Each human individual carries ∼30–60 de novo mutations that arose in the germ line of his or her parents (1). At the population level, the result is a massive influx of new mutations every generation, whose evolutionary trajectory is shaped by the joint effects of stochastic and deterministic evolutionary forces. Indeed, considerable information about human history, including changes in population size, structure, and timings of major diasporas, is embedded in extant patterns of neutral genetic variation (2). For example, genetic data from global human populations show a striking gradient of declining genetic diversity with distance from Africa, which has been attributed to serial founder effects associated with the Out-of-Africa migration and subsequent global expansion (3–5). However, in contrast to neutral variation, considerably less is known about geographic patterns of deleterious variation, the fate of which is governed by both random genetic drift and purifying selection. In PNAS, Henn et al. (6) address this question by performing whole-genome sequencing of 54 individuals from seven geographically diverse populations. The authors show that differences in demographic history among populations have influenced patterns of deleterious mutations in a manner predicted by a model of serial founder effects during geographic range expansions (Fig. 1). In addition to revealing new insights into the interaction between selection and demography during recent human evolution, the study quantifies within- and between-population variation in mutational burden, with implications for predicting individual disease risk.
Fig. 1.
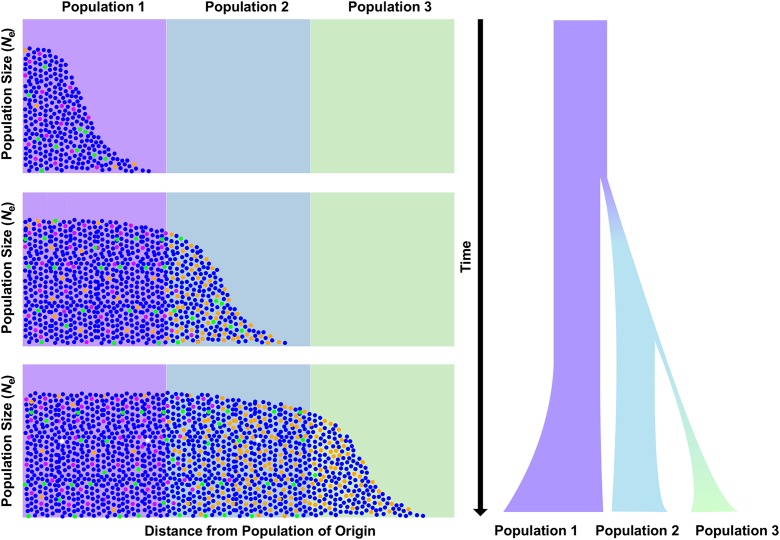
Schematic illustration of the interaction between selection and genetic drift during serial bottlenecks and range expansion. As a consequence of strong genetic drift at the edge of the expanding wave, deleterious alleles (orange) that were previously held at low frequencies by purifying selection may surf to higher frequencies. Serial bottlenecks also lead to the loss of many alleles. Together, these processes produce a pattern of declining diversity with distance from the origin of expansion, along with pushing deleterious alleles that survive toward higher frequencies.
The work of Henn et al. (6) is timely, as it addresses an issue of considerable interest, complexity, and recent controversy (7–12). Major points of contention arising from genome-scale studies include whether empirical patterns of deleterious variation vary among human populations, whether such differences lead to differences in genetic load, and the relative contributions of drift and selection in shaping these patterns. Henn et al. (6) revisit these topics armed with novel statistical methods and new genome-sequence data from geographically diverse individuals. A key novelty of this work is that the authors consider predictions of mutational effect size in their analyses. Specifically, they stratified mutations according to Genomic Evolutionary Rate Profiling (GERP) score, a method that quantifies the degree of evolutionary constraint inferred from a multispecies genome alignment (13). Henn et al. (6) observed a statistically significant elevation in the number of deleterious alleles per individual in non-African compared with African populations, as well as an increase in the number of homozygous deleterious genotypes carried by individuals as a function of distance from Africa. These observations were largely driven by mutations predicted to have modest effects, consistent with theoretical predictions (8, 14). It is important to note, however, that although statistically significant differences were detected, absolute interpopulation differences in average number of deleterious alleles per individual were small. Furthermore, there remain substantial limitations to assigning fitness effects to individual variants identified in sequencing datasets. Existing functional annotation methods are based primarily on evolutionary conservation, as well as predicted impacts on protein structure for nonsynonymous variants. However, there is relatively low concordance among sets of variants predicted to be deleterious by different algorithms, and experimental validation of these methods suggests a modest ability to distinguish clinically relevant and nearly neutral mutations (15). Furthermore, it is plausible that fitness effects vary according to both the external and genomic environment (i.e., epistasis). Despite these limitations, the work of Henn et al. (6) substantially adds to a growing body of work (reviewed in ref. 16), clearly demonstrating that patterns of deleterious mutations vary among human populations.
