The Problem: Short-Lived Heterogeneous Nuclear RNA
During the 1960s and 1970s, there was a major conundrum in eukaryotic molecular biology regarding the synthesis of mRNA in animal cells. Pulse and pulse-chase radio-labeling experiments with tritiated uridine had shown that a large fraction of nuclear RNA was degraded very rapidly after synthesis, whereas only a small fraction of the initially synthesized RNA was exported from the nucleus to the cytoplasm where it functions as far more stable mRNAs (1, 2). This short-lived nuclear RNA ranged in lengths up to several tens of kilobases, much longer than most cytoplasmic mRNAs, and was referred to as heterogeneous nuclear RNA (hnRNA) (3, 4). Pulse-chase labeling yielded results consistent with hnRNA functioning as a precursor of mRNA. hnRNA is rapidly labeled, whereas label appeared in mRNAs more slowly. Also, as for mRNA, a large fraction of hnRNA was shown to be polyadenylated at its 3′ end like mRNA, consistent with the model that RNA sequences near the 3′ end of hnRNAs are retained in shorter mRNAs exported to the cytoplasm (5, 6). Also, like mRNAs, hnRNAs were found to have the 7-methyl guanine 5′–5′ phosphotriester “cap” structure at their 5′ ends, like mRNAs. Remarkably, the methyl groups in hnRNA cap structures appeared to be conserved as RNA was exported to the cytoplasm, even though, on average, hnRNA molecules in the nucleus are at least four times longer than mRNAs in the cytoplasm (7, 8). However, it was not possible to prove rigorously from the flow of radiolabel in pulse-chase labeling experiments that hnRNAs are precursors to mRNAs. There were two principal reasons for this. First, the pool of ribonucleoside triphosphates in mammalian cells is so large that it takes more than 30 min to saturate the pools when labeled nucleosides are added to the media and equally long to dilute the intracellular pool of labeled nucleoside triphosphates by addition of a large excess of unlabeled nucleoside to the culture medium during the chase period. Consequently, a clean chase could not be achieved, and newly synthesized hnRNAs continued to be labeled as labeled mRNAs first appeared in the cytoplasm. Also, the small fraction of pulse-labeled nuclear RNA converted to more stable mRNAs, only 5–10%, added to the difficulty of proving a precursor product relationship from pulse-chase labeling experiments.
The synthesis and processing of ribosomal RNAs was much easier to analyze than processing of hnRNAs because about 50% of all nuclear RNA synthesis in growing mammalian cells in culture is synthesis of the single ∼13.7-kb pre-rRNA precursor. Moreover, this full-length pre-rRNA precursor accumulates to significant levels before it is processed into stable mature rRNAs of 18S (1,870 bases), 5.8S (156 bases), and 28S (5,034 bases), so that the fraction of pre-RNA processed into stable cytoplasmic rRNAs is ∼50%, much more than the 5–10% of pulse-labeled hnRNA processed into mRNAs. This fortuitous situation for the study of prerRNA by pulse-chase labeling made it possible to show that the rRNAs were processed by the cleavage of the pre-RNA precursor into the stable rRNAs that were transported to the cytoplasm in nearly fully mature ribosomal subunits. So-called “transcribed spacer” sequences between the stable rRNAs in the pre-rRNA precursor were rapidly degraded after they were generated by cleavage of the pre-rRNA.
Because of this precedent for the processing of a large pre-rRNA precursor into the mature rRNAs by simple cleavage of the pre-rRNA precursor, the predominant hypothesis for the synthesis of mRNAs from longer hnRNAs was that they were similarly cleaved out of larger hnRNA molecules. However, what would specify where each postulated pre-mRNA precursor is cleaved to generate mRNAs much more stable than the precursor hnRNAs? Other significant questions were raised by the observation of unstable hnRNA. For example, do the many unstable hnRNA sequences removed from hnRNAs during their processing have specific functions?
