Significance
Human osteoclast-associated receptor (OSCAR) is an immunoglobulin (Ig)-like receptor that associates with the ITAM (immunoreceptor tyrosine-based activation motif) receptor FcRγ (Fc receptor γ) to propagate an activating signal in osteoclasts, endothelial cells, and a variety of myeloid cells. The recent finding that OSCAR can bind to collagen and to surfactant protein D, which contains a collagen-like domain, presents a potentially wide array of situations in which this interaction could be targeted to alter an aberrant immune response. The structure of OSCAR bound to a collagen-like peptide describes the molecular basis of collagen recognition for a leukocyte receptor complex protein and provides a promising strategy for the development of future therapeutics aimed specifically at modulating collagen and the interactions of collagenous proteins.
Keywords: OSCAR, osteoclast, collagen, structure
Abstract
Human osteoclast-associated receptor (OSCAR) is an immunoglobulin (Ig)-like collagen receptor that is up-regulated on osteoclasts during osteoclastogenesis and is expressed in a range of myeloid cells. As a member of the leukocyte receptor complex family of proteins, OSCAR shares a high degree of sequence and structural homology with other collagen receptors of this family, including glycoprotein VI, leukocyte-associated Ig-like receptor-1, and leukocyte Ig-like receptor B4, but recognizes a unique collagen sequence. Here, we present the crystal structures of OSCAR in its free form and in complex with a triple-helical collagen-like peptide (CLP). These structures reveal that the CLP peptide binds only one of the two Ig-like domains, the membrane-proximal domain (domain 2) of OSCAR, with the middle and trailing chain burying a total of 661 Å2 of solvent-accessible collagen surface. This binding mode is facilitated by the unusual topography of the OSCAR protein, which displays an obtuse interdomain angle and a rotation of domain 2 relative to the membrane-distal domain 1. Moreover, the binding of the CLP to OSCAR appears to be mediated largely by tyrosine residues and conformational changes at a shallow Phe pocket. Furthermore, we investigated CLP peptides as inhibitors of osteoclastogenesis and found that a peptide length of 40 amino acids is required to ensure adequate inhibition of osteoclastogenesis in vitro. These findings provide valuable structural insights into the mode of collagen recognition by OSCAR and into the use of synthetic peptide matrikines for osteoclastogenesis inhibition.
Collagen is a highly versatile family of proteins consisting of more than 28 members that achieve a high degree of complexity through association of different collagen types, the use of multiple transcription initiation sites, alternative splicing patterns, posttranslational modifications, and a wide range of expression patterns (1). Despite this heterogeneity, studies of collagen over the past 60 y have revealed that collagens are commonly found as large fibrillar structures consisting of varying lengths of right-handed triple-helical structures that are formed by association of three left-handed polyproline type II helices with a one-residue stagger (2, 3). Crucial to the formation of these triple helical structures are the repetitive amino acid triplet sequences G-X-Y, of which the most frequently observed triplet is G-P-O, with O representing 4-hydroxyproline (4). In these structures the obligate glycine of the triplet is buried toward the center of the helix and is solvent inaccessible, whereas the X and Y residues are solvent accessible with the latter providing crucial stability to collagen fibrils through interchain hydrogen bonding (3). The diversity of imino and amino acids within these X and Y repetitive sequences are thought to result in changes in the helical parameters of collagen, which vary from a tight left-handed 72 to a looser 103 helical symmetry, and could provide a more flexible and accessible motif for recognition with collagen-binding proteins (5, 6).
OSCAR (human osteoclast-associated receptor) is an immunoglobulin (Ig)-like activating receptor of the leukocyte receptor complex (LRC) that associates with FcRγ and exhibits high expression levels during the development of bone-resorbing cells known as “osteoclasts” in a process called “osteoclastogenesis” (7, 8). The binding of OSCAR to collagen results in the activation of downstream signaling pathways and activation of nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 1 (NFATc1), a key transcription factor for osteoclastogenesis. Moreover, receptor activator of nuclear factor-κB ligand (RANKL) differentiation of monocytes to osteoclasts is increased in the presence of collagen-like peptides (CLPs) and can be inhibited by the addition of OSCAR antibodies (9). The description of OSCAR as a collagen-binding receptor of the LRC family places OSCAR with other collagen-binding proteins of this family, including glycoprotein VI (GPVI), leukocyte-associated Ig-like receptor-1 (LAIR-1), and the putative member LILRB4 (9–16).
In addition to its expression in osteoclasts, OSCAR has been found to be expressed in a wide variety of cells including vascular endothelial cells, macrophages, neutrophils, monocytes, and monocyte-derived dendritic cells (17, 18). Consequently, OSCAR is likely to contribute to the regulation of a wide variety of cellular functions in different cells, including roles in adhesion, activation and enhancement of proinflammatory cascades, cellular recruitment, and prevention of apoptosis. The description of an OSCAR minimal binding motif within collagen allows the potential development of a novel therapeutic peptide matrikine which could be used to control a diverse range of cellular functions, including osteoclastogenesis in rheumatoid arthritis, in which OSCAR expression has been found to be elevated as compared with healthy controls (19).
The use of synthetic peptide matrikines for therapeutic purposes currently is limited to experimental models with factors including, but not limited to, synthesis costs, stability, and dual activities affecting their translation into the clinic, as reviewed elsewhere (20, 21). To elucidate the molecular basis of the OSCAR–collagen interaction, we determined the crystal structures of the extracellular domains of OSCAR alone and in complex with a synthetic CLP and further investigated the effects of the length of this CLP on osteoclastogenesis in vitro. These structures reveal that, compared with other LRC members, OSCAR displays an unusually obtuse and rotated interdomain binding angle which facilitates the binding of CLP to a wide trench of domain 2. Furthermore, we show that the inhibitory effect on osteoclastogenesis of a therapeutic CLP containing the OSCAR minimal motif is determined by the peptide length.
Results
An Obtuse and Twisted Structure with Two Ig-Like Domains.
