Significance
The directed differentiation of human embryonic stem cells into cardiomyocytes provides a tool for understanding human heart development and disease. During the process of cardiomyocyte differentiation, tight regulation of the WNT/β-catenin signaling pathway is required. Thus, understanding which proteins are involved in promoting or repressing the WNT/β-catenin signaling pathway is crucial for identifying positive and negative regulators of cardiac development. Here we measured protein expression during a time course of cardiomyocyte differentiation. We identified a regulator of cardiac development, Disabled 2, and found that in zebrafish embryos, it negatively regulates WNT/β-catenin signaling to promote cardiomyocyte differentiation. Thus, our work reveals a highly conserved, previously unidentified process relevant for human heart development.
Keywords: quantitative proteomics, cardiomyocyte, zebrafish, embryonic stem cell, WNT/β-catenin
Abstract
To reveal the molecular mechanisms involved in cardiac lineage determination and differentiation, we quantified the proteome of human embryonic stem cells (hESCs), cardiac progenitor cells (CPCs), and cardiomyocytes during a time course of directed differentiation by label-free quantitative proteomics. This approach correctly identified known stage-specific markers of cardiomyocyte differentiation, including SRY-box2 (SOX2), GATA binding protein 4 (GATA4), and myosin heavy chain 6 (MYH6). This led us to determine whether our proteomic screen could reveal previously unidentified mediators of heart development. We identified Disabled 2 (DAB2) as one of the most dynamically expressed proteins in hESCs, CPCs, and cardiomyocytes. We used clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) mutagenesis in zebrafish to assess whether DAB2 plays a functional role during cardiomyocyte differentiation. We found that deletion of Dab2 in zebrafish embryos led to a significant reduction in cardiomyocyte number and increased endogenous WNT/β-catenin signaling. Furthermore, the Dab2-deficient defects in cardiomyocyte number could be suppressed by overexpression of dickkopf 1 (DKK1), an inhibitor of WNT/β-catenin signaling. Thus, inhibition of WNT/β-catenin signaling by DAB2 is essential for establishing the correct number of cardiomyocytes in the developing heart. Our work demonstrates that quantifying the proteome of human stem cells can identify previously unknown developmental regulators.
Directed differentiation of human embryonic stem cells (hESCs) toward definitive cardiomyocytes provides a platform for understanding human heart development and disease (1). To this end, genome-wide screens focusing on transcriptional regulation and RNA expression during time course staging of cardiac development have identified regulators of cardiac development (2, 3). Although these studies have increased our understanding of the transcriptional mechanisms active during heart development, there remains little information regarding regulation of the proteome in this same context. This led us to quantify the proteome of hESCs during a stage-specific differentiation to definitive cardiomyocytes, a primary functional cell of the heart muscle.
It is well known that tight regulation of the WNT/β-catenin signaling pathway during the process of cardiomyocyte differentiation is imperative (4, 5). Temporal overactivation or inhibition of the WNT/β-catenin signaling pathway has been shown to result in cardiac null phenotypes in vivo (4, 6, 7), because WNT/β-catenin is required to form mesoderm, and it subsequently must be repressed to form cardiomyocytes. Furthermore, overactivation of WNT/β-catenin signaling during hESC-derived cardiomyocyte differentiation results in a shift in mesoderm patterning to specify endothelium and early blood cells, but not cardiomyocytes (8). Conversely, inhibition of WNT/β-catenin has been shown to direct endocardial-like endothelial cells to differentiate toward the cardiac lineage (3). Thus, understanding which proteins are involved in promoting or repressing the WNT/β-catenin signaling pathway is crucial for resolving the ambiguities associated with cardiac development.
Here, using label-free quantitation (LFQ) proteomics, we measured protein expression patterns during a time course of hESC-derived cardiomyocyte differentiation. LFQ proteomics is a robust technology for quantifying differences in protein expression, which have been shown to correlate with differentiated cell types (9, 10). Furthermore, quantifying unmodified protein expression has the advantage of eliminating the unknown effects of posttranscriptional regulation. Using this approach, we identified regulators of cardiac development, including Disabled 2 (DAB2). We found that DAB2 negatively regulates WNT/β-catenin signaling and promotes cardiomyocyte differentiation from mesoderm-derived progenitors.