To address these questions and other aspects of mRNA synthesis and processing, many researchers turned to the study of DNA viruses that infect animal cells in culture. Detailed analysis of gene expression by bacteriophage (bacterial viruses) had revealed (and continues to reveal) insights into the mechanisms of gene expression and its control in bacteria. In the days before molecular cloning of cellular genes (around the late 1970s), it was very difficult to analyze the expression of individual cellular genes (aside from pre-rRNA) because a large number of cellular genes (>10,000) are all expressed at the same time and because it was not yet possible to obtain DNA clones of individual mammalian genes. DNA viruses express a much smaller number of genes, and the purified template DNAs encoding them could be isolated from purified virion particles. A worldwide group of researchers chose human adenovirus 2 (Ad2) and the closely related human adenovirus 5 as model systems for studying mRNA synthesis in animal cells. These viruses had the advantage that they have a genome of ∼36 kb and can be readily grown in large amounts in cultured cells so that milligram amounts of the viral genome could be isolated from purified virions. Also, at 36 kb, the Ad2 genome was similar in size and therefore potential biological complexity to the genomes of bacteriophages T7 and λ. T7 and λ had provided intriguing examples of gene control for analysis at a molecular level. It was hoped that studies of Ad2 gene expression would be similarly revealing about mechanisms controlling gene expression and mRNA synthesis in animal cells. Importantly, Ad2 had also been shown to express short-lived, high-molecular-weight nuclear RNAs containing the sequences of shorter, more stable viral mRNAs (9).
The Experiment
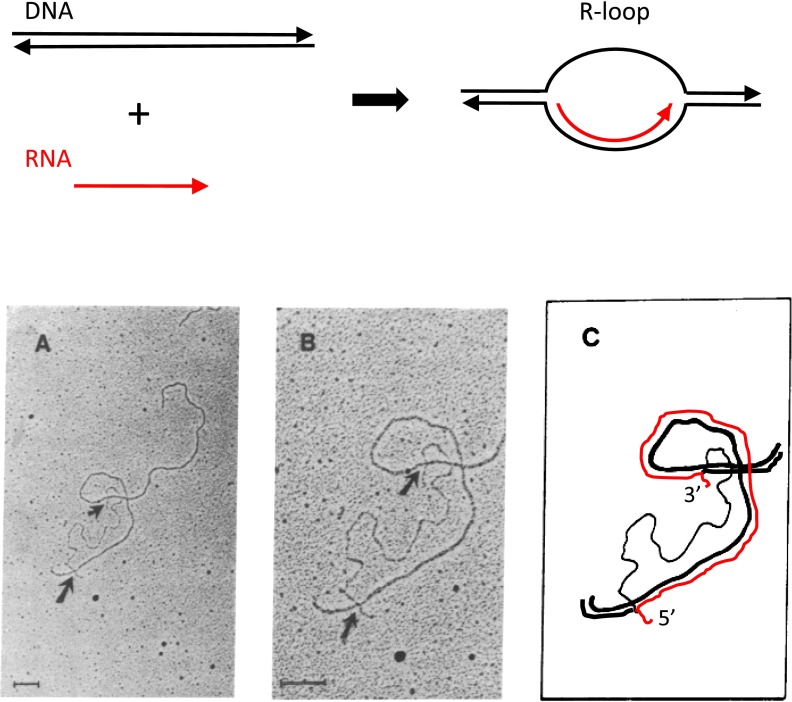
One of the first goals in the analysis of Ad2 mRNA synthesis was to map the regions of the viral genome encoding the abundantly expressed viral mRNAs for virion structural proteins expressed late in infection. This was of interest because in bacteria, DNA sequences near the transcription start sites for bacterial mRNAs function as promoters where bacterial RNA polymerase binds DNA and initiates transcription. Other bacterial DNA-binding proteins that bind sequences within a few tens of base pairs from promoters regulate the frequency of transcription initiation. It was anticipated that DNA sequences near the 5′ ends of regions encoding the abundant late viral mRNAs might also harbor promoter and other regulatory sequences in Ad2 DNA. In the 1970s, methods had been developed for observing regions of RNA base paired to a longer DNA molecule by electron microscopy (EM) of the hybrid molecules. The resolution of the method allowed mapping of the ends of the RNA-DNA hybrid region to within an accuracy of about 100 bp on the viral genome.