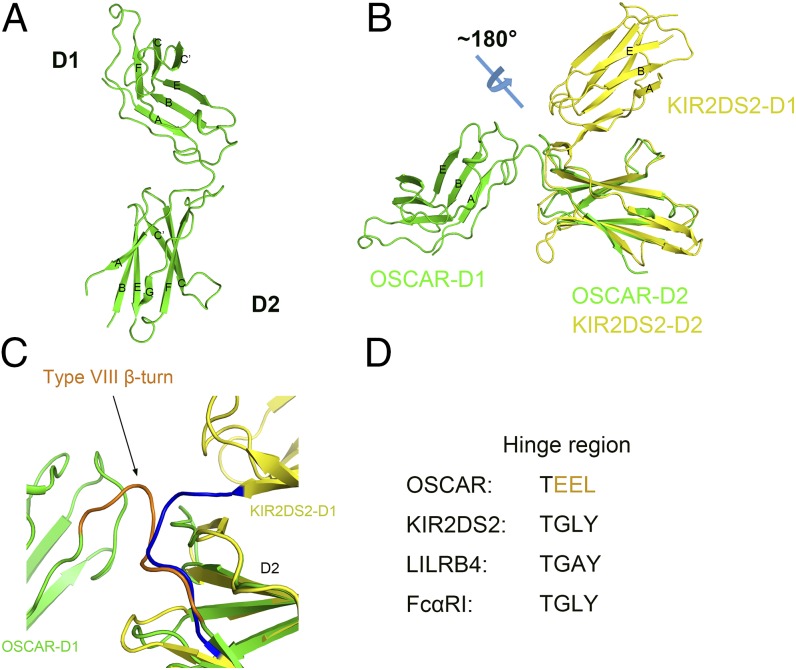
We resolved the crystal structure of the extracellular domain of OSCAR (residues 35–219) at a resolution of 2.0 Å by molecular replacement (Table S1). The crystal structure of the OSCAR apoprotein exhibits two Ig-like domains, D1 and D2, connected by the hinge region, defined by residues 35–127, 128–131, and 132–219, respectively. The Ig-like D1 domain displays a structure similar to that of the I-type structure described in GPVI D1, with a sharp bend after the first β strand that is induced by a conserved cis proline (P43). D1 also displays a single putative N-linked glycosylation site (N52) located at the end of the AB-loop, which is fully exposed to the solvent. The C-terminal D2 domain displays two putative N-linked glycosylation sites, both fully exposed to the solvent, with N149 located at the tip of B-strand, which is analogous to that of the D1 N-linked glycosylation site and N160 found at the BC-loop. Interestingly the latter of these glycosylation sites appears to be conserved in primates and, through its location oriented toward the interdomain hinge, could affect the flexibility of this region. The AB-loop of D2 (residues 142–147), unlike that in D1, has a poorly defined electron density, suggesting that it exhibits a high degree of flexibility. This poorly defined density makes it difficult to infer the Ig-like fold type of D2; however, the presence of a trans proline (P140) on the A-strand proximal to the undefined AB-loop suggests a C2-type Ig fold as seen in GPVI. A comparison of the individual domains of OSCAR with structures deposited in the Protein Data Bank (PDB) using the DALI server revealed that the D1 and D2 domains of OSCAR share the closest structural similarity to ILT3 domain 1 and ILT2 domain 4, respectively (Z-score 15.2 and 10.4 and rmsd 1.7 and 2.2 over 94 and 98 amino acids, respectively) (22). Interestingly, the OSCAR D1 domain does not display a 310 helical region in the C′ region or in the FG-loop, as found in some of the MHC-binding members of the LRC (23). Instead the region shows a prominent CC′-loop from P71 to P76 that is brought closer to the opposing FG-loop than in other members. In the OSCAR D2 domain, however, the CC′-loop extends further away from the opposing FG-loop, creating a wide trough above the GFCC′ β-sheets (Fig. 1A). The crystal structure also reveals that the D1 and D2 domains are linked at an obtuse angle with the CC′-loop of D2 rotated to a position distal to the ABE β-sheet face of D1 (Fig. 1B). This unusual orientation of the two Ig-like domains is likely results from the sharp β-turn at the interdomain linker that occurs because of hydrogen bonding between T128 and E130 (Fig. 1C). This interaction precludes any further direct hydrogen bonding between D1 (residues 35–131) and D2 (residues 132–219) at the interdomain region and is unique among LRC sequences (Fig. 1D). Moreover, the interdomain region also lacks a tryptophan (corresponding to Leu210 in OSCAR) that is conserved in other members of the LRC and has been suggested to have an important role in mediating interdomain interactions (24).
Table S1.
Crystallographic data collection and refinement statistics
| Parameters | OSCAR | OSCAR:CLP |
| Data collection | ||
| Space group | P21 | P212121 |
| Cell dimensions | ||
| a, b, c, Å | 27.22, 46.46, 66.98 | 45.94, 48.42, 118.06 |
| α, β, γ, ° | 90.00, 93.25, 90.00 | 90.00, 90.00, 90.00 |
| Resolution, Å | 50–2.00 (2.07–2.00) | 50–2.40 (2.49–2.40) |
| Rmerge | 0.079 (0.484) | 0.096 (0.669) |
| I/σI | 20.3 (4.7) | 18.2 (2.8) |
| Completeness, % | 99.8 (100.0) | 99.7 (100.0) |
| Redundancy | 5.8 (6.4) | 5.9 (6.0) |
| Refinement | ||
| Resolution, Å | 33.44–2.00 | 37.44–2.40 |
| No. reflections | 10,875 | 10,822 |
| Rwork/Rfree | 0.2230/0.2554 | 0.2252/0.2667 |
| No. atoms | ||
| Protein | 1,048 | 1,828 |
| Ligand/ion | 0 | 0 |
| Water | 67 | 28 |
| B-factors | ||
| Protein | 40.9 | 67.2 |
| Ligand/ion | ||
| Water | 41.5 | 65.3 |
| Rmsd | ||
| Bond lengths, Å | 0.009 | 0.004 |
| Bond angles, ° | 1.328 | 1.158 |
| Ramachandran analysis | ||
| Most favored, % | 95.1 | 95.2 |
| Additional allowed, % | 4.9 | 4.8 |
| Generally allowed, % | 0 | 0 |
| Disallowed, % | 0 | 0 |
Values in parentheses are for highest-resolution shell.
Fig. 1.
OSCAR crystal structure. (A) Ribbon diagram of the structure of OSCAR extracellular D1 and D2 domains with β-sheets labeled. (B) OSCAR (green) aligned to LRC protein KIR2DS2 D2 (PDB ID code 4N8V) (yellow). The ABE β-sheet face of the D1 domain in each protein is labeled, illustrating the unusual orientation of the D1 and D2 domains in comparison with other LRC members. (C) Close-up view of interdomain linker of OSCAR and KIR2DS2 illustrating the type VIII β-turn in OSCAR (brown) that results in a unique orientation of the D1 and D2 domains. (D) Sequence of the OSCAR D1–D2 interdomain linker compared with the linkers in other LRC members, which commonly exhibit the sequence G-L/A-Y.
