Significance
Developmental buffering mechanisms that stabilize phenotypes against perturbations (such as harmful mutations or environmental stress) have previously been inferred. However, it is unclear whether this “canalization” can be maintained when adaptive evolution causes phenotypes and developmental processes to change. Here, we report a loss of canalization accompanying the evolution of larger wings in high-altitude fruit flies (Drosophila melanogaster). We redeploy a classic genetics technique (mutagenesis) to show that wing development of Ethiopian flies is less robust to new mutations and that large wing size is inherited together with decanalized wing development. These results represent the first example, to our knowledge, of adaptive evolution apparently undermining the developmental buffering of a recently evolved trait. Decanalized development, thus, represents a potential “cost of adaptation.”
Keywords: developmental canalization, adaptive evolution, Drosophila melanogaster, wing, genetic robustness
Abstract
In higher organisms, the phenotypic impacts of potentially harmful or beneficial mutations are often modulated by complex developmental networks. Stabilizing selection may favor the evolution of developmental canalization—that is, robustness despite perturbation—to insulate development against environmental and genetic variability. In contrast, directional selection acts to alter the developmental process, possibly undermining the molecular mechanisms that buffer a trait’s development, but this scenario has not been shown in nature. Here, we examined the developmental consequences of size increase in highland Ethiopian Drosophila melanogaster. Ethiopian inbred strains exhibited much higher frequencies of wing abnormalities than lowland populations, consistent with an elevated susceptibility to the genetic perturbation of inbreeding. We then used mutagenesis to test whether Ethiopian wing development is, indeed, decanalized. Ethiopian strains were far more susceptible to this genetic disruption of development, yielding 26 times more novel wing abnormalities than lowland strains in F2 males. Wing size and developmental perturbability cosegregated in the offspring of between-population crosses, suggesting that genes conferring size differences had undermined developmental buffering mechanisms. Our findings represent the first observation, to our knowledge, of morphological evolution associated with decanalization in the same tissue, underscoring the sensitivity of development to adaptive change.
Canalization describes the property of some biological traits to remain constant in the face of environmental and genetic changes (1–6). This phenomenon has important implications for the relationship between genetic and phenotypic variation. By masking the phenotypic effects of genetic changes, canalization may inhibit phenotypic evolution while allowing hidden genetic variation to accumulate. If canalization is overcome by environmental and/or genetic changes, this reservoir of functional variation may then be exposed. For example, Waddington (7) selected Drosophila for a missing cross-vein trait that initially only appeared in a stressful high-temperature environment but after selection, manifested under normal conditions as well. A molecular case study of canalization was provided by Rutherford and Lindquist (8), who found that Drosophila with a disabled chaperone protein (the heat shock protein Hsp90) showed a suite of developmental abnormalities. These abnormalities varied based on genetic background and environment and could be selected for Hsp90 independence. Other studies have also found that selection in the laboratory can alter developmental stability (9–12), and Hayden et al. (13) found that in vitro directional selection on ribozyme activity led to reduced genetic and environmental robustness.
Canalization is difficult to disentangle from selective constraint, which complicates its study in natural populations. A rare potential example comes from the blowfly Lucilia cuprina, in which the evolution of insecticide resistance was accompanied by prolonged development and bristle asymmetry (14). Those disadvantages were subsequently reversed by the evolution of an unlinked modifier locus (15). Here, the initial cost of adaptation may have been because of pleiotropic decanalizing effects of the insecticide resistance mutation itself. Or given the contrast between the adaptive and decanalized phenotypes, a linked deleterious variant might have been fixed along with the resistance allele and later compensated by the modifier gene.
Canalization might evolve because of stabilizing selection favoring the same phenotypic optimum in the face of environmental and genetic variability [as shown in the case of environmental robustness (16)], or canalization might arise from inherent properties of the biological system (17). Particularly in the former scenario, it seems possible that directional selection might undermine canalization: if selection for a new phenotypic optimum alters the developmental process, then the molecular mechanisms that had previously buffered the ancestral phenotype might fail to buffer the novel phenotype. Hence, it seems possible that recently evolved traits may show reduced canalization (until new or modified buffering mechanisms can evolve), but no such example has been reported from nature.
Here, we describe a natural instance of decanalization associated with a recently evolved morphological structure, focusing on wing size and developmental stability in a high-altitude (>3,000 m) Ethiopian population of Drosophila melanogaster. Although globally distributed today, this human-commensal species probably originated in the lowlands of southern central Africa (18). The species’ arrival in Ethiopia may have roughly coincided with its crossing of the Sahara [∼10,000 y ago (19, 20)]. Highland Ethiopian flies are morphologically divergent from other D. melanogaster populations, featuring striking melanism (21), larger body size, and larger wings (Fig. 1) with distinct shape (22).
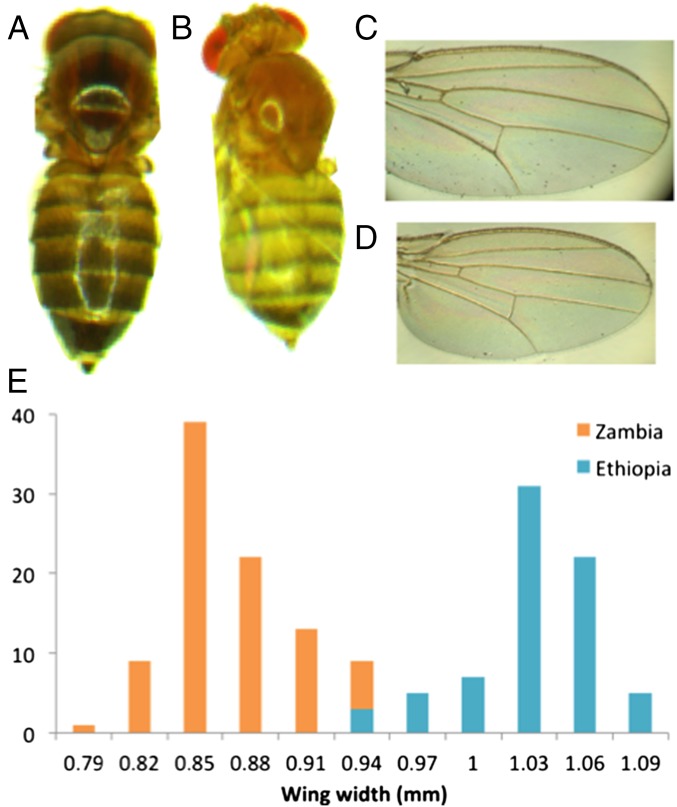
Fig. 1.
Morphological comparisons of D. melanogaster from the Ethiopian highlands and an ancestral range Zambia population. Ethiopian strains have (A vs. B) larger body size and (C vs. D) wing size. (E) The distribution of wing widths among outbred individuals shows almost no overlap between populations. Detailed size data are given in Dataset S1.