When I arrived to begin my postdoc in Phil Sharp's laboratory in the summer of 1976, Sue Berget, another postdoc who had started in the laboratory about a year earlier, was working on mapping the Ad2 genome location encoding the most abundantly expressed Ad2 mRNA, the mRNA for the major virion structural protein hexon. This was to be done by the highest-resolution method available at the time: EM of RNA-DNA hybrids. In July 1976, the David Hogness’ and Ron Davis’ laboratories had reported (10, 11) that in 70% formamide and 0.2–0.7 M NaCl, a precisely complementary RNA-DNA hybrid has a melting temperature 9 °C higher than a DNA-DNA hybrid of the same sequence. When complementary RNA was incubated with duplex DNA in 70% formamide just below the melting temperature of the DNA, because the RNA-DNA hybrid is more stable than the DNA-DNA duplex, the RNA hybridizes to its complementary DNA sequence in the DNA molecule, displacing the noncomplementary DNA strand as a single-stranded region, forming what was termed an “R-loop” (Fig. 1, Upper). The region of RNA-DNA hybrid can be measured by EM of the R-loops (Fig. 1A), and the distance from the branch-point where the RNA-DNA hybrid ended to the end of the duplex DNA molecule determined the position of the complementary region of DNA. Sue Berget applied the R-looping method with hexon mRNA, which can be readily visualized in an ethidium bromide-stained polyacrylamide gel of total polyA+ polysomal RNA isolated from Ad2-infected cells late in infection (ref. 12, figure 1). The band of hexon mRNA was cut from the gel, eluted, and hybridized under R-loop conditions to Ad2 DNA restriction fragments from the region of the Ad2 genome where hexon mRNA hybridized. The approximate region of the Ad2 genome encoding hexon had been determined earlier by in vitro translation of RNAs from infected cells hybridized to filter-bound Ad2 DNA restriction fragments (13). R-loops of hexon mRNA with Ad2 restriction fragments EcoRI A, EcoRI B, and HindIII A were observed and analyzed, mapping the region encoding most of the length of the hexon mRNA to 50.1–73.6 map units on the Ad2 genome (100 map units is ∼36 kb) (ref. 12, figure 4 A–F).
Fig. 1.
Single-stranded RNA tails observed at both 5′ and 3′ ends of R-loops of Ad2 hexon mRNA and the Ad2 HindIII A fragment. (Upper) R-loop formation near the temperature of DNA melting in 70% formamide, 0.4 M NaCl. Arrowheads indicate 3′ ends of strands. Adapted from ref. 10. (Lower) R-loop in Ad2 HindIII A restriction fragment formed with purified hexon mRNA. (A) EM of R-loop showing all of the Hind III A DNA fragment with an R-loop at the lower end. (B) Higher magnification of the R-loop. In A and B, the upper arrow points to a single-stranded region at the 3′ end of the RNA, and the bottom arrow points to a single-stranded region at the 5′ end. In these formamide spreads, single-stranded DNA appears thinner and has a more irregular path than double-stranded DNA or RNA-DNA hybrid, which appear similar. (C) Interpretation of EM in B. The hexon mRNA is represented in red and each strand of the Hind III A DNA in black. Regions where the red RNA is parallel to the black DNA strand represent base paired regions of RNA-DNA hybrid. Regions where the two black DNA strands are parallel represent DNA-DNA duplex. A, B, and C adapted from ref. 12.
However, careful examination of R-loops formed between hexon mRNA and the HindIII A fragment, which included the region encoding most of the length of the hexon mRNA, revealed an unexpected result: because of their skill in the tricky art of preparing “formamide spreads” that allow visualization of single-stranded regions of nucleic acids, Sue Berget and Claire Moore, who assisted Sue with EM, observed single-stranded tails of RNA of ∼100–200 nucleotides at both the 5′ end (relative to the hexon mRNA) and the 3′ end of the R-loops (Fig. 1 A–C). The single-stranded RNA at the 3′ end of the R-loop could be explained by the polyA tail on hexon mRNA that is added following transcription and consequently is not complementary to Ad2 DNA. However, what could be the explanation for the ∼200 bases of RNA at the 5′ end of the message that did not hybridize with viral DNA?