CLP Binding to OSCAR D2.
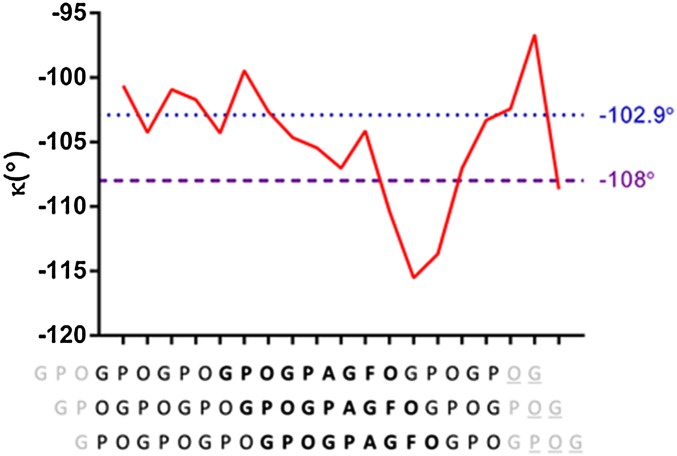
The CLP designed for crystallization with OSCAR was based on the findings of the minimal binding motif of collagen for OSCAR recognition (GXOGPX′GFX′) published by Barrow et al. (9). The peptide was optimized for crystallization by flanking the guest central binding motif with fewer host GPO repeats. The crystal structure of a complex of the CLP and the extracellular domain of the apoprotein was resolved using molecular replacement methods at a resolution of 2.4 Å (Table S1). In the crystal structure of the OSCAR/CLP complex the (GPO)3GPOGPAGFO(GPO)2G peptides form a right-handed triple helix that is fairly straight and ∼65 Å long, with some flexibility in the C-terminal residues. The CLP triple helix exhibits a structural conformation with a one-residue stagger and interchain hydrogen bonding similar to previously resolved structures in which the individual peptides have been designated as the “leading,” “middle,” and “trailing” chains (25). Moreover, the individual CLP helical properties approximate those expected in collagen, with the diversity of imino and amino acids resulting in a variation from a tight left-handed 72 conformation to a looser 103 helical symmetry in non–imino-rich regions (Fig. S1) (6).
Fig. S1.
Helical parameters of CLP in complex with OSCAR. The variation of the internal triple-helical twist (κ) (y axis) of the CLP is shown through the length of the CLP in relation to the local staggered sequence of the peptide (x axis). The two dashed horizontal lines indicate the internal twist values of ideal helices: −102.86° for a left-handed 72 helix (blue) and −108° (purple) for a left-handed 103 helix. Data are plotted so that each point in the diagram corresponds to the twist rotation between the two consecutive helical triplets below. Terminal and missing residues (gray and gray underlined, respectively) are not included in the plot (6).
The OSCAR/CLP complex structure reveals that the CLP binds to the D2 domain of OSCAR in a rather lopsided valley that has a floor shaped by the GFCC′ β-sheets (Fig. 2A). All the residues of the minimal binding motif are positioned over this valley, with the helix being closer to the shoulder created by the FG-loop than to the opposing shoulder of the CC′-loop. A sequence comparison of OSCAR isoforms (Fig. S2) and OSCAR orthologs (Fig. S3) reveals a high degree of conservation throughout the valley surface, suggesting a conserved mode of collagen recognition.
Fig. 2.
Crystal structure of the OSCAR/CLP complex. (A) OSCAR is shown in cartoon representation with semitransparent surface rendering. CLP is shown in cartoon representation with the N and C terminals labeled (the leading chain is shown in blue, the middle chain in yellow, and the trailing chain in magenta). (B) OSCAR (gray) bound to the trailing and middle chains of CLP shown as a cartoon. The N and C terminals of the CLP are labeled. OSCAR residues within 4.5 Å of CLP residues are colored orange. (Upper) All residues of CLP forming interactions with the OSCAR surface are shown in stick representation. (Lower) Residues forming substantial interactions between OSCAR and CLP are shown in stick representation. Dashed lines indicate hydrogen bonds. (C) Surface representation of apoprotein OSCAR and CLP-bound OSCAR. The trailing chain of the CLP is shown in cartoon representation, and labeled Phe17 is shown in CLP-bound OSCAR in stick representation. Residues delineating the Phe pocket are labeled and colored red, illustrating the conformational changes induced upon CLP binding.
Fig. S2.
Amino acid sequence alignment of OSCAR isoforms. Identical residues are shown in white text against a red background; similar residues are shown in red text; and dissimilar residues are shown in black text. Regions of identical and similar residues are highlighted with blue boxes. OSCAR residues involved in collagen recognition indicated by asterisks show a stringent conservation in all OSCAR isoforms (UniProt Q8IYS5). The alignment also illustrates differences in C-terminal sequences that result in only isoforms 2, 3, and 6 exhibiting a predicted transmembrane region.
Fig. S3.
Amino acid sequence alignment of OSCAR orthologs. Identical residues are shown in white text against a red background; similar residues are shown in red text; and dissimilar residues are shown in black text. Regions of identical and similar residues are highlighted with blue boxes. Human OSCAR (UniProt Q8IYS5) residues involved in collagen recognition are indicated by asterisks, illustrating a relatively stringent conservation across all OSCAR orthologs. Rat, Rattus norvegicus; Dog, Canis lupus familiaris; Pig, Sus scrofa; galago (Galgo), Otolemur garnettii; Marmoset, Callithrix jacchus; Orangutan, Pongo abelii; Gibbon, Nomascus leucogenys; Chimpanzee, Pan troglodytes; Mouse, Mus musculus (REFSEQ accession nos: NM_001184973.1, XM_014118576.1, XM_003127401.4, XM_003801596.2, XM_002762466.2, XM_002829721.2, XM_003259762.2, XM_001163782.4 and GenBank accession no. BC137776.1, respectively).
In total 15 OSCAR residues interact with the middle and trailing strands of the CLP and bury a total of 661 Å2 of solvent-accessible collagen surface. Interactions are largely of a hydrophobic nature with stabilizing polar interactions. Direct interactions occur between the CLP middle chain and OSCAR residues Y166 and P204 and between the trailing chain and residues Y200 and Y208 (Fig. 2B). Interaction with at least two chains of the collagen peptide is a common feature of previously resolved protein structures and explains the requirement for the peptide to form a triple-helical conformation for binding. Except for O18, all the residues of the collagen minimal binding motif from at least one of the leading or middle peptide chains interact with OSCAR residues. Furthermore, the hydroxyproline next to the N-terminal start of the guest central binding motif, O9, displays several interactions with OSCAR residues (Fig. S4).