Results
Quantification of the Proteome During hESC-Directed Differentiation Toward Cardiomyocytes.
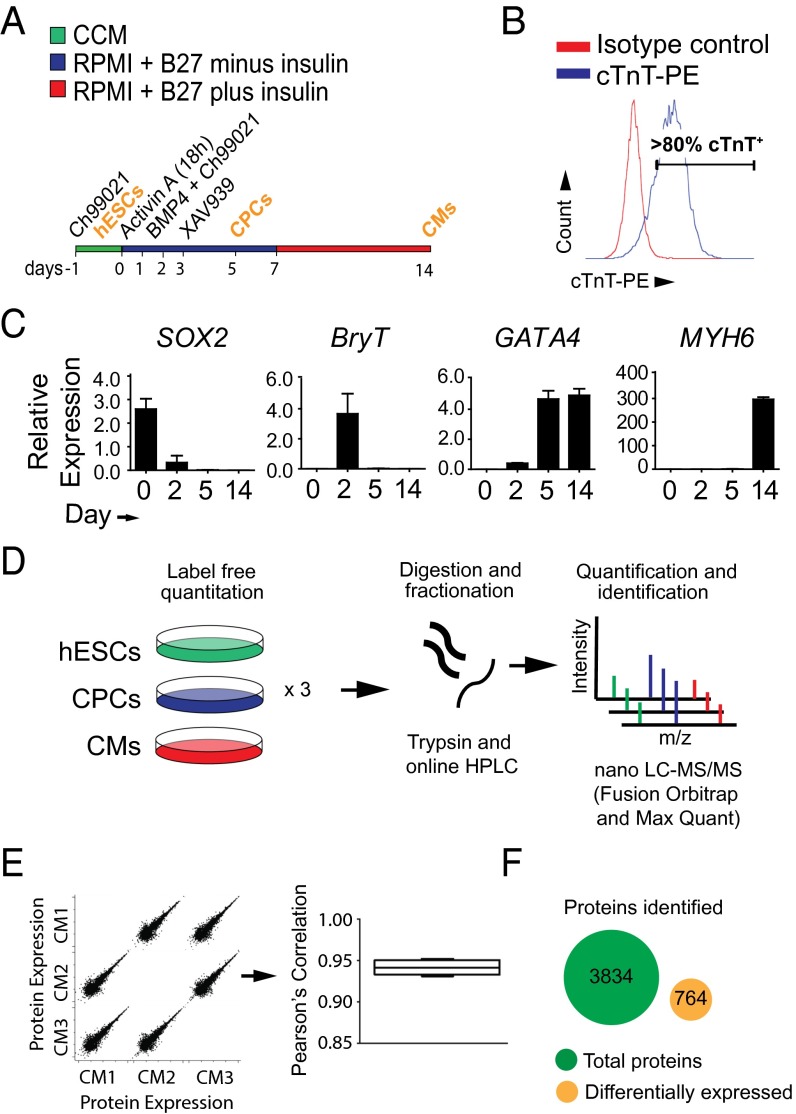
To reveal molecular mediators and markers of cardiac development, we quantified the proteome of hESCs as they differentiated toward cardiac progenitor cells (CPCs) and definitive beating cardiomyocytes. hESCs were induced to differentiate with activin A and bone morphogenetic protein 4 (BMP4), in combination with sequential small molecule activation and inhibition of WNT/β-catenin signaling. Cells were then harvested as pluripotent hESCs (day 0), CPCs (day 5), or cardiomyocytes (day 14) (3, 5, 8) (Fig. 1A). We have previously shown that as cells differentiate toward CPCs, they express cardiac transcription factors, such as GATA binding protein 4 (GATA4), T-box 5 (TBX5), and NK2 homeobox 5 (NKX2.5) (3, 8) (Fig. 1A). CPCs then in turn differentiate predominately toward cardiomyocytes and express the cardiac structural genes [cardiac troponin T (cTnT)] and [myosin heavy chain 6 (MYH6)], with a minority of cells showing characteristics of the fibroblast/smooth muscle (2, 3). Thus, we assessed our cultures by fluorescence-activated cell sorting (FACS) analysis to determine cardiomyocyte purity and differentiation efficiency, and verified that >80% of the cardiomyocyte population was cTnT-positive (Fig. 1B).