One possibility was that for some reason, the RNA-DNA hybrid formed between the 5′ end of the hexon mRNA and the complementary region of Ad2 DNA in the HindIII A fragment was less stable than the RNA-DNA hybrid formed by most of the length of the hexon mRNA, allowing the complementary DNA strands at the 5′ end of the R-loop to base pair, extruding the 5′ end of the mRNA. To test this idea, hexon mRNA was hybridized to the coding single strand of the HindIII A fragment. In such a hybrid, the complementary DNA strand would not be present to displace the 5′ end of the mRNA. This experiment was greatly simplified by the recent report from James Casey and Norman Davidson (Phil Sharp's postdoctoral advisor) (14) that hybridization of a complementary RNA to a DNA fragment above the melting temperature of the DNA in 80% formamide and 0.4 M NaCl, but below the 9 °C higher melting temperature of the RNA-DNA hybrid, allowed formation of RNA-DNA hybrid in the absence of hybridization of the complementary DNA strands. Consequently, Sue Berget hybridized the purified hexon mRNA to the HindIII A Ad2 DNA fragment under these “Casey–Davidson” conditions and examined the products by EM. As expected from the Casey–Davidson results, RNA-DNA hybrids were observed in the absence of DNA-DNA hybrids (ref. 12, figure 4 D–F). Remarkably, the 5′ end of the RNA was observed as a single-stranded region of ∼200 nucleotides that branched off from the region of RNA-DNA duplex, just as observed for the 3′ poly(A) tail at the other end of the RNA. This result proved unequivocally that the 5′ ∼200 nt of the hexon mRNA was not complementary to the Ad2 DNA sequence just 5′ to the region from 50.1 to 73.6 map units that encoded most of the length of the hexon mRNA.
My recollection of the situation at this point is that a paper was submitted to PNAS reporting the observation that the 5′ end of the hexon mRNA was not encoded by the region of Ad2 DNA encoding most of the length of the hexon mRNA. However, reviewers of the paper felt that it was important to determine the origin of the ∼200-nt sequence at the 5′ end of hexon mRNA. This seemed like a tall order indeed, because it seemed possible that this sequence could be supplied from a host cell RNA added to the 5′ end of the virus encoded protein coding region of the hexon mRNA. However, after much discussion in the laboratory and with other colleagues on the fifth floor of the Cancer Research Center at the Massachusetts Institute of Technology (MIT) where this work was performed, it was decided that a potential source of this “5' tail” sequence might be from a region of the Ad2 genome transcriptionally upstream from the region encoding the main body of the hexon mRNA. This idea immediately suggested a method to search for an upstream region encoding the 5′ leader: hybridization of hexon mRNA under Casey–Davidson conditions to the large EcoRI A fragment that extended from the left end of the genome to within a few hundred base pairs of the region that had been mapped to encode the main body of the hexon mRNA. If the 5′ tail sequence was encoded by an upstream region of DNA, one would expect to observe a single-stranded loop of DNA extending from the left end of the region hybridized to the main body of the hexon mRNA to the upstream sequence hybridized to the 5′ tail.
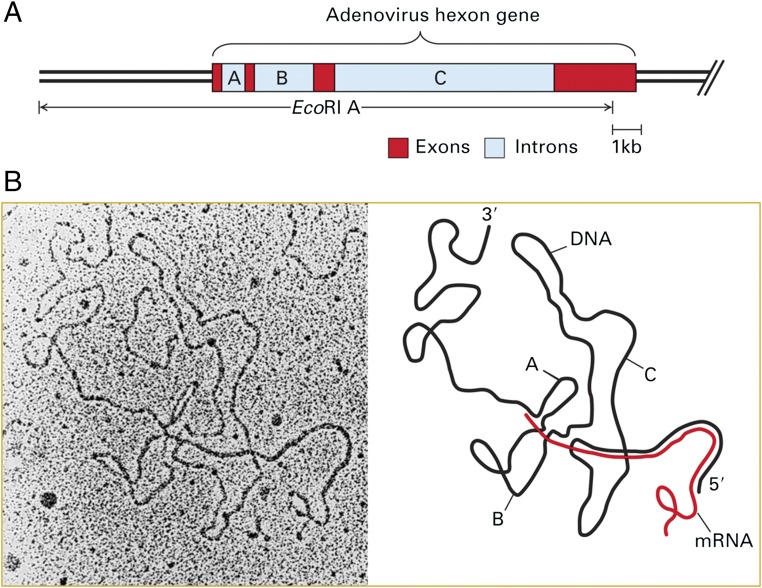
Sue Berget performed this hybridization in the spring of 1977 and performed the high formamide spreading technique developed by Ron Davis and Norman Davidson for visualization of both double-stranded and single-stranded DNA by EM. Phil, Sue Berget, and Claire Moore went down to the EM in the MIT Cancer Center a couple of floors below the Sharp laboratory. I was working on my own projects that day and anxiously awaited news of the results. After a couple of hours or so, Phil came back into the laboratory looking somewhat stunned, a very unusual expression for Phil. “Did you see a loop?,” I asked him anxiously. By this time I had been in the Sharp laboratory for about 9 mo, and I had never heard Phil use anything but the mildest forms of profanity. Nonetheless, Phil excitedly responded: “There are three [profanity] loops!” This is shown in the now famous EM of ref. 12 (figure 4G) taken that afternoon and reproduced in Fig. 2 here.