Past studies have shown that the Drosophila wing provides a convenient visible readout of development, allowing, for example, the study of variation unmasked by specific mutations (23, 24). This study repurposes mutagenesis as a generalized genetic perturbation to assess whether wing size evolution in Ethiopian D. melanogaster has undermined the stability of wing development. Initially, our observation of frequent wing abnormalities in Ethiopian inbred strains motivated the hypothesis of decanalized wing development. Mutagenesis experiments confirmed that de novo mutations were far more likely to produce wing defects in the Ethiopian strains than in the smaller-winged Zambia population (whereas a control trait showed no such difference), implying less buffered development of Ethiopian wings. A final mutagenesis experiment confirmed that wing size and decanalization were inherited together in the advanced generation offspring of an Ethiopia–Zambia cross, implying that alleles conferring larger Ethiopian wings contributed to destabilized development.
Results
Ethiopian Wing Size and Wing Abnormalities.
Under the same laboratory conditions, the Ethiopia–Fiche population (EF) displays greater size than other populations of D. melanogaster (Fig. 1 and Dataset S1), indicating a genetic basis to this trait difference. Larger flies have often been observed in cooler environments (25), but the larger size of highland Ethiopian flies relative to temperate populations may indicate additional size-related selective pressures at high altitude (such as desiccation) (26). The increased area of Ethiopian wings relative to body mass implies reduced wing loading (Dataset S1), which might aid flight in thin, cool air.
The distributions of wing size in EF vs. an ancestral range population (Zambia–Siavonga; ZI) are almost entirely nonoverlapping (Fig. 1 and Dataset S1), with phenotypic differentiation quantified by a QST value (27–29) of 0.985. By comparison, the average value for genetic differentiation (as indexed by FST) (30, 31) between genomes from these populations (18, 32) is 0.151, and only 0.052% of genomic windows have an FST greater than QST for wing width (Methods). Regardless of whether natural selection targeted wing size or a correlated trait, the observed magnitude of phenotypic divergence relative to genomic differentiation suggests that wing size has evolved because of directional selection acting on this or a correlated trait.
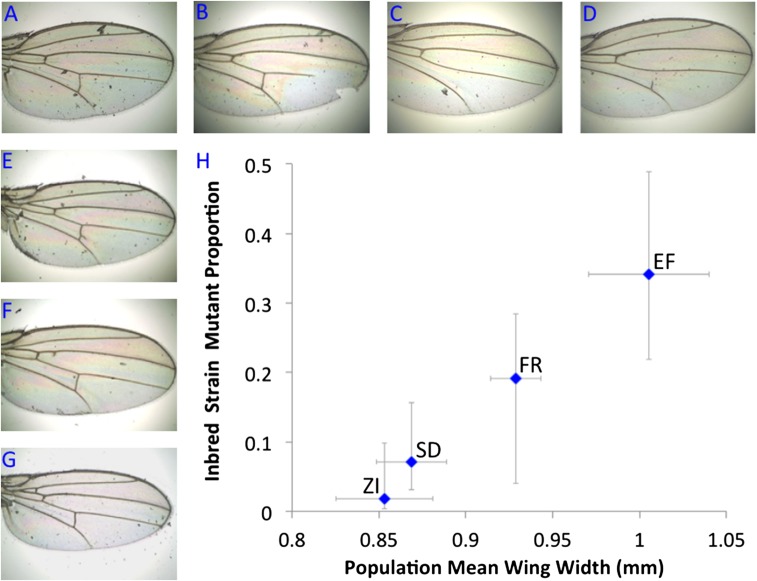
On inbreeding many strains from Ethiopia and elsewhere, we noticed that highland inbred lines displayed visible wing vein abnormalities much more often than other populations (Fig. 2 and Dataset S2). Overall, 15 of 44 EF inbred strains showed wing abnormalities compared with just 1 of 54 ZI inbred strains. Data from other populations also supported a relationship between larger wings and more frequent anomalies (Fig. 2 and Dataset S2). All wing abnormalities persisted when three of the mutant EF strains were raised at their native air pressure, indicating that these defects were not the product of a stressful higher-pressure environment.
Fig. 2.
Ethiopian inbred strains show greatly elevated frequencies of visible wing vein abnormalities. These defects including (A–C) truncated longitudinal veins, cross-veins (C) missing or (D) incomplete, (E and F) extraneous vein material, and (G) ectopic cross-vein. (H) The relationship between larger wings and inbred wing vein abnormalities suggested by the comparison of Ethiopia (EF) with southern African ancestral range populations (ZI and SD, the latter indicating South Africa–Dullstroom) is also supported by a temperate European population (France–Lyon; FR), which shows moderately large wings and somewhat elevated wing abnormalities. Horizontal bars indicate 1 SD for wing width among a population’s outbred crosses. Vertical bars indicate the binomial 95% CI around the proportion of inbred strains displaying abnormalities. Detailed results are given in Dataset S2.
Inbreeding is known to represent a genetic perturbation in Drosophila—recessive deleterious variants are abundant in natural populations (33) and may become homozygous in inbred strains. Therefore, we hypothesized that wing development might be more susceptible to perturbation (i.e., less canalized) in highland Ethiopian populations relative to lowland ancestral range populations. By contrast, the elevated rate of wing abnormalities in Ethiopia is unlikely to result from a higher genetic load of deleterious variants than the Zambia population. The moderately lower genetic diversity of Ethiopian samples seems to reflect a population bottleneck along with a relatively recent colonization rather than long-term isolation with lower population size (18). After a handful of generations has passed after such a bottleneck, the genetic load of recessive deleterious variants should actually be lower than in the ancestral population (34).
Thus, we postulated that, in smaller-winged ancestral range populations, long-term stabilizing selection may have canalized wing development against environmental and genetic variability. However, these ancestral buffering mechanisms might be undermined if directional selection changed the outcome of wing development, which happened in Ethiopia. Ethiopian wing size evolution may have been too recent for natural selection to restore these potentially intricate buffering mechanisms. Based on this model, we sought to explicitly test whether Ethiopian wing development is decanalized.
Mutagenesis Test for Canalization and Its Loss.