Fig. S4.
Contacts between OSCAR and CLP. Schematic view of all contacts within 4.5 Å between OSCAR and CLP, illustrating the dominant roles of OSCAR residues Y166, Y176, Y200, Y208, and R213 in the interaction. Hydrogen bonds are shown in red, and residues strictly conserved in the minimal binding motif are shown in black boxes (9).
CLP Binding Induced Conformational Changes in the OSCAR Phe Pocket.
Interestingly, the trailing chain phenylalanine (F17) anchors in a positively charged shallow Phe pocket delineated by R133, R213, S214, and V216. Comparison of the free and CLP-bound OSCAR structures revealed that R213 undergoes a modest conformational change upon binding, moving ∼3.9 Å toward the Phe anchor (Fig. 2C). CLP binding also results in the reorientation of E168, which, in shifting 4.0 Å to facilitate hydrophilic contacts with R213, ties the loop of residues forming the Phe pocket. In this pocket we hypothesize that a cation–π interaction occurs between the buried phenylalanine of the CLP and R213. The R213 residue is located in the most favorable position for such an interaction, being perpendicular from the charged atom to the π-plane. Cation–π interactions between several other crystal structures of CLPs and their receptors also have been observed. In this case, however, the Phe pocket lacks a large aromatic residue such as tryptophan or histidine that hydrogen bonds with the collagen backbone (26–28).
Tyrosine-Mediated OSCAR/CLP Interaction.
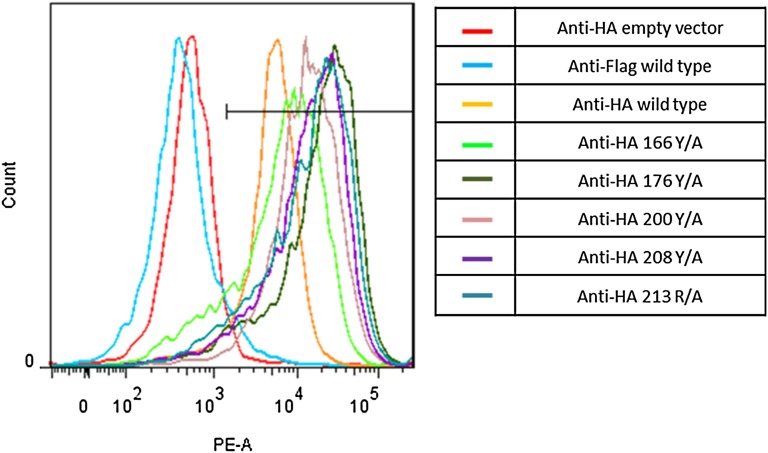
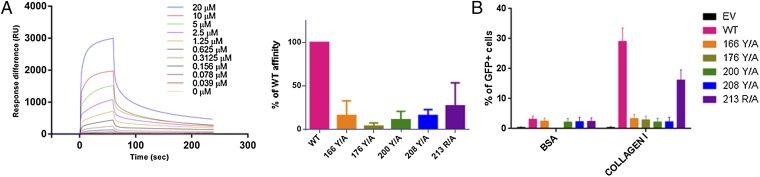
A functional characterization of the OSCAR residues displaying the most significant interactions with the CLP was carried out with WT OSCAR and mutant protein constructs expressed at a comparable level in an OSCAR-CD3ζ NFAT-GFP reporter cell line (Fig. S5) and with soluble extracellular domain mutant proteins expressed in Escherichia coli (9, 29). These mutants were investigated against WT for their binding properties to immobilized collagen I using surface plasma resonance (SPR) (Fig. 3A) and FACS platforms (Fig. 3B), respectively. All data corroborate the crystal structure analysis showing that the tyrosine residues Tyr166, Tyr176, Tyr200, and Tyr208 have highly significant interactions with collagen. This interaction is exemplified by a reduction in OSCAR signaling and GFP activation in NFAT-GFP reporter cells and by a reduction in binding affinity in SPR experiments with OSCAR mutants as compared with WT OSCAR. Specifically, the percentage of activated NFAT-GFP reporter cells, when cultured immobilized on collagen I, was reduced significantly from an average ± SEM of 29.1 ± 4.4% seen with WT OSCAR (n = 4) to averages of 3.3 ± 1.4%, 2.9 ± 1.2%, 2.3 ± 1.1%, and 2.3 ± 1.5% with 166 (n = 3), 176 (n = 4), 200 (n = 4), and 208 (n = 4) alanine screening mutations of tyrosine residues, respectively. This reduction in cellular activation is likely the result of a reduction in affinity of OSCAR mutants for the minimal binding motif of collagen as shown with SPR experiments (Fig. S6). Interestingly, the disruption of the Phe pocket of the OSCAR binding site by an alanine mutation (Arg213) was less pronounced than the aforementioned mutations, suggesting the Phe pocket has a less prominent role in OSCAR recognition of collagen I. This finding supports previously published work, which illustrates that a phenylalanine-to-alanine mutation in the minimal collagen binding motif causes only a partial reduction in OSCAR binding. Moreover, it can explain why a collagen motif from surfactant protein D, which lacks a phenylalanine, is able to bind to OSCAR and result in cellular signaling (9, 13).
Fig. S5.
Surface expression of WT and mutant OSCAR on NFAT-GFP reporter cells. Fluorescence intensity profile of stably transduced single-cell clones stained with anti-HA or anti-FLAG isotype control illustrates comparable levels of surface expression on WT and mutant cells. Low levels of fluorescence intensity are seen in empty vector-transfected NFAT-GFP reporter cells and with isotype control staining of WT cells.
Fig. 3.
Mutational analysis of OSCAR binding to collagen I. (A, Right) Sensogram of WT OSCAR binding to immobilized collagen I. The dissociation constant (Kd) (±SEM) is ∼26 ± 9 nM (n = 3). (Left) SPR measurements of the interaction of OSCAR WT and mutants with collagen I. Results are expressed as the percentage of the WT interaction. n = 3 independent experiments; error bars represent SEM. (B) Response of WT and mutant OSCAR-CD3ζ NFAT-GFP reporter cells relative to immobilized BSA and collagen I. FACS data are expressed as a percentage of GFP+ cells. n ≥ 3 independent experiments; error bars represent SEM.
Fig. S6.