Fig. 2.
EM of hybrid between purified hexon mRNA and the transcribed strand of the Ad2 EcoRI A DNA fragment. (A) Diagram of the positions of hexon mRNA exons (red) and the introns between them (A, B, and C in light blue) in the left ∼25 kb of the Ad2 genome. (B) EM of hybrid between hexon mRNA and the EcoRI A coding strand. In the interpretation shown on the right, the mRNA is shown in red and DNA in black. Regions where the red RNA is parallel to the black DNA strand represent base paired regions of RNA-DNA hybrid. Adapted from ref. 12.
With characteristic insight and creativity, Phil Sharp immediately recognized the relevance of these observations to the problem of hnRNA and its processing into shorter cellular mRNAs. Here was an explanation to the puzzle of how the 5′ cap and the RNA sequence adjacent to the poly(A) tail in hnRNAs could both be retained as a long precursor hnRNA is processed into a shorter mRNA. The Discussion of the paper points out that “During the late stage of the lytic cycle the r strand of Ad2 is transcribed into long transcripts that originate in the left third of the genome and terminate near the right end... The region of the genome coding for the body of the hexon mRNA and the sequences in these three short RNA segments in the 5' tail of this mRNA are probably included in this long transcript. Thus, a plausible model for the synthesis of the mature hexon mRNA would be the intramolecular joining of these short segments to the body of the hexon mRNA during the processing of a nuclear precursor to generate the mature mRNA.” The Discussion went on to point out that this mechanism of mRNA processing, i.e., splicing together what we now call exons in a long, initially transcribed mRNA precursor that is colinear with genomic DNA sequence, followed by degradation of the sequences between exons that were removed (now called introns), could account for much of the short-lived, long hnRNA observed in pulse-labeling analyses since the early 1960s. This initial discovery of spliced hexon mRNA was followed rapidly by S1 nuclease protection assays (15) of nuclear RNA from Ad2-infected cells showing the existence of the postulated pre-mRNA that is colinear with viral DNA sequence from the transcription start site at ∼6 kb from the left end of the genome (16) through the viral sequences that are spliced together, strongly supporting the model of mRNA processing by RNA splicing of a long primary transcript (17). This was followed by EM analysis of hybrids between single-stranded viral DNA restriction fragments and nuclear RNA isolated during the late phase of infection, revealing intermediates expected from hnRNA processing by splicing together exons and removal of introns: long Ad2 RNAs with one, two, or three of the leader introns removed (18).
Implications of the Discovery: Insights into Gene Evolution and the Complexity of Gene Expression in Multicellular Organisms
Shortly after the discovery of the adenovirus RNA splicing reported in ref. 12 and contemporaneously in ref. 19, the methods for molecular cloning of cellular genomic DNA and complementary DNA copies of mRNAs were developed to the point that the sequences of abundant cellular mRNAs such as β-globin, chicken ovalbumin, and Ig light chain could be compared with the genomic DNA encoding them. Of course, these results showed that the vast majority of human genes contain introns and encode spliced mRNAs. It is therefore obvious that pre-mRNA splicing is required for most mammalian gene expression.