Fig. 1.
LFQ proteomics during directed differentiation of hESCs into cardiomyocytes. (A) Schematic of differentiation protocol: hESCs (day 0), CPCs (day 5), and definitive cardiomyocytes (CMs; day 14). (B) FACS analysis of day 14 hESC-derived CMs stained for cTnT (y-axis, total cell counts %; x-axis, cells stained for cTnT). (C) qRT-PCR during CM differentiation at stages of hESCs (day 0), mesoderm (day 2), CPCs (day 5), and CMs (day 14) for pluripotency marker SOX2, pan-mesodermal marker BryT, cardiac transcription factor GATA4, and the cardiac structural protein MYH6. n = 4 biological replicates. Data are mean ± SEM. (D) Schematic of the experimental design for LFQ proteomics, with assays of hESCs, CPCs, and CMs. (E) Relative protein expression (LFQ intensity) quantified by measuring tryptic digestions of cell lysates using nano LC-MS/MS. n = 3 biological replicates per time point; average Pearson’s correlation, 0.94. (F) Peptides corresponding to 3,834 proteins measured as in E.
As a readout of normal cardiac differentiation, we assessed markers of pluripotency, [SRY-box2 (SOX2)], mesoderm formation [Brachyury T (BryT)], CPCs (GATA4), and cardiomyocytes (MYH6) by quantitative reverse-transcriptase PCR (qRT-PCR) over the time course of differentiation (Fig. 1C). SOX2 is highly expressed in hESCs, whereas expression decreases to baseline levels as cells differentiate toward precardiac mesoderm. As cells exit pluripotency, the pan-mesodermal marker BryT is expressed on day 2, whereas during specification of CPCs, BryT expression returns to baseline levels. The increased expression of the cardiac transcription factor GATA4 is observed at day 5, followed by cardiac structural proteins MHY6 and cTnT at day 14 (Fig. 1B). These data indicate successful differentiation of hESCs toward cardiomyocytes with normal transitioning among hESCs, mesoderm, CPCs, and definitive cardiomyocytes.
Using LFQ proteomics, we measured changes in protein expression during a time course of hESC cardiomyocyte differentiation by quantifying protein extracted from hESCs (day 0), CPCs (day 5), and cardiomyocytes (day 14) (Fig. 1D). Normalized quantities of protein were tryptically digested and fractionated using reverse-phase chromatography before measurement by mass spectrometry. Protein quantification was reproducible, with an average Pearson’s correlation of 0.94 across all samples (Fig. 1E). A total of 3,834 proteins were identified and quantified, of which 764 were differentially regulated among hESCs, CPCs, and cardiomyocytes (Fig. 1F and Datasets S1 and S2).
LFQ Proteomics Identifies DAB2 as a Putative Regulatory Protein for Cardiomyocyte Development.
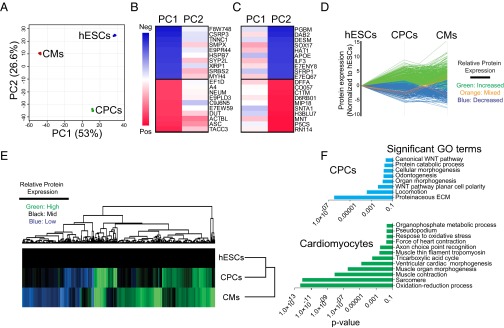
To identify system-level changes in metabolic or signaling pathways during differentiation, we performed principal component analysis (PCA), followed by normalization of protein expression of CPCs and cardiomyocytes to hESCs (Fig. 2). PCA demonstrated distinct patterns of protein expression in hESCs, CPCs, and cardiomyocytes (Fig. 2A). The first principal component (PC1) distinguished cardiomyocytes from hESCs and CPCs, whereas the second (PC2) distinguished CPCs from cardiomyocytes and hESCs. We subsequently assessed which proteins contributed to PC1 and PC2 based on positive and negative PC loadings (Fig. 2 B and C and Dataset S3). After normalizing CPCs and cardiomyocytes to hESCs (Fig. 2D), we performed unbiased hierarchical clustering to confirm that global protein expression can distinguish these three cellular fates (Fig. 2E).