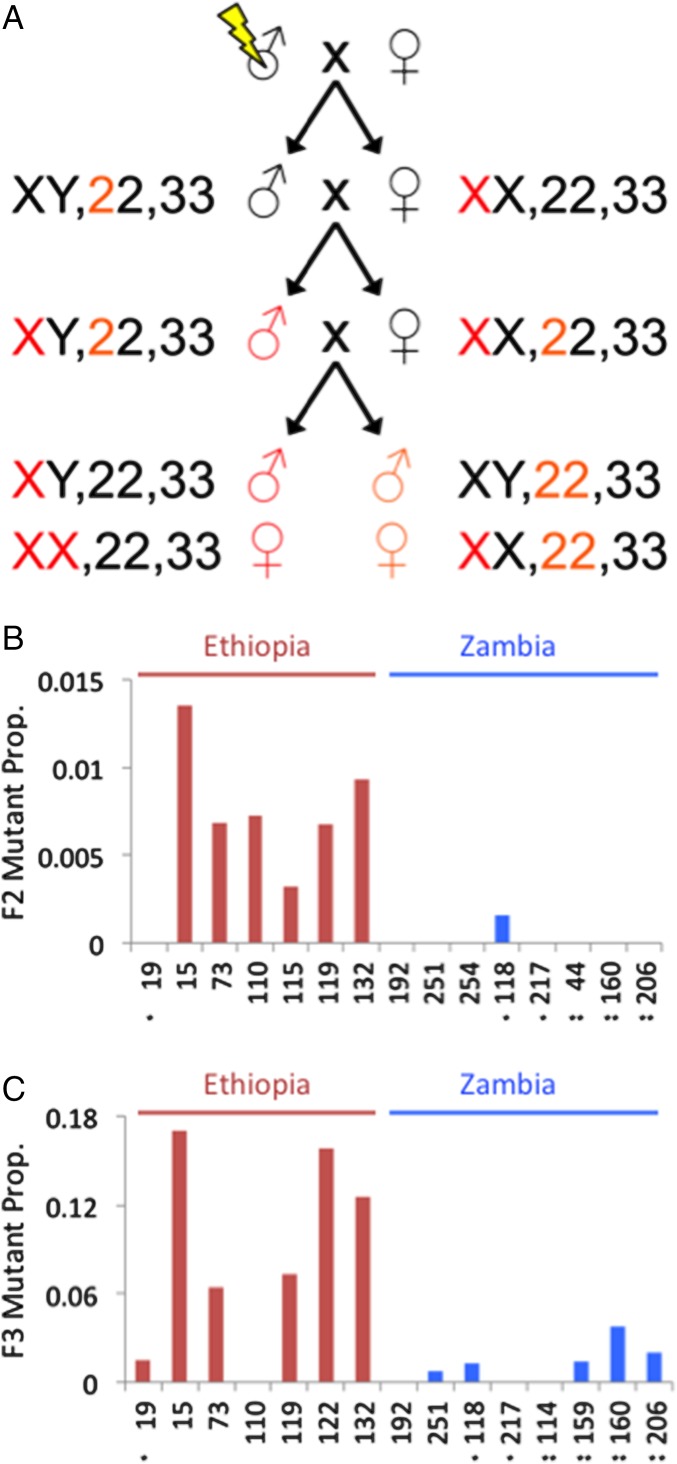
Developmental canalization can be quantified in terms of environmental robustness or genetic robustness (4). Whereas an environmental perturbation, such as extreme temperature, might be more stressful for one population or genotype than another, artificial mutagenesis (as an experimentally controlled form of genetic perturbation) offers a clear method to test for differences in genetic robustness. If Ethiopian wing development is decanalized, newly occurring mutations should be more likely to cause novel wing defects for this population relative to Zambia. We, therefore, mutagenized males from multiple EF and ZI inbred strains lacking wing abnormalities (based on examining 100 flies from each strain) and then monitored their F2 and F3 offspring for wing vein anomalies caused by new mutations (Methods and Fig. 3).
Fig. 3.
Design and outcomes of the mutagenesis test for (de)canalization. (A) The crossing scheme for mutagenized flies is depicted, including the three major chromosome pairs of Drosophila. Colored chromosomes highlight potential modes of inheritance leading to observable recessive mutations in F2 males and F3 flies of either sex (only a subset of potential genotypes are depicted). The observed frequencies of wing vein defects are then shown for each tested inbred strain for (B) F2 males and (C) F3 females. The large excess of de novo wing mutants for Ethiopian strains indicates reduced canalization of wing development relative to Zambian strains. *Number of segregating inversions detected within each inbred strain.
A dramatic population effect on mutant frequencies was clear from the offspring of mutagenized flies. Among F2 males, the Ethiopian strains had 26 times more wing vein mutants (Fig. 3 and Table S1). Only one Zambia strain showed any mutants, whereas all but one of the Ethiopia strains had higher abnormality rates (Mann–Whitney P = 0.0039). Higher mutant frequencies were observed among F3 females for both populations, but Ethiopian strains still averaged seven times more wing vein mutants than Zambian strains (Mann–Whitney P = 0.0139).
Table S1.
Frequency of mutant phenotypes in the offspring of mutagenized WT flies
| Inbred line | Total flies | Wing vein mutants | Wing vein mutant frequency | Eye color mutants | Eye color mutant frequency |
| F2 male | |||||
| EF15N | 593 | 8 | 0.01349 | ||
| EF19N | 429 | 0 | 0 | ||
| EF73N | 440 | 3 | 0.00682 | ||
| EF110N | 550 | 4 | 0.00727 | ||
| EF115N | 312 | 1 | 0.00321 | ||
| EF119N | 296 | 2 | 0.00676 | ||
| EF132N | 755 | 7 | 0.00927 | ||
| ZI117N | 197 | 0 | 0 | ||
| ZI118N | 635 | 1 | 0.00157 | ||
| ZI144N | 430 | 0 | 0 | ||
| ZI160N | 385 | 0 | 0 | ||
| ZI192N | 245 | 0 | 0 | ||
| ZI206N | 460 | 0 | 0 | ||
| ZI251N | 534 | 0 | 0 | ||
| ZI254N | 613 | 0 | 0 | ||
| F3 female | |||||
| EF15N | 282 | 48 | 0.17021 | 1 | 0.00355 |
| EF19N | 402 | 6 | 0.01493 | 2 | 0.00498 |
| EF73N | 375 | 24 | 0.064 | 1 | 0.00267 |
| EF110N | 354 | 0 | 0 | 3 | 0.00847 |
| EF119N | 411 | 30 | 0.07299 | 0 | 0 |
| EF122N | 513 | 81 | 0.15789 | 2 | 0.00390 |
| EF132N | 652 | 82 | 0.12577 | 1 | 0.00153 |
| ZI114N | 303 | 0 | 0 | 1 | 0.00330 |
| ZI117N | 165 | 0 | 0 | 2 | 0.01212 |
| ZI118N | 474 | 6 | 0.01266 | 4 | 0.00844 |
| ZI159N | 432 | 6 | 0.01389 | 2 | 0.00463 |
| ZI160N | 426 | 16 | 0.03756 | 1 | 0.00235 |
| ZI192N | 273 | 0 | 0 | 3 | 0.01099 |
| ZI206N | 300 | 6 | 0.02 | 2 | 0.00667 |
| ZI251N | 405 | 3 | 0.00741 | 1 | 0.00247 |
To verify that decanalization in Ethiopian strains pertains to wing development specifically and does not reflect a general susceptibility to mutagenic perturbation, we also checked F3 flies for eye color mutants. In contrast to the wing data, we observed that Ethiopian strains had nonsignificantly lower rates of these mutations than Zambian strains (Table S1). The 95% confidence intervals (95% CIs) for population ratios of eye color mutant frequencies (EF/ZI eye mutant frequency = 0.5809; 95% CI = 0–2.7807) and wing vein mutant frequencies (EF/ZI vein mutant frequency = 6.8073; 95% CI = 5.4029–8.2117) were nonoverlapping.