The discovery of pre-mRNA splicing and the corollary that most genes of multicellular organisms are split into pieces, i.e., exons, separated by longer introns, was startling and unexpected. It has had profound consequences for our understanding of the complexity of gene expression and the evolution of new genes in multicellular organisms. Considerable evidence (20) supports the original suggestion (21) that exons often encode portions of the linear primary amino acid sequence of proteins that fold into structural domains. Over evolutionary time, recombination between introns of distinct genes and the transposition of exons to new regions in the genome associated with the movement of transposons have resulted in the evolutionary process called “exon shuffling.” Exon shuffling generates new proteins with novel combinations of protein domains. If the new protein is beneficial, the new sequence encoding it can be selected and evolve into a new gene. For example, tissue plasminogen activator involved in blood clotting, the Neu receptor involved in control of epithelial cell differentiation and replication, and epidermal growth factor that binds to receptors on epidermal cells stimulating cell replication all contain EGF domains that make different protein-protein interactions through surfaces of the compact, highly stable domain. Exon shuffling presumably resulted in the insertion of an EGF domain-encoding exon into an intron of an ancestral form of each of these genes. Moreover, because exons encode protein domains and because sites of RNA splicing are generally designated by on the order of 60 bp surrounding the splice site, insertion of a new exon and its neighboring sequences anywhere within an intron generates a primary transcript that will splice the inserted exon into an mRNA, regardless of changes in the length and sequence of the newly generated introns on either side of it. Because exons encode protein domains, the chance that the newly generated protein will fold into stable protein domains that may perform a beneficial function is far greater than when recombination between bacterial genes without introns generates amino acid sequence fusions between portions of protein domains that are much less likely to fold into a functional structure. Consequently, RNA splicing appears to have had a profound effect on the expansion of the proteome in higher organisms and the evolution of large, multidomain proteins that are far more common in higher eukaryotes than in organisms with few introns.
In addition to the discovery of RNA splicing, analysis of Ad2 mRNAs showed that the initial primary transcript from the Ad2 major late promoter can be processed into at least a dozen mRNAs encoding different viral proteins. Consequently, RNA splicing was immediately discovered to generate alternatively processed mRNAs from a single primary transcript. This is the general rule for cellular mRNAs as well. Extensive sequencing of complementary DNAs from mammalian tissues and comparison with the genomic DNA sequence have revealed that 95% or more mammalian genes express alternatively spliced mRNAs that encode related proteins that differ by the sequence encoded by an alternative exon. This allows a greater number of protein “isoforms” to be encoded in the genomes of multicellular organisms than the number of protein-coding genes and a further expansion of the proteome in higher eukaryotes. Moreover, regulation of alternative splicing, first revealed in the 1977 PNAS Classic paper by Berget et al. (12), is essential for normal mammalian development. This is true for the human central nervous system where alterations in the activities of RNA-binding proteins that regulate alternative RNA-splicing interfere with the proper function of the cerebral cortex (22). Mutations in or abnormal functions of RNA-binding proteins that have relatively subtle effects on the ratio of alternatively spliced mRNAs are associated with several neurological disorders leading to abnormal cognitive function. Thus, cerebral function, required for this and all other scientific research, depends on the proper regulation of alternative RNA splicing first revealed in reference 12.
Footnotes
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
This article is part of the special series of PNAS 100th Anniversary articles to commemorate exceptional research published in PNAS over the last century. See the companion article, “Spliced segments at the 5′ terminus of adenovirus 2 late mRNA,” on page 3171 in issue 8 of volume 74.