Fig. 2.
Global proteomics analyses reveal enrichment of WNT/β-catenin signaling. (A) PCA of hESCs, CPCs, and cardiomyocytes (CMs). (B and C) Heat map analyses of the top-20 contributing proteins (negative and positive PC loadings) for PC1 (B) and PC2 (C). (D) Protein expression of CPCs and CMs normalized to that of hESCs. (E) Unbiased hierarchical clustering of total protein expression normalized to that of hESCs. (F) Significant GO terms associated with CPCs and CMs.
We used Gene Ontology (GO) term enrichment to investigate the functional relationships between differentially expressed proteins (Fig. 2F and Dataset S4). During the transition between hESCs and CPCs, proteins involved primarily in regulation of the WNT/β-catenin pathway and cellular morphogenesis were represented. In contrast, GO terms associated with proteins that regulate muscle and ventricular morphogenesis, the development of sarcomeres, and metabolic processes were enriched in cardiomyocytes. These data indicate that our LFQ proteomic approach can identify known, and also possibly unknown, regulatory proteins associated with human cardiomyocyte differentiation.
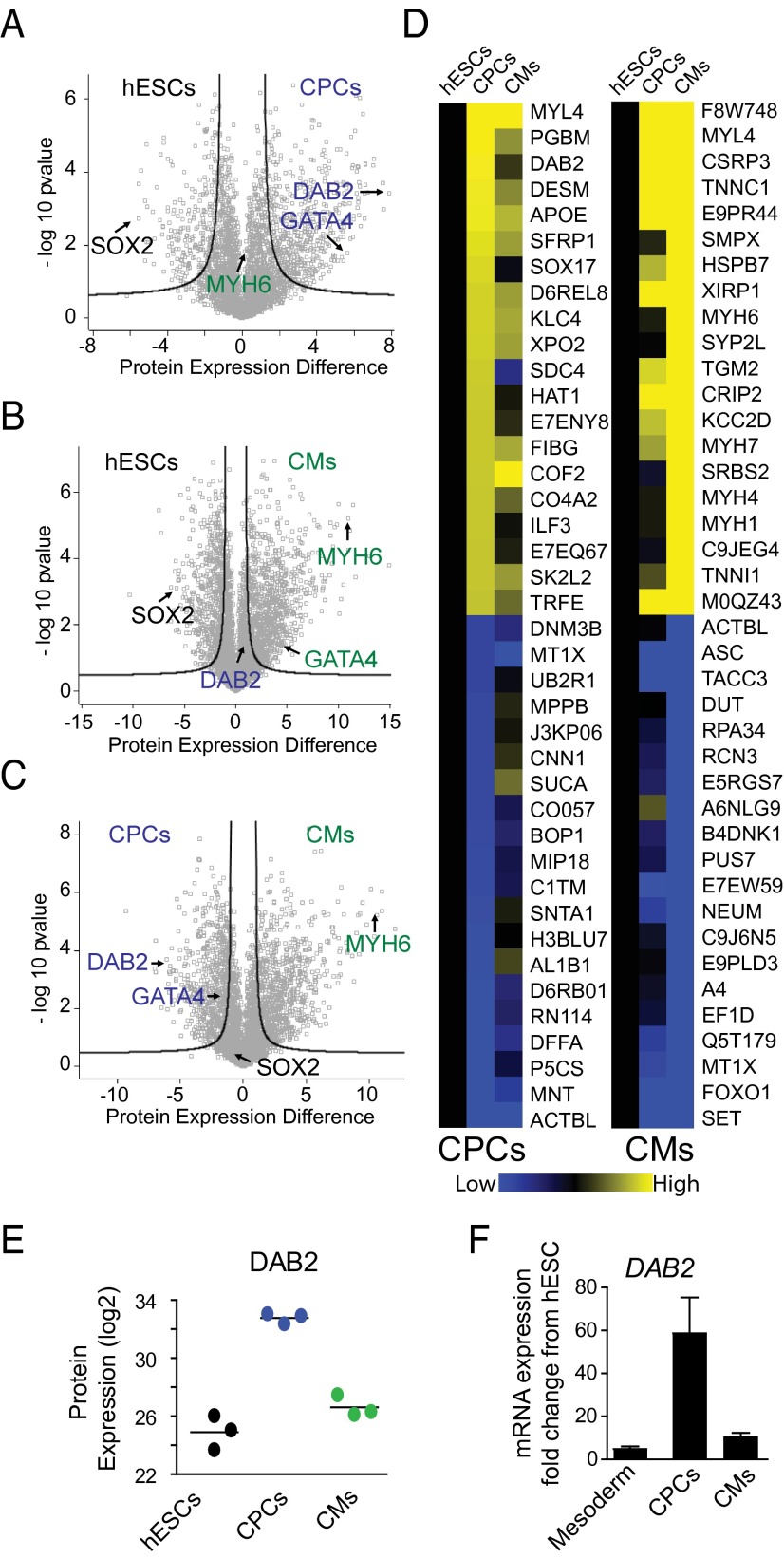
To visualize changes in protein expression during cardiomyocyte differentiation, we compared hESCs with CPCs (Fig. 3A) or cardiomyocytes (Fig. 3B) and compared CPCs with cardiomyocytes (Fig. 3C), graphing differences in protein expression vs. significance (P value). This method identified known and unknown stage-specific markers of differentiation, including SOX2, GATA4, and MYH6 (Fig. 3 A–C). Furthermore, protein expression levels of these markers were temporally correlated with mRNA expression (Fig. 3 A and B; cf. Fig. 1C).
Fig. 3.