To better understand the origins of these differences, Henn et al. (6) performed spatially explicit forward simulations to show that a model of serial founder effects during range expansion can recapitulate features of deleterious variation observed in the empirical data. To quantify the impacts of simulated demographic events on deleterious versus neutral variation, the authors devised a statistic called RH, defined as (Hnue – Hdel)/Hneu, where Hneu indicates heterozygosity at neutral sites and Hdel indicates heterozygosity at sites predicted to be deleterious based on their GERP score. This statistic was then compared across populations to determine whether selection had distinct impacts relative to drift in different populations. The authors found that RH was significantly lower in non-African relative to African populations across a wide range of GERP scores. A diminished strength of selection relative to drift as a consequence of a demographic bottleneck is a classic prediction of population genetic theory. Specifically, Ohta (14) showed that the product of the effective population size (Ne) and the selection coefficient (s) determines the behavior of a deleterious mutation. When s << 1/4Ne, mutations will behave as effectively neutral. During a bottleneck, more mutations meet this condition, and drift allows some to reach high frequencies and occasionally fix in the population (Fig. 1). The true demographic history of human populations is certainly more complex than the serial bottleneck model studied by Henn et al. (6), including ancient introgression with other hominin species, long-distance dispersal, population replacement, and repeated admixture among human lineages (17). Nonetheless, the fact that a simple model captures key features of the data underscores the importance of serial bottlenecks in shaping patterns of deleterious variation.
Henn et al. (6) also addressed the more difficult question of whether genetic load varies among the study populations. The significance of genetic load—the aggregate fitness reduction as a result of deleterious mutations—has been controversial since its conception by Haldane (18). Formalized by Crow (19), genetic load (L) is quantified by comparing the average fitness in the population () to that of a theoretical genotype with the maximum possible fitness (generally scaled to 1), such that L = 1 – . Henn et al. (6) show that estimates of L are highly sensitive to model parameters, such as the distributions of fitness and dominance effects. These quantities are among the least understood in population genetics, although mutation accumulation experiments in model organisms (20) and large-scale sequencing of rare mutations are providing paths forward in this regard. Given the uncertainty about the distribution of dominance effects, Henn et al. (6) evaluated two extreme models to place bounds on their estimates:
The work of Henn et al. is an important step in narrowing the divide between seemingly contradictory observations and conclusions regarding patterns of deleterious mutations among human populations.
a model in which the fitness effects of all deleterious mutations are completely recessive and a model in which all deleterious mutations are perfectly additive. The authors found that load increases with distance from Africa for the recessive model (by ∼10–30%), but is approximately equal across populations for the additive model, consistent with earlier work (11, 12). As the true distribution of dominance effects undoubtedly lies somewhere in between, these results support the interpretation that genetic load differs modestly among populations.