SPR mutational analysis of OSCAR binding to collagen I. Interactions of soluble WT and mutant OSCAR proteins with immobilized collagen I were measured by SPR. Sensograms are representative of three independent experiments with curves representing decreasing concentrations of soluble OSCAR proteins: blue, 4 μM; red, 2 μM; green, 1 μM; purple, 0.5 μM; orange, 0.25 μM; black, 0.125 μM; gray, 0.0625 μM. Approximate affinities ± SEM: WT, 26 ± 9 nM; 166Y/A, 249 ± 71 nM; 176Y/A, 353 ± 42 nM (n = 2); 200Y/A, 305 ± 73 nM; 208Y/A, 154 ± 34 nM; 213R/A, 138 ± 38 nM.
Design of Peptide Inhibitors of Osteoclastogenesis.
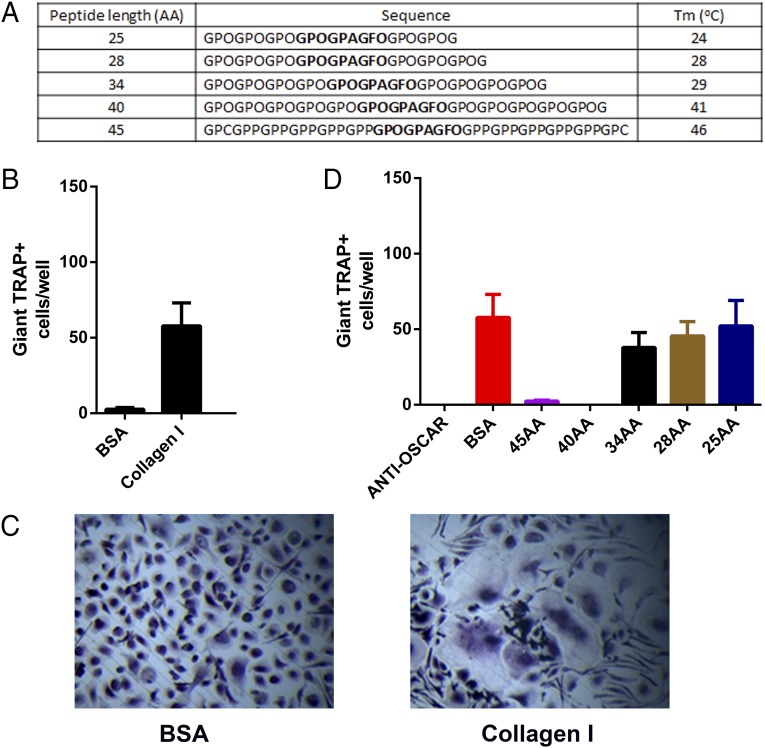
The interaction between OSCAR and two of the peptide chains in the triple-helical structure of the CLP reveals the importance of the triple-helical structure for binding. To assess the potential use of this CLP as a therapeutic matrikine targeted at inhibiting osteoclastogenesis in diseases such as rheumatoid arthritis, we designed an array of CLPs (Fig. 4A) and investigated their effects on inhibition of osteoclastogenesis in vitro. Following the culture of CD14+ monocytes on immobilized collagen I or BSA for 14 d at 37 °C in the presence of RANKL and macrophage colony-stimulating factor (M-CSF), osteoclasts were found to differentiate on immobilized collagen but not on BSA (Fig. 4 B and C). The addition of 10 μM of a CLP 40 or 45 amino acids (AA) in length throughout the culture was sufficient to inhibit this osteoclast growth to levels similar to those seen with an anti-OSCAR antibody (Fig. 4D and Fig. S7A). However, no reduction in osteoclastogenesis was observed in the presence of a CLP with a length of 25, 28, or 34 AA (Fig. 4D and Fig. S7A). Furthermore, the thermostability of these five CLP peptides was analyzed by circular dichroism (CD) spectroscopy. We found that the 45- and 40-AA peptides (with melting temperatures of 46 and 41 °C, respectively) exhibit better thermostability than do the 34-, 28-, and 25-AA peptides (with melting temperatures of 29, 28, and 24 °C, respectively) (Fig. S7B). These results indicate that, although a stable triple-helical peptide is illustrated with a length of 25 AA in our crystal structure, a longer peptide length with a higher thermostability is required to inhibit osteoclastogenesis effectively at body temperature.
Fig. 4.
OSCAR CLPs as inhibitors of osteoclastogenesis from human monocytes. (A) Rational design of CLP inhibitors of osteoclastogenesis based on sequence length and stability. (B) Osteoclastogenesis from human monocytes cultured on immobilized BSA or collagen I at day 14. n = 3 independent experiments; error bars represent SEM. (C) Tartrate-resistant acid phospatase (TRAP) staining of day 14 osteoclast cultures. (D) Effect of CLP on osteoclastogenesis from human monocytes cultured on immobilized collagen I at day 14. CLP inhibitors and BSA were added at a concentration of 10 μM throughout the culture, and medium was changed every 3 d. Anti-OSCAR antibody was added at a concentration of 2.5 μg/mL. n = 3 independent experiments; error bars represent SEM.
Fig. S7.
OSCAR CLP inhibitors of osteoclastogenesis from human monocytes. (A) TRAP staining of day 14 osteoclast cultures. CLP inhibitors and BSA were added at a concentration of 10 μM throughout the culture, and medium was changed every 3 d. Anti-OSCAR antibody was added at a concentration of 2.5 μg/mL. The positive osteoclasts with three or more nuclei were stained pink. (B) Thermostability measurements of CLP peptides by CD spectroscopy at 225 nM. The melting temperatures (Tm) are calculated at the state of 50% unfolding.
Discussion
The LRC complex encodes a variety of highly homologous Ig-like molecules that have evolved individual strategies for binding to a diverse set of ligands. The finding that several of these receptors are able to bind to collagen has led to research exploring whether there is a shared collagen-binding region on these receptors, as seen with the utilization of the D1–D2 hinge region by MHC-binding members (23). An intriguing investigation into the mode of collagen recognition of LAIR-1, through NMR titration experiments, revealed a putative collagen-binding site on the single Ig-like domain of LAIR-1 (15). The conservation of several residues in the same region of GPVI, which binds to a shared motif on collagen, led the authors to hypothesize that GPVI and LAIR-1 may use a conserved region for collagen recognition, a region at odds with a previous model of GPVI collagen binding (15, 16). Here we present the crystal structure of an LRC member in complex with a CLP. This complex reveals the binding site of the CLP to be located in the D2 domain of OSCAR over a wide trench that has a floor shaped by the GFCC′ β-sheets with the opposing shoulders formed by the FG-loop in combination with the polyproline II helix and the CC′-loop, respectively. Importantly, the putative collagen-binding area of LAIR-1 aligns unerringly with that of the OSCAR CLP-binding site, with the direction of the CLP also aligned to that predicted for LAIR-1. Our data strongly support the LAIR-1 putative collagen-binding region and lend support to the possibility that GPVI and other collagen-binding members of the LRC may use a wide trench region on the Ig-like domains flanked by CC′- and FG-loops to bind to collagen. This hypothesis regarding binding topology mirrors a more general prediction about collagen–protein interaction made more than 15 y ago, which predicted that the diversity of residues encoded in such trenches may confer specificity to certain types or regions of collagens (30).