System-wide proteomics analyses identify DAB2 as a putative regulator of cardiac development. (A–C) Volcano plots of protein expression comparing hESCs with CPCs (A) and cardiomyocytes (CMs; B) and comparing CPCs with CMs (C). Highlighted proteins are the pluripotency marker SOX2, cardiac transcription factor GATA4, cardiac structural protein MYH6, and putative cardiomyocyte developmental regulator DAB2. (D) Heat map analyses of the top-40 differentially expressed proteins in CPCs (Left) and CMs (Right). (E and F) Protein expression (LFQ intensity; E) and mRNA expression (F) of DAB2 during hESC–cardiomyocyte differentiation. n = 3 biological replicates measured by mass spectrometry and n ≥4 biological replicates for qRT-PCR. Data are mean ± SEM.
To further reveal regulators of cardiac development, we examined proteins that were dynamically expressed throughout the time course of hESC cardiomyocyte differentiation. To this end, we analyzed the top-40 differentially expressed proteins from CPCs and cardiomyocytes (20 up-regulated and 20 down-regulated for each cell type). Heat map analyses demonstrated dynamic expression levels, with many nonoverlapping proteins regulated between CPCs and cardiomyocytes (Fig. 3D). From these analyses, we identified a WNT/β-catenin inhibitor and mitogen-responsive phosphoprotein, Disabled 2 (DAB2), as one of the most dynamically expressed proteins across hESC cardiomyocyte differentiation (Fig. 3 A–D). DAB2 protein expression was markedly increased between hESCs and CPCs (mean ± SEM LFQ intensity, 7.86 ± 0.25), with levels approaching baseline in cardiomyocytes (mean ± SEM LFQ intensity, 1.71 ± 0.51). Moreover, DAB2 was dynamically expressed at both the protein and transcript levels over the time course of cardiomyocyte differentiation (Fig. 3 E and F). As predicted by the proteomic results, we found a robust increase in DAB2 mRNA expression between hESCs and CPCs, but not between hESCs and mesoderm or cardiomyocytes (Fig. 3F). Collectively, these data demonstrate that our LFQ proteomics can identify putative regulators of cardiac development.
DAB2 Loss of Function Results in Increased WNT/β-Catenin Signaling.