A subset of the inbred strains analyzed above was found to harbor segregating inversion polymorphisms (up to two per strain) (Table S2). The ZI strains’ greater presence of inversions could contribute to some of our results by preventing recessive mutations from becoming homozygous. However, several points argue against inversions as an explanation for our mutagenesis results. First, our F2 male experiment is designed to detect hemizygous X-linked mutations and should be less influenced by the exclusively autosomal inversions detected among these strains (although we did not confirm X-linked inheritance of these mutations). Second, although F3 results could potentially be more affected by inversion presence (Fig. 3A), the inversions observed for Zambia (Table S2) would be predicted to insulate this population against eye color mutants (in contrast to our results). Third, for both the F2 and F3 wing mutant experiments, ZI strains with an inversion actually averaged higher wing mutant frequencies than inversion-free ZI strains (Fig. 3), again suggesting the absence of an important protective effect of inversions against homozygous mutations in these experiments. Fourth, we found that wing size and decanalization cosegregated in the offspring of a cross between Ethiopian and Zambian strains with no detected inversions as described below.
Table S2.
Inversion PCR testing results for strains used in the F2 and/or F3 mutagenesis experiments
| Inbred line | Polymorphic inversions | Fixed inversions |
| EF15N | None | None |
| EF19N | In(3R)K | None |
| EF73N | None | None |
| EF110N | None | None |
| EF115N | None | None |
| EF119N | None | None |
| EF122N | None | None |
| EF132N | None | None |
| ZI44N | In(2L)t and In(2R)NS | In(3L)OK |
| ZI114N | In(2L)t and In(3L)OK | None |
| ZI118N | In(2L)t | None |
| ZI159N | In(2L)t and In(3L)OK | None |
| ZI160N | In(2L)t and In(3L)OK | None |
| ZI192N | None | None |
| ZI206N | In(2L)t and In(2R)NS | None |
| ZI217N | In(3L)OK | In(2L)t |
| ZI251N | None | None |
| ZI254N | None | None |
Linking Wing Size Evolution with Decanalization.
Is the above-shown decanalization of Ethiopian wing development connected to wing size evolution? To address this question, we used a genetic mapping population composed of 16th-generation offspring between Ethiopian and Zambian strains (Methods). If the same genes caused wing size evolution and decanalization in Ethiopian D. melanogaster, then larger-winged flies from the mapping population should produce more wing mutant offspring after mutagenesis relative to smaller-winged individuals. In contrast, if wing size evolution and decanalization were genetically unrelated, then after 15 generations of recombination between the Ethiopian and Zambian parental genomes, there should be little relationship between wing size and canalization in the mapping population.
We, therefore, measured wing length of 830 F16 individuals from the mapping population and then selected the largest 15% and the smallest 15% of males and (virgin) females. Large-winged males were mutagenized and then mated with large-winged females, and small-winged males were mated with small-winged females after mutagenesis. F3 female offspring of these matings were then assessed for wing abnormality frequency. We found that large-winged matings yielded over twice as many mutants, with 3.4% of offspring showing wing abnormalities (24 of 713) compared with 1.5% for small-winged matings (10 of 688). In both groups, the observed abnormalities encompassed a wide range of unique wing vein phenotypes.
The above results seem inconsistent with the null hypothesis that wing size and decanalization are encoded by independent loci. We performed detailed simulations to assess the significance of this difference, while allowing for residual linkage in these F16 individuals between Ethiopian wing size loci and decanalization loci (which are separate groups of loci under the null hypothesis). Specifically, we evaluated whether our observed ratio of wing mutant frequencies from large- vs. small-winged flies (i.e., 2.32) exceeds the predictions of random linkage in the null simulations. Each individual’s genomic blocks of Ethiopian and Zambian ancestry were tracked to the F16 generation, from which specified numbers of large- and small-winged individuals were mutagenized and mated, and their F3 offspring were monitored for wing abnormalities. Wing size and decanalization loci were located randomly across the genome. Each individual’s wing abnormality probability was based on the pure population Ethiopia and Zambia F3 mutant rates (given above), the individual’s genotype at each simulated decanalization locus, and a genetic model of decanalization. Because wing abnormality rates were lower in the genetically admixed mapping population than the midpoint of the pure population rates, we focused on models in which decanalization was recessive (two Ethiopian alleles at a locus required to raise an individual’s mutant probability) and/or multiplicative across loci. Although some models produced an average ratio as high as 1.31 because of linkage, none matched our observed ratio at least 5% of the time (Table S3). Hence, our results are poorly fit by models of chance linkage between independent wing size and decanalization loci. Instead, we suggest that Ethiopian wing size loci may directly contribute to decanalization, potentially by modifying the developmental process and undermining ancestral buffering mechanisms.
Table S3.
Higher mutant frequency of large-winged flies is not expected under the null hypothesis of independent wing size and decanalization loci
| Decanalization loci | RA ratio | RA P value | RM ratio | RM P value | CM ratio | CM P value |
| 1 | 1.31 | 0.0490 | 1.31 | 0.0484 | 1.22 | 0.0141 |
| 5 | 1.29 | 0.0275 | 1.29 | 0.0425 | 1.22 | 0.0124 |
| 10 | 1.28 | 0.0236 | 1.29 | 0.0423 | 1.22 | 0.0129 |
| 20 | 1.28 | 0.0207 | 1.29 | 0.0430 | 1.23 | 0.0130 |
| 50 | 1.28 | 0.0205 | 1.29 | 0.0410 | 1.23 | 0.0120 |
For the intercross mutagenesis experiment, the incompatibility of our findings with null models of separate wing size and decanalization loci is documented below. These results stem from detailed simulations of the experimental process in terms of each fly’s Ethiopian and Zambian ancestry along the genome (Methods). Fifteen scenarios were investigated regarding the genetic architecture of the population difference in canalization. Cases involving 1, 5, 10, 20, and 50 decanalization loci were simulated. Because the mutant frequencies of both large- and small-winged F16 intercross flies were less than the average frequency of the parental populations (in the text), we concluded that canalization was not encoded in a purely additive manner within and between loci. Instead, we analyzed models in which canalization was recessive (i.e., two Ethiopian alleles required for an elevated mutant probability), multiplicative across loci, or both. These models are specified as recessive additive (RA), recessive multiplicative (RM), and codominant multiplicative (CM). These models define the mutant probability for each fly relative to a fly that had all Ethiopian alleles at decanalization loci (and hence, would match the Ethiopian F3 mutant probability estimated from our initial mutagenesis experiment) or all Zambian alleles. For each scenario, the average mutant frequency ratio between large- and small-selected flies is given along with the proportion of 10,000 simulations that gave a larger value than the empirically observed ratio of 2.316 (representing a P value for this model). All P values are below 0.05, suggesting that our data are poorly fit by a model of separate decanalization and wing size loci.