The unusual orientation of the wide D2 trench relative to the D1 domain of OSCAR that facilitates CLP binding is unique among the published crystal structures of other LRC proteins and highlights the diversity of binding modes observed in LRC proteins. The D1 domain of OSCAR has a structural topology similar to that of the D2 domain and therefore might be expected to bind collagen. However, the narrow topology of the D1 trench—the result of an extended FG-loop and a relatively upright CC′-loop induced by the presence of two proline residues (P37 and P42) at the beginning and end of the loop—may preclude collagen binding across the D1 GFCC′ β-sheets. The topology of the D1 domain in other LRC collagen-binding proteins appears to be more favorable to collagen binding and to make the D1 domain a more likely candidate collagen-binding site than it is in OSCAR. Furthermore, a large conformational change would be required for the D2 domain of these proteins to interact with collagen at the corresponding region, in a manner analogous to a collagen hug seen with the Staphylococcus aureus collagen binding adhesion CNA (31). It also is likely that such an interaction may be restricted because of the lack of a large D1–D2 domain linker in LRC proteins. Moreover, the back-to-back dimer observed in the asymmetric unit of the GPVI crystal structure would not favor binding of collagen to D2 of GPVI.
A comparison of the binding features of OSCAR with other published protein/CLP crystal structures reveals that they have complex modes of collagen recognition. We have loosely categorized these binding modes, based on the structural features of the collagen-binding interface, into four groups: loop-shaped surface (integrin α2β1, DDR2, CIIC1, MASP-1, C1s, CIIJ1); sheet-shaped surface (CNA, HSP47, OSCAR); helix-shaped surface (SPARC); and composite-shaped surfaces (VWF, MMP-1) (Fig. S8). Despite large structural differences in their binding modes, all these molecules share a preference for charged and aromatic amino acids at the collagen-binding site (Table S2). These charged residues are able to form crucial hydrophilic interactions with the CLP and also have been found to stabilize some ion-mediated collagen interactions. The large aromatic amino acids, on the other hand, are largely responsible for forming a dense network of hydrophobic interactions with the CLP. An assessment of these shared features with those of the binding sites predicted for GPVI and LAIR-1 shows that both the predicted binding sites contain charged and aromatic amino acids and that their binding modes would be expected to fall into the loop surface and β-sheet surface groups, respectively (Table S2). Although our data strongly suggest a conserved mode of collagen recognition for LRC complex family members, the plethora of binding regions and mechanisms used by LRC proteins and the diverse range collagen-binding proteins raises the possibility that LRC members have several modes of collagen recognition. Therefore further studies into the structural recognition of collagen by LRC proteins are required to validate our hypothesized mode of collagen recognition.
Fig. S8.
Comparison of CLP binding modes. Crystal structures of 11 proteins in complex with CLP (leading chain, blue; middle chain, yellow; trailing chain, magenta) are shown in cartoon representation. Residues within 4.5 Å of CLP residues are colored orange. Proteins are grouped on the basis of the structural features of the CLP binding site. (A) Loop-shaped binding site (PDB ID codes: 1DZI, 4BKL, 2WUH, 3POB, 4LOR). (B) Sheet-shaped binding site (PDB ID codes: 5CJB, 2F6A, 4AU2). (C) Helix-shaped binding site (PDB ID code 2V53). (D) Composite-shaped binding site (PDB ID codes: 4DMU, 4AUO).
Table S2.
Protein residues that contact CLPs
 |
Protein residues that form contacts under 4.5 Å with a CLP in 11 crystal structures are categorized based on their aromatic (blue), charged (green), or other side chains. In the absence of a crystal structure for GPVI and LAIR-1, the residues found to interact with CLP in NMR or mutational studies are included.
The current crystal structures combined with our structure-based design of CLP inhibitors of osteoclastogenesis reinforce the importance of peptide structure and stability in the development of therapeutic matrikines and help define the minimal active sequence of an inhibitory OSCAR CLP. Moreover, these results provide a basis for the rational development of therapeutic matrikines aimed at fine tuning the cross-talk between osteoimmunology and vascular immunology systems to control aberrant osteoclastogenesis in conditions such as rheumatoid arthritis. However, our current understanding of the role of OSCAR in various cell types and of the expression patterns of OSCAR isoforms under normal and atypical conditions is limited and warrants further investigation to reduce the chance of off-target side effects.
Materials and Methods
Protein Expression and Purification.
The extracellular region of OSCAR (35-219 National Center for Biotechnology Information accession no. NP_570127.3) was synthesized (Generay) and then cloned into a pET21A expression vector without a His-tag for WT protein crystallization. Protein was expressed in BL21 cells, and inclusion bodies were refolded and purified as described previously (32).
Collagen, Peptide Synthesis, and Complex Formation.
Human collagen I was purchased from Sigma-Aldrich. Peptides were synthesized by the standard solid-phase method, and purity was analyzed by HPLC (GL Biochem). A triple-helical forming peptide composed of the minimal binding motif, (GPO)3GPOGPAGFO(GPO)2G, was used for crystallization. The complex form was isolated from the apoprotein by size-exclusion chromatography using Superdex 75 100/300GL.
Crystallization and Structure Determination.
Crystals of the OSCAR apoprotein were grown at 18 °C in a reservoir solution of 0.2 M potassium fluoride, 20% (wt/vol) polyethylene glycol 3350 (Hampton Research). Crystals of the OSCAR/CLP complex were grown at 4 °C in a reservoir solution of 5% polyvinyl alcohol type II, 0.2 M potassium acetate (pH 7), 0.1 M Hepes NaOH (Molecular Dimensions). X-ray data were collected at the Shanghai Synchrotron Radiation Facility beamline BL17U and were processed and scaled using HKL2000 (33). Data collection and processing statistics are summarized in Table S1. The structure of OSCAR was solved by molecular replacement using Phaser (34) from the CCP4 program suite (35) with the N-terminal domain (PDB ID code 1G0X) and C-terminal domain (PDB ID code 1B6U) as the search models, respectively (36, 37). The OSCAR/CLP complex also was solved by molecular replacement, with the structure of the refined OSCAR and CLP structure (PDB ID code 3B0S) as a search models (38). Initial restrained rigid-body refinement and manual model building were performed using REFMAC5 and COOT, respectively (39, 40). Further rounds of refinement were performed using the phenix.refine program implemented in the PHENIX package with coordinate refinement, isotropic ADP refinement, and bulk solvent modeling (41). The stereochemical quality of the final model was assessed with the program PROCHECK (42).