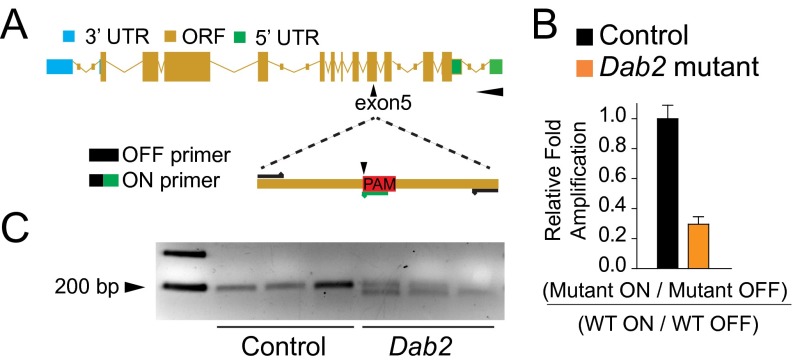
We sought to understand the role of DAB2 in vivo by using a highly conserved in vivo model of heart development (11, 12) in zebrafish (Danio rerio). Previous studies have shown that during early-stage cardiac specification, dab2 is expressed within a critical region for CPC specification, the ventral mesoderm, as well as the pronephros (13, 14), but during late-stage heart development, dab2 is expressed not in the heart but primarily within regions of the developing endothelium (15). These data corroborate the dynamic expression patterns of DAB2 during hESC cardiomyocyte differentiation. Thus, we created a loss-of-function mutant using clustered regularly interspaced short palindromic repeats (CRISPR) coupled with the CRISPR-associated protein 9 (Cas9; CRISPR/Cas9) gene editing system to target the Dab2 locus (Fig. S1).
Fig. S1.
CRISPR/Cas9 inhibition of zebrafish Dab2. (A) Schematic of the zebrafish Dab2 locus, with the target site indicated by a black arrowhead at exon 5, and an illustration of the primer design to flank (OFF) and overlap (ON) the PAM. (B) qRT-PCR showing relative fold amplification between uninjected (control) and sgRNA- and Cas9 RNA-injected zebrafish embryos. (C) T7 endonuclease assay of control and Dab2 CRISPR/Cas9-injected zebrafish embryos. Independent biological replicates are shown; n = 3. The single ∼200-bp bands indicate the WT sequence, and two bands denote mutated sequences at the PAM site. Related to Fig. 4.
To introduce mutations, we injected one-cell stage zebrafish with single guide RNA (sgRNA) and Cas9 RNA. The presence of insertions or deletions (indels) was validated by reverse genetics. In brief, quantitative PCR was conducted on genomic DNA harvested from uninjected and Dab2-injected zebrafish embryos at 24 h postfertilization (hpf) using the primers shown in Fig. S1A and described in Materials and Methods (16). These data indicate that following injection of sgRNA and Cas9, ∼70% of the total zebrafish alleles surveyed showed indels proximal to the protospacer adjacent motif (PAM) (Fig. S1B), which we term Dab2 mutants. As an ancillary method, a T7 endonuclease assay confirmed correct targeting (Fig. S1C).
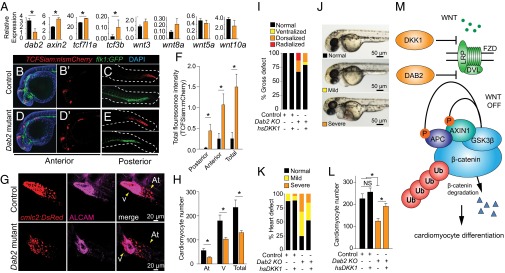
DAB2 is a known inhibitor of the WNT/β-catenin signaling pathway (17, 18) and has been shown to regulate endothelial development in zebrafish (15). Thus, we sought to understand whether our Dab2 mutants showed increased levels of WNT/β-catenin signaling coupled with delayed development of the endothelium. To test this, we performed qRT-PCR analysis on a panel of WNT-responsive genes in control and Dab2 mutants at 24 hpf (Fig. 4A). Dab2 transcript abundance was significantly reduced by Dab2 mutagenesis, whereas expression of WNT/β-catenin target genes, such as axin-related protein 2 (axin2), transcription factor 7-like 1a (tcf7I1a), and transcription factor 3b (tcf3b), was significantly up-regulated. As a secondary method, we crossed a WNT/β-catenin reporter (Siam:mCherry) with an endothelial reporter (flk1:GFP) and assayed control and Dab2 mutants at 24 hpf. Loss of Dab2 significantly increased WNT/β-catenin reporter expression primarily in regions of the forebrain, midbrain, hindbrain, anterior pronephros, and posterior vasculature (Fig. 4 B–E, quantified in F). Consistent with previous reports (15), Dab2 mutants also appeared delayed in the development of posterior vasculature (Fig. 4 C and E). These data correspond with previous findings indicating that Dab2 regulates WNT/β-catenin signaling and endothelial development.