Discussion
Above, we investigate whether size evolution in highland Ethiopian D. melanogaster was accompanied by a destabilization of wing development. We first observed that, relative to small-winged populations from the species’ ancestral range, populations that recently evolved larger wings tended to produce more wing vein abnormalities after inbreeding, consistent with a greater susceptibility to the genetic stress of inbreeding. We then repurposed the classical genetics technique of mutagenesis to formally test for a difference in robustness to genetic perturbation between natural populations. We found that, indeed, Ethiopian flies showed a much greater susceptibility to novel mutations with regard to wing vein development (but not eye pigmentation). A similar mutagenesis experiment on genetically admixed flies confirmed that wing size and decanalization loci cosegregated, suggesting a direct contribution of Ethiopian wing size evolution to the loss of developmental canalization. We note that genetic robustness may be viewed as a form of epistasis (e.g., a mutation may have negative fitness consequences in an Ethiopia genetic background but not in a Zambia genetic background) and that previous studies have successfully applied mutagenesis to examine epistatic interactions (35, 36).
These results provide the clearest evidence to date for a loss of canalization in a natural population and the first case, to our knowledge, in which directional selection had altered the same morphological structure (with wing size either a target of selection or a correlated response). Here, we have leveraged Drosophila as a laboratory model and specifically, the utility of the fly wing as a visually accessible developmental readout. Although our study does not pursue the molecular mechanisms of canalization and its loss, the natural difference in developmental buffering that we have found—along with the tools of Drosophila genetics—may pave the way for a more thorough understanding of canalization’s mechanisms. Such studies may also lead to a more detailed understanding of the interaction between size evolution and canalization in the wings of Ethiopian D. melanogaster.
At present, it is unclear how often the evolution of form may destabilize formerly canalized developmental processes. Decanalization may be viewed as a potential cost of adaptive evolution in addition to the pleiotropic effects of beneficial mutations on unrelated traits and the fixation of linked deleterious mutations. Additional mutagenesis experiments should be undertaken in experimentally tractable systems to determine how frequently recent morphological evolution has led to a breakdown of developmental canalization. In addition to its implications for the interplay between evolutionary and developmental processes, this phenomenon could also be relevant to rates of disease affecting evolutionarily labile tissues relative to anciently conserved structures.
Methods
Phenotypic Analysis.
All population samples used in this study were previously described (18, 32). In the case of inbred strains, the original isofemale lines were inbred by sibling mating for eight generations. Size measurements for population comparisons were performed on outbred flies from crosses between independent pairs of inbred strains from the same population sample. These flies were kept at controlled temperature (20 °C) and humidity (70%) throughout development. Larval density was controlled by placing 20 virgin females with 20 males in half-pint bottles and allowing females to oviposit for 48 h. Medium was prepared in batches consisting of 4.5 L water, 500 mL cornmeal (Quaker yellow), 500 mL molasses (Grandma’s unsulfured), 200 mL powdered yeast (MP Biochemical Brewer’s), 54 g agar (Genesee Drosophila type II), 20 mL propionic acid, and 45 mL 10% Tegosept solution in 95% ethanol (concentrations vol/vol).
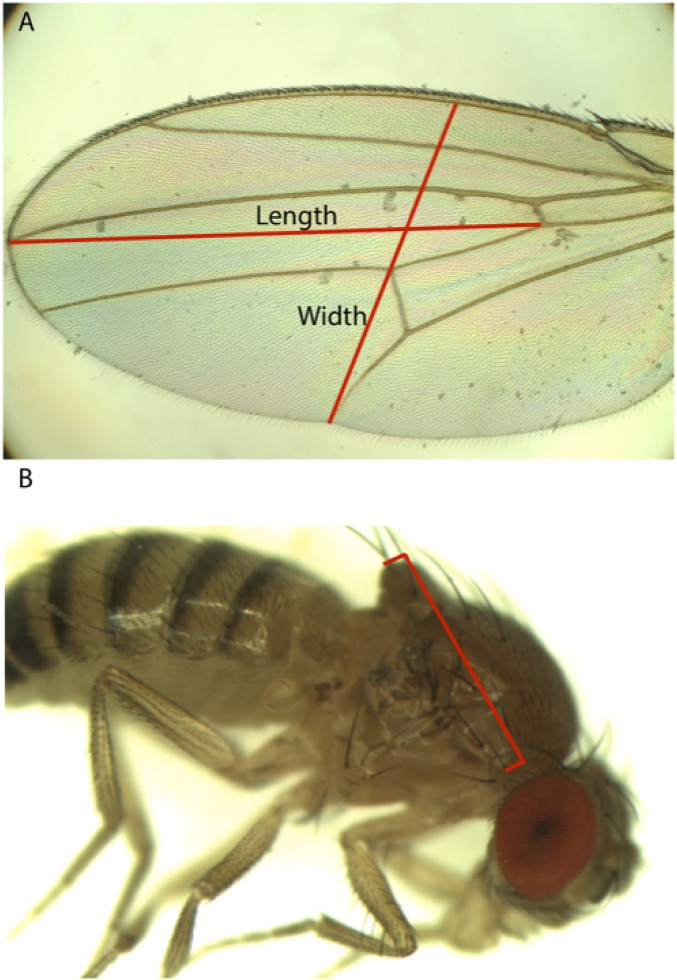
As a proxy for overall body size, we quantified thorax length in 3- to 5-d-old F1 adult females raised as described above. For each independent cross, 10–20 females were photographed with a digital camera attached to a stereo dissecting microscope (AmScope MU1000 and SM-4BX), and the thorax was measured from the base of the anterior humeral bristle to the posterior tip of the scutellum. For wing size, we similarly examined 3- to 5-d-old F1 adult females from crosses generated as described above. For five females per cross, a wing was removed and photographed at 50× magnification using a digital camera attached to a compound microscope (Olympus BH-2). The length and depth of each wing were then measured using ImageJ (version 1.48). For wing length, we measured a straight line drawn from the intersection of the anterior cross-vein and L4 longitudinal vein to where the L3 longitudinal vein intersects the wing margin. For depth, we measured a straight line from the intersection of the L5 longitudinal vein and the posterior wing margin passing through the intersection of the posterior cross-vein and L4 and terminating at the anterior wing margin. Examples of these measurements are illustrated in Fig. S1.
Fig. S1.
Measurements used for (A) wing dimensions and (B) thorax length.
Wing abnormalities were identified by visually examining each slide-mounted wing for qualitative defects in wing vein architecture—namely incomplete veins, extraneous vein material, and missing or extra cross-veins (Fig. 2). To confirm that Ethiopian wing defects were not a product of development at an unfamiliar air pressure, we raised three EF inbred strains that showed high-frequency wing abnormalities (EF16N, EF43N, and EF117N) at their native air pressure of 70 kPa [the EF sample originates from 3,050 m above sea level (32)]. Eggs from each strain were laid inside a bell jar, with air pressure controlled by a vacuum regulator (Kem Scientific DVR 200). After adults had eclosed, 15 flies from each strain were removed from the vacuum and assayed for wing defects.
Genetic and Phenotypic Differentiation Between Populations.
