabbit Alexa Fluor 488 secondary Ab (Life Technologies). Following Ab incubation, cells were stained with DAPI (Invitrogen) for 10 min and then mounted on a slide with ProLong Gold Antifade Mountant (Life Technologies). Fluorescent images were acquired at room temperature using an LSM 710 confocal microscope (Zeiss) with 63× magnification, and images were acquired with Zen2009 software (Zeiss) at the University of Iowa Central Microscopy Research Facility. Images were further analyzed with ImageJ software (NIH).
Real-Time PCR.
RNA from mouse primary B cells was extracted with an RNeasy lipid extraction kit (Qiagen) and cDNA synthesized using SuperScript III polymerase (Invitrogen). RT-PCR was performed on an ABI PRISM 7900 Sequence Detection System (Applied Biosystems). Primers were purchased from Applied Biosystems (Mm00501607_m1 for CREB, Mm01257351_g1 for Mcl-1, and Mm99999915_g1 for GAPDH). Expression data were analyzed using the comparative Ct method (36) and normalized to GAPDH expression.
CREB Ubiquitination Assay.
B cells were lysed with IP Lite buffer as above and the ubiquitinated fraction was isolated with a Pierce Ubiquitin Enrichment kit (Life Technologies) following the manufacturer’s protocol. Resulting samples were blotted with anti-CREB Ab (Cell Signaling) for ubiquitinated CREB.
Luciferase Reporter Assay for CREB-Mediated Transcription.
A CREB reporter gene assay kit was purchased from BPS Bioscience. The 1 × 108 primary B cells isolated from littermate WT and B-Traf3−/− mice were activated with 20 μg of LPS (Sigma) and 10 ng/mL IL-4 (R&D Systems) for 24 h. Cells were transfected with 120 ng of luciferase reporter plasmid DNA by 200 V electroporation for 50 ms as previously described (20) and rested for 24 h. Cell lysates were analyzed for firefly and Renilla luciferase activities with the Dual-Glo Luciferase Reporter Assay kit (Promega) on a TD-20/20 Luminometer (Turner Designs) following the manufacturer’s protocol. Firefly luminescence from the CRE luciferase reporter vector was normalized to luminescence from the noninducible luciferase vector, with Renilla luminescence normalization controlling for transfection efficiency. Data were reported as fold change in luminescence over that of WT B cells.
Transfection of Human and Mouse B Cells with siRNA.
B cells were transfected using electroporation following a protocol previously described for primary T cells (37). The 5–10 × 106 cells were resuspended in 100 μL electroporation buffer. For TRAF3−/− mouse B cells, we used 2S buffer and for human B cells we used 1SM buffer. A total of 300 pmol of siRNA was added and cells were electroporated using the Nucleofector II electroporation system (Lonza). The X-001 program was used for mouse B cells and U-015 for human B cells. Cells were resuspended in 20% FCS-supplemented B-cell media and analyzed 24–48 h later.
Production of NLS Mutant TRAF3.
The putative NLS site in mouse TRAF3 was mutated using mouse TRAF3 cDNA as a template, and overlap extension PCR with Phusion HF (New England Biolabs) with the following primers: forward point mutant primer: 5′-CTA CGC TCG TCG TGC ACA GGA GGC CGT CAT GGG G-3; reverse point mutant primer: 5′-CTG TGC ACG ACG AGC GTA GTC ACG GAT CTT CCA GAT-3′; forward outboard primer: 5′-CTG TGC AAC CCG AAG CAG AC-3′; and reverse outboard primer: 5′-CAG ATC CGA AGT ATC CAC TAT GAC- 3′. The product was cloned into the inducible pOPRSV.neo vector, with 3× FLAG in frame at the 3′ end (36). The original protein sequence of the putative TRAF3 NLS site is RDYKRRKQ, and the mutated sequence is RDYARRAQ.
Sequence of the mutant NLS-containing plasmid was confirmed at the University of Iowa Genomic Core Facility. The plasmid was linearized using the restriction enzyme Ahd1, and TRAF3−/− CH12.LX cells, stably transfected with a plasmid coding the Lac repressor (35), were transfected with the mutant NLS plasmid using the previously described procedure (20). The cells were plated and grown under G418 and hygromycin selection, and growth-positive wells were harvested for nuclear/cytoplasmic separations following IPTG induction of expression of the mutant TRAF3.
Flow Cytometry.