References
- 1.Scherrer K, Latham H, Darnell JE. Demonstration of an unstable RNA and of a precursor to ribosomal RNA in HeLa cells. Proc Natl Acad Sci USA. 1963;49:240–248. doi: 10.1073/pnas.49.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris H, Watts JW. The relationship between nuclear and cytoplasmic ribonucleic acid. Proc R Soc Lond B Biol Sci. 1962;156:109–121. doi: 10.1098/rspb.1962.0031. [DOI] [PubMed] [Google Scholar]
- 3.Soeiro R, Birnboim HC, Darnell JE. Rapidly labeled HeLa cell nuclear RNA. II. Base composition and cellular localization of a heterogeneous RNA fraction. J Mol Biol. 1966;19(2):362–372. doi: 10.1016/s0022-2836(66)80010-4. [DOI] [PubMed] [Google Scholar]
- 4.Soeiro R, Vaughan MH, Warner JR, Darnell JE., Jr The turnover of nuclear DNA-like RNA in HeLa cells. J Cell Biol. 1968;39(1):112–118. doi: 10.1083/jcb.39.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edmonds M, Vaughan MH, Jr, Nakazato H. 1971. Polyadenylic acid sequences in the heterogeneous nuclear RNA and rapidly-labeled polyribosomal RNA of HeLa cells: Possible evidence for a precursor relationship. Proc Natl Acad Sci USA 68(6):1336–1340. [DOI] [PMC free article] [PubMed]
- 6.Darnell JE, Philipson L, Wall R, Adesnik M. Polyadenylic acid sequences: Role in conversion of nuclear RNA into messenger RNA. Science. 1971;174(4008):507–510. doi: 10.1126/science.174.4008.507. [DOI] [PubMed] [Google Scholar]
- 7.Rottman F, Shatkin AJ, Perry RP. Sequences containing methylated nucleotides at the 5′ termini of messenger RNAs: Possible implications for processing. Cell. 1974;3(3):197–199. doi: 10.1016/0092-8674(74)90131-7. [DOI] [PubMed] [Google Scholar]
- 8.Perry RP, Kelley DE. Kinetics of formation of 5′ terminal caps in mRNA. Cell. 1976;8(3):433–442. doi: 10.1016/0092-8674(76)90156-2. [DOI] [PubMed] [Google Scholar]
- 9.Wall R, Philipson L, Darnell JE. Processing of adenovirus specific nuclear RNA during virus replication. Virology. 1972;50(1):27–34. doi: 10.1016/0042-6822(72)90342-x. [DOI] [PubMed] [Google Scholar]
- 10.White RL, Hogness DS. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]
- 11.Thomas M, White RL, Davis RW. Hybridization of RNA to double-stranded DNA: Formation of R-loops. Proc Natl Acad Sci USA. 1976;73(7):2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berget SM, Moore C, Sharp PA. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci USA. 1977;74(8):3171–3175. doi: 10.1073/pnas.74.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis JB, Atkins JF, Anderson CW, Baum PR, Gesteland RF. Mapping of late adenovirus genes by cell-free translation of RNA selected by hybridization to specific DNA fragments. Proc Natl Acad Sci USA. 1975;72(4):1344–1348. doi: 10.1073/pnas.72.4.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casey J, Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berk AJ, Sharp PA. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- 16.Weber J, Jelinek W, Darnell JE., Jr The definition of a large viral transcription unit late in Ad2 infection of HeLa cells: Mapping of nascent RNA molecules labeled in isolated nuclei. Cell. 1977;10(4):611–616. doi: 10.1016/0092-8674(77)90093-9. [DOI] [PubMed] [Google Scholar]
- 17.Berget SM, Berk AJ, Harrison T, Sharp PA. Spliced segments at the 5′ termini of adenovirus-2 late mRNA: A role for heterogeneous nuclear RNA in mammalian cells. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):523–529. doi: 10.1101/sqb.1978.042.01.055. [DOI] [PubMed] [Google Scholar]
- 18.Berget SM, Sharp PA. Structure of late adenovirus 2 heterogeneous nuclear RNA. J Mol Biol. 1979;129(4):547–565. doi: 10.1016/0022-2836(79)90468-6. [DOI] [PubMed] [Google Scholar]
- 19.Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell. 1977;12(1):1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 20.Liu M, Grigoriev A. Protein domains correlate strongly with exons in multiple eukaryotic genomes: Evidence of exon shuffling? Trends Genet. 2004;20(9):399–403. doi: 10.1016/j.tig.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert W. Why genes in pieces? Nature. 1978;271(5645):501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 22.Darnell RB. RNA protein interaction in neurons. Annu Rev Neurosci. 2013;36:243–270. doi: 10.1146/annurev-neuro-062912-114322. [DOI] [PMC free article] [PubMed] [Google Scholar]