Genomes used in FST comparisons were from the Drosophila Genome Nexus (32). A phenotype QST value that exceeds the range of observed FST values may indicate a trait subject to spatially varying selection (27, 28). An important assumption of such comparisons is that the neutral variance of QST is no larger than that of FST (29). We sought to fulfill this requirement in two ways. First, the number of (homozygous) genomes analyzed for FST (5 EF genomes and 10 ZI genomes) did not exceed the number of strains phenotyped from each population. Second, the genomic distribution of FST between each pair of populations was obtained from short windows, averaging ∼500 bp and scaled by diversity as previously implemented (18). FST for these windows should have a conservatively higher neutral variance than QST for any monogenic trait with a mutational target size greater than the window length, whereas polygenic traits like the one studied here are likely to have even lower neutral variance for QST.
Mutagenesis and Analysis of Canalization.
Mutagenesis of WT inbred strains was conducted by exposing 20 males (starved for 12 h) from 10 inbred lines from each population to an ethyl methane sulfonate solution (25 mM ethyl methane sulfonate in 1% sucrose) for 24 h. Lines were selected based on the absence of observed vein mutants after examination of >100 female flies. After mutagenesis, males were crossed to virgin females from the same inbred strain and then removed after 5 d. This timing means that males should pass on postmeiotic mutations, which should each contribute to a single gamete and should not be shared between siblings. F1 offspring were then allowed to interbreed, yielding F2 males potentially subject to X-linked de novo mutations (Fig. 3), which were assayed for wing abnormalities. F2 flies then interbred to yield F3 females and males, which were assayed for wing abnormalities encoded by X-linked or autosomal mutations and also assayed for eye pigmentation mutants to provide a control independent of wing development. Mutants were identified visually as any color clearly distinguishable from the WT, such as brown-like or vermilion-like phenotypes. Scoring was done blindly with respect to strain identities. To determine whether wing vein and eye color mutant frequency differences between the two populations were distinct, 95% CIs for population ratios were estimated (37).
For all strains used in mutagenesis testing, we used PCR to test whether any of nine common inversions were segregating within the inbred strain: In(1)A, In(1)Be, In(2L)t, In(2R)NS, In(3L)Ok, In(3L)P, In(3R)K, In(3R)Mo, and In(3R)P. Primers were based on those used by Corbett-Detig et al. (38), with modifications to accommodate primer site polymorphisms (primer and amplification details are given in Tables S4 and S5). Primer pairs to detect both standard and inverted arrangements were used, with appropriate positive and negative controls (Tables S4 and S5), to differentiate segregating from fixed inversions. Although this inversion testing was done approximately 1 y after the inbred strain mutagenesis experiments, we expect that any inversion polymorphism that could be lost to drift probably would have been purged during the original inbreeding of these strains.
Table S4.
PCR primers
| Inversion and orientation | Forward primer | Forward sequence | Reverse primer | Reverse sequence |
| In(1)A | ||||
| Standard | 1AStdDistF2 | ATTGCAGTCCTGGCTAGTTC | 1AStdDistR2 | GAGCCGAAAGAAAWGCGTAG |
| Inverted | 1AInvProxF2 | GCATAGTYTTGGTCCGTCGT | 1AInvProxR | TTCGCCTCGCCAGATTTAT |
| In(1)Be | ||||
| Standard | 1BeStdDistF | TGTAGGGCCTTTTCACCAAC | 1BeStdDistR | ACCATATTGACGGGCATTGT |
| Inverted | 1BeInvDistF | GCCAGGATGACCGTGAGTAT | 1BeInvDistR | GTCACCACCTGTTGCCTTTT |
| In(2L)t | ||||
| Standard | 2LtInvStdProxF2 | GTCTTTGCAAACTCGATTTTCG | 2LtStdProxR2 | TTCCCACCGCACAGAGTTG |
| Inverted | 2LtInvStdProxF2 | GTCTTTGCAAACTCGATTTTCG | InA151 | TATTTTGGTGGCCTGTTTCAG |
| In(2R)NS | ||||
| Standard | 2RNSStdDistF2 | CCCTTAGACAAACAGGAAGC | 2RNSStdDistR2 | ACATTGGACAGTAGCCTCTG |
| Inverted | 2RNSInvProxF3 | CAGCATTAATCCGCACACC | 2RNSInvProxR3 | GCTAAACGCTTCCGATTGAG |
| In(3L)Ok | ||||
| Standard | 3LOkStdProxF1InvDistR1 | GGCTTACATCCAAAGGTC | 3LOkStdProxR2 | CACTTGGATTGCTTTGCTTG |
| Inverted | 3LOkInvDistF2 | CCAGAGTAAGGCTCTGTAAAC | 3LOkStdProxF1InvDistR1 | GGCTTACATCCAAAGGTC |
| In(3L)P | ||||
| Standard | P1 | AGAACCGCAAACGAACACTT | P2 | ATCTCCTGCCCACACTCACAT |
| Inverted | P1 | AGAACCGCAAACGAACACTT | 3LPInvDistR2 | GAGTTTGCATGTGCAGCAG |
| In(3R)K | ||||
| Standard | 3RKStdProxF | TTTTAGCCAACGCAATAGGG | 3RKStdProxR | AGCCCGTGTGGTAATCGTAG |
| Inverted | 3RKInvDistF2 | CGGTTATTCGCGTAATCC | 3RKInvDistR2 | CCTTTCGACCTTTTGACAGG |
| In(3R)Mo | ||||
| Standard | 3RMoStdDistF | AACTTCCAGCACGTGGGATA | 3RMoStdDistR | CGCCCTATCCGAGGCTTAG |
| Inverted | 3RMoInvDistF | ACCTCACTGCGGATGAAGAG | 3RMoInvDistR2 | TCCATGGCAATACSTTCACA |
| In(3R)P | ||||
| Standard | 3RPStdProxF2 | CATTAGGAAGCAGCCATGAC | 3RPStdProxR | CAAGCCATACAGTTCCATAAATCCA |
| Inverted | 3RPInvDistF2 | TGTTGTCGTCGGACTGCATC | 3RPInvDistR2 | GTGCGTGTGAATGAGCTTTTG |
Primers ending in F1, F2, F3, R1, R2, and R3 were designed to minimize primer site polymorphisms or accommodate them by degenerate nucleotides. All other primers are the same as those used in the work by Corbett-Detig et al. (38). Names of strains used as positive and negative controls for each primer set are given.
Table S5.