Characterization of Mutants.
MAXIsorp (Nunc) flat-bottomed plates (96-well) were coated overnight at 4 °C with collagen and control proteins at concentrations of 20 μg/mL in 10 mM acetic acid. For NFAT-GFP reporter cells assays WT and mutant chimeric reporter constructs of N-terminal HA-tagged OSCAR-PDGFR transmembrane fusion proteins with the cytoplasmic tail of the human CD3ζ chain were synthesized (Generay) based on a previous publication (9). Stably transduced single-cell clones were selected by 2.5 μg/mL puromycin and were sorted for high cell-surface expression staining with HA-specific murine mAb (Sigma Aldrich) by FACS (FACSAria; BD Biosciences). Stably transduced reporter cells (2.5 × 105/mL) were incubated on immobilized proteins at 37 °C in 5% CO2 for 48 h and then were analyzed for GFP expression by flow cytometry.
SPR Experiments.
SPR binding studies were performed using a BIAcore2000 system (BIAcore). Approximately 600–1,000 response units (RU) of acid-soluble human collagen type I was immobilized on a CM5 sensor chip (GE Healthcare) using the amine coupling kit. OSCAR proteins were diluted in buffer [150 mM NaCl, 0.005% (vol/vol) Tween 20, and 10 mM Hepes, pH 7.4] and increasing concentrations ranging from 62.5 to 4,000 nM were passed over the surface at a constant flow rate of 30 μL/min for 60 s. Dissociation was monitored for 200 s. The data were analyzed using BIAevaluation Software 4.1.
Osteoclast Cultures.
Osteoclasts were cultured from CD14+ monocytes MACS-sorted from human peripheral blood monocytes as described previously (9, 43). Briefly, 96-well tissue-culture plates were coated with BSA or collagen I as described above, and unbound protein was removed by washing with PBS followed by consecutive blocking steps with 2% BSA and complete α-MEM. CD14+ monocytes (5 × 104) were added to each well of the coated tissue-culture plates and were cultured at 37 °C, 5% CO2, with 30 ng/mL RANKL, 30 ng/mL M-CSF (PeproTech), and 10 μM of inhibitor or 2.5 μg/mL of anti-OSCAR antibody (Santa Cruz Biotechnology). The mean number of giant TRAP+ cells with three or more nuclei was established from three wells following fixation in 4% formalin and subsequent staining for TRAP with a TRAP-staining kit (Sigma-Aldrich).
Thermostability Measurements Using CD.
The thermostability of CLP peptides was analyzed by CD spectroscopy. Peptides were dissolved at 50 μM in PBS (pH 7.0), were heated to 60 °C for 15 min to dissociate, and then were stored at 4 °C overnight. CD spectra at 225 nm were measured on a Chirascan spectrometer (Applied Photophysics) using a thermostatically controlled cuvette at temperature intervals of 1 °C at a rate of 1 °C/min between 8 and 95 °C. The denaturation curves were generated by nonlinear fitting with GraphPad Prism.
Figures and Analysis.
The figures were made with PYMOL (www.pymol.org). Accessible surfaces were calculated with the CCP4 program AREAIMOL (35).
Acknowledgments
Collagen superhelix parameters were calculated with a CNS script kindly provided by Jordi Bella (University of Manchester). An NFAT-GFP reporter construct was generously provided by Hisashi Arase (Osaka University). This work was supported by the 973 project, China Ministry of Science and Technology Grants 2013CB531502 and 2014CB542503; the National Natural Science Foundation of China (NSFC) Grant 31390432; and Strategic Priority Research Program of the Chinese Academy of Sciences Grant XDB08020100. G.F.G. is a Leading Principal Investigator of the Innovative Research Group of the NSFC supported by NSFC Grant 81321063. Y.S. is supported by the Excellent Young Scientist Program of the Chinese Academy of Sciences (CAS) and the Youth Innovation Promotion Association, CAS Grant 2015078.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Human osteoclast-associated receptor (OSCAR) apoprotein and in complex with a collagen-like peptide has been deposited in the Protein Data Bank (PDB ID codes 5CJ8 and 5CJB, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522572113/-/DCSupplemental.