Primary B cells isolated from littermate WT and B-Traf3−/− mice were stained for intracellular Mcl-1 expression using Cytofix/Cytoperm reagents (BD Bioscience). Anti-CD16/32 Ab (eBioscience) and rat serum were used for blocking. Cells were stained with anti-Mcl1 Ab (Cell Signaling) followed by anti-rabbit Alexa Fluor 488 secondary Ab. To assess viability, cells were stained with propidium iodide (Sigma). Flow cytometric analysis was performed on an Accuri C6 Flow Cytometer (BD Bioscience) and results were analyzed with FlowJo software (TreeStar).
Acknowledgments
We thank Dr. Ahmad Halwani for production of the FLAG-tagged human WT and LP1 TRAF3 plasmids and acknowledge the use of the University of Iowa Central Microscopy and Genomic Research Facilities, core resources supported by the vice president for Research and Economic Development; the Holden Comprehensive Cancer Center; and the Carver College of Medicine. This material is based upon work supported in part by facilities and equipment provided by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development. This work was supported by NIH Grants AI028847, CA97274, CA086862, and a Senior Research Career Scientist award from the Department of Veterans Affairs (to G.A.B.). N.M. was supported by NIH Awards T32AI007485 and T32GM007337.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514586113/-/DCSupplemental.
References
- 1.Hildebrand JM, et al. Roles of tumor necrosis factor receptor associated factor 3 (TRAF3) and TRAF5 in immune cell functions. Immunol Rev. 2011;244(1):55–74. doi: 10.1111/j.1600-065X.2011.01055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie P, Stunz LL, Larison KD, Yang B, Bishop GA. Tumor necrosis factor receptor-associated factor 3 is a critical regulator of B cell homeostasis in secondary lymphoid organs. Immunity. 2007;27(2):253–267. doi: 10.1016/j.immuni.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore CR, et al. Specific deletion of TRAF3 in B lymphocytes leads to B-lymphoma development in mice. Leukemia. 2012;26(5):1122–1127. doi: 10.1038/leu.2011.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.San Miguel JF. Introduction to a series of reviews on multiple myeloma. Blood. 2015;125(20):3039–3040. doi: 10.1182/blood-2015-01-613596. [DOI] [PubMed] [Google Scholar]
- 5.Bushell KR, et al. Genetic inactivation of TRAF3 in canine and human B-cell lymphoma. Blood. 2015;125(6):999–1005. doi: 10.1182/blood-2014-10-602714. [DOI] [PubMed] [Google Scholar]
- 6.Zhang B, et al. An oncogenic role for alternative NF-κB signaling in DLBCL revealed upon deregulated BCL6 expression. Cell Reports. 2015;11(5):715–726. doi: 10.1016/j.celrep.2015.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otto C, et al. Genetic lesions of the TRAF3 and MAP3K14 genes in classical Hodgkin lymphoma. Br J Haematol. 2012;157(6):702–708. doi: 10.1111/j.1365-2141.2012.09113.x. [DOI] [PubMed] [Google Scholar]
- 8.Nagel I, et al. Biallelic inactivation of TRAF3 in a subset of B-cell lymphomas with interstitial del(14)(q24.1q32.33) Leukemia. 2009;23(11):2153–2155. doi: 10.1038/leu.2009.149. [DOI] [PubMed] [Google Scholar]
- 9.Gardam S, Sierro F, Basten A, Mackay F, Brink R. TRAF2 and TRAF3 signal adapters act cooperatively to control the maturation and survival signals delivered to B cells by the BAFF receptor. Immunity. 2008;28(3):391–401. doi: 10.1016/j.immuni.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Mackay F, Schneider P. Cracking the BAFF code. Nat Rev Immunol. 2009;9(7):491–502. doi: 10.1038/nri2572. [DOI] [PubMed] [Google Scholar]
- 11.Woodland RT, et al. Multiple signaling pathways promote B lymphocyte stimulator dependent B-cell growth and survival. Blood. 2008;111(2):750–760. doi: 10.1182/blood-2007-03-077222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie P, Kraus ZJ, Stunz LL, Liu Y, Bishop GA. TNF receptor-associated factor 3 is required for T cell-mediated immunity and TCR/CD28 signaling. J Immunol. 2011;186(1):143–155. doi: 10.4049/jimmunol.1000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lalani AI, et al. Myeloid cell TRAF3 regulates immune responses and inhibits inflammation and tumor development in mice. J Immunol. 2015;194(1):334–348. doi: 10.4049/jimmunol.1401548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin WW, Hildebrand JM, Bishop GA. A complex relationship between TRAF3 and non-canonical NF-κB2 activation in B lymphocytes. Front Immunol. 2013;4:477. doi: 10.3389/fimmu.2013.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urbich C, et al. Upregulation of TRAF-3 by shear stress blocks CD40-mediated endothelial activation. J Clin Invest. 2001;108(10):1451–1458. doi: 10.1172/JCI13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y, et al. Spatiotemporal pattern of TRAF3 expression after rat spinal cord injury. J Mol Histol. 2014;45(5):541–553. doi: 10.1007/s10735-014-9575-2. [DOI] [PubMed] [Google Scholar]