PCR conditions used to test for nine inversion polymorphisms
| Inversion and orientation | +Control | −Control | Anneal temperature (°C) | Elongation time (s) | Experimental length (bp) | PCR details |
| In(1)A | ||||||
| Standard | y;cnbwsp | MW63-1 | 51 | 60 | 1,530 | * |
| Inverted | MW63-1 | y;cnbwsp | 61 | 45 | 330 | † |
| In(1)Be | ||||||
| Standard | y;cnbwsp | RG11N | 52 | 45 | 530 | * |
| Inverted | RG11N | y;cnbwsp | 51 | 30 | 560 | * |
| In(2L)t | ||||||
| Standard | y;cnbwsp | MW28-2–3 | 50 | 60 | 580 | * |
| Inverted | MW28-2–3 | y;cnbwsp | 50 | 30 | 280 | * |
| In(2R)NS | ||||||
| Standard | y;cnbwsp | MW28-2–3 | 60 | 90 | 3,060 | † |
| Inverted | MW28-2–3 | y;cnbwsp | 64 | 30 | 520 | ‡ |
| In(3L)Ok | ||||||
| Standard | y;cnbwsp | MW27-3 | 51 | 60 | 1,800 | † |
| Inverted | MW27-3 | y;cnbwsp | 51 | 60 | 2,300 | † |
| In(3L)P | ||||||
| Standard | y;cnbwsp | BL1204 | 53 | 30 | 380 | * |
| Inverted | BL1204 | y;cnbwsp | 65 | 30 | 360 | ‡ |
| In(3R)K | ||||||
| Standard | y;cnbwsp | MW56-2–3 | 52 | 90 | 1,230 | * |
| Inverted | MW56-2–3 | y;cnbwsp | 49 | 30 | 260 | * |
| In(3R)Mo | ||||||
| Standard | y;cnbwsp | RAL437 | 52 | 30 | 250 | * |
| Inverted | RAL437 | y;cnbwsp | 52 | 30 | 400 | * |
| In(3R)P | ||||||
| Standard | y;cnbwsp | ZH27-2–3 | 64 | 240 | 5,620 | † |
| Inverted | ZH27-2–3 | y;cnbwsp | 51 | 60 | 980 | * |
PCR conditions are given. PCR details are in the footnotes.
GoTaq (Promega). Initial denature at 95 °C for 2 min; 35 cycles of denature for 30 s at 95 °C, anneal for 30 s, and elongation at 72 °C; and final elongation at 72 °C for 5 min.
Phusion with GC Buffer (New England BioLabs). Denature at 98 °C for 30 s; 35 cycles of denature at 98°C for 10 s, anneal for 30 s, and elongation at 72 °C; and elongation at 72 °C for 10 min.
Phusion with HF Buffer (New England BioLabs). Denature at 98 °C for 30 s; 35 cycles of denature at 98 °C for 10 s, anneal for 30 s, and elongation at 72 °C; and elongation at 72 °C for 10 min.
Mutagenesis and Analysis of an Advanced Generation Cross.
A subsequent mutagenesis experiment was performed using flies from an advanced generation offspring of crosses between Ethiopian and Zambian strains (inbred lines EF73N and ZI418N, which were found to be free of known inversions). This genetic mapping population was established by mating eight EF73N virgin females with eight ZI418N males and separately mating equal numbers of flies for the reciprocal cross; 125 F1 females and 125 F1 males from each of the reciprocal crosses were added to a single-population cage, and these 500 flies gave rise to the F2 generation. Subsequent generations were maintained with population sizes of around 1,000 to yield a diversity of recombination breakpoints between parental genomes. In the F16 generation, 400 virgin females and 430 males were assayed for wing length. For each sex, the largest 15% and smallest 15% for wing length were sorted, and the intermediate flies were discarded. Males from the extreme groups were mutagenized as described above and then mated with females of the same wing size category. Crosses involving mutagenized flies were conducted as described for the previous experiment. This experiment did not yield enough F2 males for a statistical analysis of wing abnormalities. We, therefore, focused on F3 females for the comparison of wing mutant frequencies between the offspring of larger- and smaller-winged flies from the mapping population. We then tested whether the mutant frequency of large- vs. small-winged cage flies differed beyond null expectations for separate size and decanalization loci as detailed below.
Although flies in the above experiment have undergone up to 16 generations of recombination between the Ethiopian and Zambian genetic backgrounds, it is still possible that some linkage between independent wing size and decanalization loci might persist in these F16 mutagenized flies and their F3 offspring. To interpret our results against an informed null model, we simulated the full experimental process while tracking the ancestry of each fly (i.e., Ethiopian vs. Zambian parental strain) along chromosomes. Mirroring our experiment, the simulations began with 1,000 F1s (equal numbers from reciprocal Ethiopia × Zambia crosses) and continued with nonoverlapping generations until the F16 generation. Selection of the 15% largest-winged flies and the 15% smallest-winged flies was based on the recorded genotypes of simulated individuals at randomly located wing size loci. Ten wing size loci were simulated with equal additive effects. The selected F16 females and F16 mutagenized males then interbred to produce the F3 females examined for wing abnormalities (with numbers of flies matching those used in our experiment).
A wide range of genetic architecture of decanalization was simulated to account for each simulated F3 individual’s probability of being a wing mutant. Between 1 and 50 equally causative decanalization loci were simulated. Because the mapping population yielded a wing mutant rate closer to the Zambia pure population F3 mutant frequency, Ethiopian decanalization alleles seemed unlikely to act codominantly and additively across loci. Instead, to mirror this empirical observation, we focused on models in which decanalization alleles were recessive and/or multiplicative across loci. In all cases, an individual having all Ethiopian decanalization alleles was simulated as having the same mutant probability as the observed Ethiopia F3 average, and conversely, an individual with no Ethiopian decanalization alleles would match the Zambia F3 rate. The inheritance of decanalization alleles from the parental to the F3 generation after mutagenesis (with recombination) was simulated. Mutant probabilities for the three genetic models of decanalization were implemented as follows:
where LTOT and LHOM_ETH are the total number of decanalization loci and the number for which this individual is homozygous for Ethiopian ancestry, respectively, whereas RETH and RZAM are the wing mutant frequencies observed for F3 flies in the initial mutagenesis experiment:
where ATOT is the total number of decanalization alleles (equal to 2LTOT), and AETH is the number of Ethiopian decanalization alleles carried by this individual. Each F3 female was assigned a wing mutant with this probability, and the proportions of mutants for the large- and small-wing selection groups are noted. In each of 10,000 simulated replicates, we test how each of these scenarios could produce a wing mutant frequency ratio as extreme as that observed from the empirical data.
The above mutant probabilities were used instead of assuming a specific genetic basis for the observed wing mutants. We note that relatedness within the experiment is unlikely to produce the same mutant in different F3 females. The large- and small-winged experimental groups each involved ∼200 F1 flies (each carrying unique de novo mutations generated after male meiosis) and 400 F2 flies. If we assume that an observed wing defect results from a single autosomal recessive allele, the chance that the two alleles of a second fly also trace back to the same F1 ancestor is equal to
The binomial probability that any of 700 other F3 females fulfills the above criterion (and thus, shares the mutation because of relatedness) is then 0.0098. Thus, of 34 wing mutations observed in this experiment, very few, if any, should result from shared ancestry with another mutant under this scenario. Concordantly, the wing mutants observed among F3 females were quite diverse, with no specific phenotype predominating.
Supplementary Material
Acknowledgments
We thank Ian Dworkin for helpful advice, Robert Kreber for assistance with mutagenesis, and Russell Corbett-Detig and Charis Cardeno for help in identifying and testing inversion PCR primers. This work was supported by the National Institutes of Health through Grant R01 GM111797 (to J.E.P.) and a Ruth L. Kirschstein National Research Service Award, F32 GM106594 (to J.B.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515964113/-/DCSupplemental.