References
- 1.Ricard-Blum S. The collagen family. Cold Spring Harb Perspect Biol. 2011;3(1):a004978. doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramachandran GN, Kartha G. Structure of collagen. Nature. 1954;174(4423):269–270. doi: 10.1038/174269c0. [DOI] [PubMed] [Google Scholar]
- 3.Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science. 1994;266(5182):75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 4.Ramshaw JA, Shah NK, Brodsky B. Gly-X-Y tripeptide frequencies in collagen: A context for host-guest triple-helical peptides. J Struct Biol. 1998;122(1-2):86–91. doi: 10.1006/jsbi.1998.3977. [DOI] [PubMed] [Google Scholar]
- 5.Kramer RZ, Bella J, Mayville P, Brodsky B, Berman HM. Sequence dependent conformational variations of collagen triple-helical structure. Nat Struct Biol. 1999;6(5):454–457. doi: 10.1038/8259. [DOI] [PubMed] [Google Scholar]
- 6.Bella J. A new method for describing the helical conformation of collagen: Dependence of the triple helical twist on amino acid sequence. J Struct Biol. 2010;170(2):377–391. doi: 10.1016/j.jsb.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 7.Kim N, Takami M, Rho J, Josien R, Choi Y. A novel member of the leukocyte receptor complex regulates osteoclast differentiation. J Exp Med. 2002;195(2):201–209. doi: 10.1084/jem.20011681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koga T, et al. Costimulatory signals mediated by the ITAM motif cooperate with RANKL for bone homeostasis. Nature. 2004;428(6984):758–763. doi: 10.1038/nature02444. [DOI] [PubMed] [Google Scholar]
- 9.Barrow AD, et al. OSCAR is a collagen receptor that costimulates osteoclastogenesis in DAP12-deficient humans and mice. J Clin Invest. 2011;121(9):3505–3516. doi: 10.1172/JCI45913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moroi M, Jung SM. Platelet glycoprotein VI: Its structure and function. Thromb Res. 2004;114(4):221–233. doi: 10.1016/j.thromres.2004.06.046. [DOI] [PubMed] [Google Scholar]
- 11.Lebbink RJ, et al. Collagens are functional, high affinity ligands for the inhibitory immune receptor LAIR-1. J Exp Med. 2006;203(6):1419–1425. doi: 10.1084/jem.20052554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L, Barclay AN. New assay to detect low-affinity interactions and characterization of leukocyte receptors for collagen including leukocyte-associated Ig-like receptor-1 (LAIR-1) Eur J Immunol. 2009;39(4):1167–1175. doi: 10.1002/eji.200839188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrow AD, Palarasah Y, Bugatti M, Holehouse AS. OSCAR is a receptor for surfactant protein D that activates TNF-alpha release from human CCR2+ inflammatory monocytes. J Immunol. 2015;194(7):3317–3326. doi: 10.4049/jimmunol.1402289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz HS, et al. Collagen induces maturation of human monocyte-derived dendritic cells by signaling through osteoclast-associated receptor. J Immunol. 2015;194(7):3169–3179. doi: 10.4049/jimmunol.1402800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brondijk TH, et al. Crystal structure and collagen-binding site of immune inhibitory receptor LAIR-1: Unexpected implications for collagen binding by platelet receptor GPVI. Blood. 2010;115(7):1364–1373. doi: 10.1182/blood-2009-10-246322. [DOI] [PubMed] [Google Scholar]
- 16.Horii K, Kahn ML, Herr AB. Structural basis for platelet collagen responses by the immune-type receptor glycoprotein VI. Blood. 2006;108(3):936–942. doi: 10.1182/blood-2006-01-010215. [DOI] [PubMed] [Google Scholar]
- 17.Merck E, et al. Ligation of the FcR gamma chain-associated human osteoclast-associated receptor enhances the proinflammatory responses of human monocytes and neutrophils. J Immunol. 2006;176(5):3149–3156. doi: 10.4049/jimmunol.176.5.3149. [DOI] [PubMed] [Google Scholar]
- 18.Goettsch C, et al. Quantitative proteomics reveals novel functions of osteoclast-associated receptor in STAT signaling and cell adhesion in human endothelial cells. J Mol Cell Cardiol. 2012;53(6):829–837. doi: 10.1016/j.yjmcc.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Herman S, et al. Induction of osteoclast-associated receptor, a key osteoclast costimulation molecule, in rheumatoid arthritis. Arthritis Rheum. 2008;58(10):3041–3050. doi: 10.1002/art.23943. [DOI] [PubMed] [Google Scholar]
- 20.Ricard-Blum S, Salza R. Matricryptins and matrikines: Biologically active fragments of the extracellular matrix. Exp Dermatol. 2014;23(7):457–463. doi: 10.1111/exd.12435. [DOI] [PubMed] [Google Scholar]
- 21.Monboisse JC, Oudart JB, Ramont L, Brassart-Pasco S, Maquart FX. Matrikines from basement membrane collagens: A new anti-cancer strategy. Biochim Biophys Acta. 2014;1840(8):2589–2598. doi: 10.1016/j.bbagen.2013.12.029. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Rosenstrom P. 2010. Dali server: Conservation mapping in 3D. Nucleic Acids Res 38(Web Server issue):W545–549.
- 23.Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 2003;4(9):913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- 24.Cheng H, et al. Crystal structure of leukocyte Ig-like receptor LILRB4 (ILT3/LIR-5/CD85k): A myeloid inhibitory receptor involved in immune tolerance. J Biol Chem. 2011;286(20):18013–18025. doi: 10.1074/jbc.M111.221028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Structural basis of collagen recognition by integrin alpha2beta1. Cell. 2000;101(1):47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- 26.Hohenester E, Sasaki T, Giudici C, Farndale RW, Bächinger HP. Structural basis of sequence-specific collagen recognition by SPARC. Proc Natl Acad Sci USA. 2008;105(47):18273–18277. doi: 10.1073/pnas.0808452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carafoli F, et al. Crystallographic insight into collagen recognition by discoidin domain receptor 2. Structure. 2009;17(12):1573–1581. doi: 10.1016/j.str.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brondijk TH, Bihan D, Farndale RW, Huizinga EG. Implications for collagen I chain registry from the structure of the collagen von Willebrand factor A3 domain complex. Proc Natl Acad Sci USA. 2012;109(14):5253–5258. doi: 10.1073/pnas.1112388109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296(5571):1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 30.Rich RL, et al. Trench-shaped binding sites promote multiple classes of interactions between collagen and the adherence receptors, alpha(1)beta(1) integrin and Staphylococcus aureus cna MSCRAMM. J Biol Chem. 1999;274(35):24906–24913. doi: 10.1074/jbc.274.35.24906. [DOI] [PubMed] [Google Scholar]
- 31.Zong Y, et al. A ‘Collagen Hug’ model for Staphylococcus aureus CNA binding to collagen. EMBO J. 2005;24(24):4224–4236. doi: 10.1038/sj.emboj.7600888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garboczi DN, Hung DT, Wiley DC. HLA-A2-peptide complexes: Refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci USA. 1992;89(8):3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Read RJ. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 10):1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- 35.Bailey S. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Chapman TL, Heikema AP, West AP, Jr, Bjorkman PJ. Crystal structure and ligand binding properties of the D1D2 region of the inhibitory receptor LIR-1 (ILT2) Immunity. 2000;13(5):727–736. doi: 10.1016/s1074-7613(00)00071-6. [DOI] [PubMed] [Google Scholar]
- 37.Maenaka K, Juji T, Stuart DI, Jones EY. Crystal structure of the human p58 killer cell inhibitory receptor (KIR2DL3) specific for HLA-Cw3-related MHC class I. Structure. 1999;7(4):391–398. doi: 10.1016/s0969-2126(99)80052-5. [DOI] [PubMed] [Google Scholar]
- 38.Okuyama K, Miyama K, Mizuno K, Bächinger HP. Crystal structure of (Gly-Pro-Hyp)(9) : Implications for the collagen molecular model. Biopolymers. 2012;97(8):607–616. doi: 10.1002/bip.22048. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 43.Agrawal A, Gallagher JA, Gartland A. Human osteoclast culture and phenotypic characterization. Methods Mol Biol. 2012;806:357–375. doi: 10.1007/978-1-61779-367-7_23. [DOI] [PubMed] [Google Scholar]