In summary, the work of Henn et al. (6) is an important step in narrowing the divide between seemingly contradictory observations and conclusions regarding patterns of deleterious mutations among human populations. It is becoming increasingly clear that some population genetic characteristics of deleterious mutations vary among populations, but as is the case with neutral mutations, differences among populations are small relative to differences among individuals. Nonetheless, there remains much to learn about the dynamics of nonneutral variation in nonequilibrium populations, particularly for more realistic, complex, and spatially explicit models of human history. In addition to evolutionary questions, the study of deleterious mutations has practical implications for understanding and predicting human disease. As we enter the era of personalized medicine, it is important to consider which measures of damaging mutations best capture individual disease risk. While genetic load is a useful construct in theoretical population genetics, its utility for studying human disease is limited. Studies like that of Henn et al. (6) bring us closer to the goal of characterizing the individual burden of deleterious mutations, as well as understanding the complex evolutionary processes that produced it.
Footnotes
The authors declare no conflict of interest.
See companion article on page E440.
References
- 1.Shendure J, Akey JM. The origins, determinants, and consequences of human mutations. Science. 2015;349(6255):1478–1483. doi: 10.1126/science.aaa9119. [DOI] [PubMed] [Google Scholar]
- 2.Schraiber JG, Akey JM. Methods and models for unravelling human evolutionary history. Nat Rev Genet. 2015;16(12):727–740. doi: 10.1038/nrg4005. [DOI] [PubMed] [Google Scholar]
- 3.Ramachandran S, et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc Natl Acad Sci USA. 2005;102(44):15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Excoffier L, Foll M, Petit RJ. Genetic consequences of range expansions. Annu Rev Ecol Evol Syst. 2009;40:481–501. [Google Scholar]
- 5.Peischl S, Dupanloup I, Kirkpatrick M, Excoffier L. On the accumulation of deleterious mutations during range expansions. Mol Ecol. 2013;22(24):5972–5982. doi: 10.1111/mec.12524. [DOI] [PubMed] [Google Scholar]
- 6.Henn BM, et al. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci USA. 2016;113:E440–E449. doi: 10.1073/pnas.1510805112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohmueller KE. The distribution of deleterious genetic variation in human populations. Curr Opin Genet Dev. 2014;29:139–146. doi: 10.1016/j.gde.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Peischl S, Excoffier L. Expansion load: Recessive mutations and the role of standing genetic variation. Mol Ecol. 2015;24(9):2084–2094. doi: 10.1111/mec.13154. [DOI] [PubMed] [Google Scholar]
- 9.Lohmueller KE, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451(7181):994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu W, Gittelman RM, Bamshad MJ, Akey JM. Characteristics of neutral and deleterious protein-coding variation among individuals and populations. Am J Hum Genet. 2014;95(4):421–436. doi: 10.1016/j.ajhg.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simons YB, Turchin MC, Pritchard JK, Sella G. The deleterious mutation load is insensitive to recent population history. Nat Genet. 2014;46(3):220–224. doi: 10.1038/ng.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Do R, et al. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat Genet. 2015;47(2):126–131. doi: 10.1038/ng.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLOS Comput Biol. 2010;6(12):e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 1992;23:263–286. [Google Scholar]
- 15.Miosge LA, et al. Comparison of predicted and actual consequences of missense mutations. Proc Natl Acad Sci USA. 2015;112(37):E5189–E5198. doi: 10.1073/pnas.1511585112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henn BM, Botigué LR, Bustamante CD, Clark AG, Gravel S. Estimating the mutation load in human genomes. Nat Rev Genet. 2015;16(6):333–343. doi: 10.1038/nrg3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickrell JK, Reich D. Toward a new history and geography of human genes informed by ancient DNA. Trends Genet. 2014;30(9):377–389. doi: 10.1016/j.tig.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haldane JBS. The effect of variation of fitness. Am Nat. 1937;71(735):337–349. [Google Scholar]
- 19.Crow JF. Some possibilities for measuring selection intensities in man. Hum Biol. 1958;30(1):1–13. [PubMed] [Google Scholar]
- 20.Halligan DL, Keightley PD. Spontaneous mutation accumulation studies in evolutionary genetics. Annu Rev Ecol Evol Syst. 2009;40:151–172. [Google Scholar]