References
- 1.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150(3811):563–565. [Google Scholar]
- 2.Schmalhausen II. Factors of Evolution: The Theory of Stabilizing Selection. Blaskiston; Philadelphia: 1949. [Google Scholar]
- 3.Scharloo W. Canalization: Benetic and developmental aspects. Annu Rev Ecol Syst. 1991;22:65–93. [Google Scholar]
- 4.Wagner GP, Booth G, Bagheri-Chaichian H. A population genetic theory of canalization. Evolution. 1997;51(2):329–347. doi: 10.1111/j.1558-5646.1997.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 5.Dworkin I. Canalization, cryptic variation, and developmental buffering: A critical examination and analytical perpective. In: Hall BK, editor. Variation—A Central Concept in Biology. Academic; Waltham, MA: 2005. pp. 131–158. [Google Scholar]
- 6.Flatt T. The evolutionary genetics of canalization. Q Rev Biol. 2005;80(3):287–316. doi: 10.1086/432265. [DOI] [PubMed] [Google Scholar]
- 7.Waddington CH. Genetic assimilation of an acquired character. Evolution. 1953;7(2):118–126. [Google Scholar]
- 8.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396(6709):336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 9.Leamy L, Atchley W. Directional selection and developmental stability: Evidence from fluctuating asymmetry of morphometric characters in rats. Growth. 1985;49(1):8–18. [PubMed] [Google Scholar]
- 10.Kindred B. Selection for canalisation in mice. Genetics. 1967;55(4):635–644. doi: 10.1093/genetics/55.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pélabon C, Hansen TF, Carter AJR, Houle D. Response of fluctuating and directional asymmetry to selection on wing shape in Drosophila melanogaster. J Evol Biol. 2006;19(3):764–776. doi: 10.1111/j.1420-9101.2005.01054.x. [DOI] [PubMed] [Google Scholar]
- 12.Pélabon C, Hansen TF, Carter AJR, Houle D. Evolution of variation and variability under fluctuating, stabilizing, and disruptive selection. Evolution. 2010;64(7):1912–1925. doi: 10.1111/j.1558-5646.2010.00979.x. [DOI] [PubMed] [Google Scholar]
- 13.Hayden EJ, Weikert C, Wagner A. Directional selection causes decanalization in a group I ribozyme. PLoS One. 2012;7(9):e45351. doi: 10.1371/journal.pone.0045351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke GM, McKenzie JA. Developmental stability of insecticide resistant phenotypes in blowfly; a result of canalizing natural selection. Nature. 1987;325(6102):345–346. [Google Scholar]
- 15.McKenzie JA, Game AY. Diazinon resistance in Lucilia cuprina; mapping of a fitness modifier. Heredity (Edinb) 1987;59(3):371–381. [Google Scholar]
- 16.Zhang X-S, Hill WG. Mutation-selection balance for environmental variance. Am Nat. 2008;171(3):394–399. doi: 10.1086/527503. [DOI] [PubMed] [Google Scholar]
- 17.de Visser JAGM, et al. Perspective: Evolution and detection of genetic robustness. Evolution. 2003;57(9):1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 18.Pool JE, et al. Population Genomics of sub-saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 2012;8(12):e1003080. doi: 10.1371/journal.pgen.1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Stephan W. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet. 2006;2(10):e166. doi: 10.1371/journal.pgen.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thornton K, Andolfatto P. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics. 2006;172(3):1607–1619. doi: 10.1534/genetics.105.048223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bastide H, Yassin A, Johanning EJ, Pool JE. Pigmentation in Drosophila melanogaster reaches its maximum in Ethiopia and correlates most strongly with ultra-violet radiation in sub-Saharan Africa. BMC Evol Biol. 2014;14:179. doi: 10.1186/s12862-014-0179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitchers W, Pool JE, Dworkin I. Altitudinal clinal variation in wing size and shape in African Drosophila melanogaster: One cline or many? Evolution. 2013;67(2):438–452. doi: 10.1111/j.1558-5646.2012.01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Debat V, Debelle A, Dworkin I. Plasticity, canalization, and developmental stability of the Drosophila wing: Joint effects of mutations and developmental temperature. Evolution. 2009;63(11):2864–2876. doi: 10.1111/j.1558-5646.2009.00774.x. [DOI] [PubMed] [Google Scholar]
- 24.Chari S, Dworkin I. The conditional nature of genetic interactions: The consequences of wild-type backgrounds on mutational interactions in a genome-wide modifier screen. PLoS Genet. 2013;9(8):e1003661. doi: 10.1371/journal.pgen.1003661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Partridge L, French V. Thermal evolution of ectotherm body size: Why get big in the cold. In: Johnston IA, Bennet AF, editors. Animals and Temperature: Phenotypic and Evolutionary Adaptation. Cambridge Univ Press; Cambridge, United Kingdom: 1996. pp. 265–292. [Google Scholar]
- 26.Kennington WJ, Killeen JR, Goldstein DB, Partridge L. Rapid laboratory evolution of adult wing area in Drosophila melanogaster in response to humidity. Evolution. 2003;57(4):932–936. doi: 10.1111/j.0014-3820.2003.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 27.Lande R. Neutral theory of quantitative genetic variance in an island model with local extinction and colonization. Evolution. 1992;46(2):381–389. doi: 10.1111/j.1558-5646.1992.tb02046.x. [DOI] [PubMed] [Google Scholar]
- 28.Spitze K. Population structure in Daphnia obtusa: Quantitative genetic and allozymic variation. Genetics. 1993;135(2):367–374. doi: 10.1093/genetics/135.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller JR, Wood BP, Hamilton MB. FST and QST under neutrality. Genetics. 2008;180(2):1023–1037. doi: 10.1534/genetics.108.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution. 1965;19(3):395–420. [Google Scholar]
- 31.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132(2):583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lack JB, et al. The Drosophila Genome Nexus: A population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population. Genetics. 2015;199(4):1229–1241. doi: 10.1534/genetics.115.174664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenberg R, Crow JF. A comparison of the effect of lethal and detrimental chromosomes from Drosophila populations. Genetics. 1960;45(8):1153–1168. doi: 10.1093/genetics/45.8.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirkpatrick M, Jarne P. The effects of a bottleneck on inbreeding depression and the genetic load. Am Nat. 2000;155(2):154–167. doi: 10.1086/303312. [DOI] [PubMed] [Google Scholar]
- 35.Peters AD, Keightley PD. A test for epistasis among induced mutations in Caenorhabditis elegans. Genetics. 2000;156(4):1635–1647. doi: 10.1093/genetics/156.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You L, Yin J. Dependence of epistasis on environment and mutation severity as revealed by in silico mutagenesis of phage t7. Genetics. 2002;160(4):1273–1281. doi: 10.1093/genetics/160.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrés AM, Hernández MA. Two-tailed approximate confidence intervals for the ratio of proportions. Stat Comput. 2014;24(1):65–75. [Google Scholar]
- 38.Corbett-Detig RB, Cardeno C, Langley CH. Sequence-based detection and breakpoint assembly of polymorphic inversions. Genetics. 2012;192(1):131–137. doi: 10.1534/genetics.112.141622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.