- 17.Wen AY, Sakamoto KM, Miller LS. The role of the transcription factor CREB in immune function. J Immunol. 2010;185(11):6413–6419. doi: 10.4049/jimmunol.1001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blois JT, Mataraza JM, Mecklenbrauker I, Tarakhovsky A, Chiles TC. B cell receptor-induced CREB activation in B lymphocytes requires novel protein kinase Cδ. J Biol Chem. 2004;279(29):30123–30132. doi: 10.1074/jbc.M402793200. [DOI] [PubMed] [Google Scholar]
- 19.Jin J, et al. Proinflammatory TLR signalling is regulated by a TRAF2-dependent proteolysis mechanism in macrophages. Nat Commun. 2015;6:5930. doi: 10.1038/ncomms6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie P, Hostager BS, Bishop GA. Requirement for TRAF3 in signaling by LMP1 but not CD40 in B lymphocytes. J Exp Med. 2004;199(5):661–671. doi: 10.1084/jem.20031255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bishop GA, Xie P. Multiple roles of TRAF3 signaling in lymphocyte function. Immunol Res. 2007;39(1-3):22–32. doi: 10.1007/s12026-007-0068-1. [DOI] [PubMed] [Google Scholar]
- 22.Lange A, et al. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J Biol Chem. 2007;282(8):5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA. 2009;106(25):10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keats JJ, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12(2):131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fulda S. Molecular pathways: Targeting death receptors and smac mimetics. Clin Cancer Res. 2014;20(15):3915–3920. doi: 10.1158/1078-0432.CCR-13-2376. [DOI] [PubMed] [Google Scholar]
- 26.Wang JM, et al. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol Cell Biol. 1999;19(9):6195–6206. doi: 10.1128/mcb.19.9.6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perciavalle RM, Opferman JT. Delving deeper: MCL-1's contributions to normal and cancer biology. Trends Cell Biol. 2013;23(1):22–29. doi: 10.1016/j.tcb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson BE, Mochon E, Boxer LM. Induction of bcl-2 expression by phosphorylated CREB proteins during B-cell activation and rescue from apoptosis. Mol Cell Biol. 1996;16(10):5546–5556. doi: 10.1128/mcb.16.10.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Best JL, et al. Identification of small-molecule antagonists that inhibit an activator: Coactivator interaction. Proc Natl Acad Sci USA. 2004;101(51):17622–17627. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miliara S, Gkouskou KK, Sharp TV, Eliopoulos AG. SUMOylation is required for optimal TRAF3 signaling capacity. PLoS One. 2013;8(11):e80470. doi: 10.1371/journal.pone.0080470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4(9):690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 32.Craxton A, Draves KE, Gruppi A, Clark EA. BAFF regulates B cell survival by downregulating the BH3-only family member Bim via the ERK pathway. J Exp Med. 2005;202(10):1363–1374. doi: 10.1084/jem.20051283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackay F, Figgett WA, Saulep D, Lepage M, Hibbs ML. B-cell stage and context-dependent requirements for survival signals from BAFF and the B-cell receptor. Immunol Rev. 2010;237(1):205–225. doi: 10.1111/j.1600-065X.2010.00944.x. [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto KM, Frank DA. CREB in the pathophysiology of cancer: Implications for targeting transcription factors for cancer therapy. Clin Cancer Res. 2009;15(8):2583–2587. doi: 10.1158/1078-0432.CCR-08-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hostager BS, Bishop GA. Cutting edge: Contrasting roles of TNF receptor-associated factor 2 (TRAF2) and TRAF3 in CD40-activated B lymphocyte differentiation. J Immunol. 1999;162(11):6307–6311. [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Chicaybam L, Sodre AL, Curzio BA, Bonamino MH. An efficient low cost method for gene transfer to T lymphocytes. PLoS One. 2013;8(3):e60298. doi: 10.1371/journal.pone.0060298. [DOI] [PMC free article] [PubMed] [Google Scholar]