Fig. 4.
Dab2 promotes cardiomyocyte development by negatively regulating WNT/β-catenin signaling. (A) qRT-PCR of dab2 and a panel of known WNT/β-catenin target genes (axin2, tcf7I1a, and tcf3b) and ligands (wnt3, wnt8a, wnt5a, and wnt10a) in control (black bars) and Dab2 mutants (orange bars) at 24 hpf. (B–E) Representative brightest point projection confocal image (Z series) of control (B and C) and Dab2 mutants (D and E) at 24 hpf. TCFSiam:nlsmCherry is shown in red, flk1:GFP in green, and DAPI in blue. Panels shown are anterior merged (B and D), anterior red (B′ and D′), and posterior red and green (C and E). (F) Quantification of total fluorescence intensity between control and Dab2 mutants at 24 hpf. n ≥3 biological replicates. *P ≤ 0.05, Student’s t test. Data are mean ± SEM. (G) Representative brightest point projection confocal (Z series) images of control (Top) and Dab2 mutants (Bottom) at 48 hpf. cmlc2:DsRed-nuc is in red (Left), and ALCAM is in magenta (Middle) (H) Quantification of atrial (At) and ventricular (V) cardiomyocytes. (I) Percent gross morphological defect scoring in control and Dab2 mutants after overexpression of DKK1 scored at 48 hpf. (J) Representative images of normal, mild, and severe heart defects at 48 hpf. (K) Percent heart defects in control and Dab2 mutants after overexpression of DKK1 scored at 48 hpf. (L) Quantification of total cardiomyocyte number. *P ≤ 0.05, Student’s t test. n = 30–45 per group for heat shock and scoring for panels I and K. For quantification of cardiomyocyte nuclei, n = 4–6 fish hearts per condition analyzed by z-series confocal microscopy. Data are mean ± SEM. (M) Model: DAB2 inhibition of WNT/β-catenin signaling promotes cardiomyocyte differentiation.
Dab2 Promotes Cardiomyocyte Differentiation in Part by Negatively Regulating WNT/β-Catenin Signaling.
To assess whether Dab2 regulates cardiomyocyte differentiation, we assessed control and Dab2 mutants carrying a transgene (cmlc2:DsRed-nuc) for cardiomyocyte nuclei at 48 hpf. In the absence of Dab2, cardiomyocyte numbers were significantly reduced in both ventricles and atria compared with controls (Fig. 4 G and H). Thus, Dab2 is a mediator of in vivo cardiomyocyte development. Because Dab2 negatively regulates WNT/β-catenin signaling and cardiomyocyte differentiation, we hypothesized that during late-stage cardiomyocyte differentiation, overexpression of an upstream inhibitor of the WNT/β-catenin signaling pathway, dickkopf 1 (DKK1), would rescue our phenotype. To this end, heterozygous (hsDKK1:GFP+/−;cmlc2:DsRed-nuc+/−) control and Dab2 mutants were heat-shocked to induce transgene expression at the 16–18 somite stage and assessed at 48 hpf for gross morphology and cardiac abnormalities. Gross morphology was scored using a previously described phenotype scale (19), and cardiac abnormalities (normal, mild, or severe) were scored as shown in Fig. 4J. In the heat-shock controls, there was no indication of broad organismal or cardiac defects between DKK1+ and DKK1− treatment groups (Fig. 4 I–K). In contrast, compared with controls, Dab2 mutagenesis resulted in broad organismal and cardiac defects, whereas this phenotype was partially rescued by overexpression of DKK1 (Fig. 4 I–K).
To determine whether overexpression of DKK1 could restore Dab2-mediated cardiomyocyte deficiencies, we assessed heat-shocked hsDKK1:GFP+/−;cmlc2:DsRed-nuc+/− control and Dab2 mutants as before and quantified total cardiomyocytes at 48 hpf. The controls showed no change in cardiomyocyte number associated with overexpression of DKK1 (control DKK−, 226.2 ± 22.0; control DKK1+, 256.0 ± 18.4; P = 0.345), whereas Dab2 mutants consistently showed a significant decrease in total cardiomyocyte number compared with controls. In contrast, following overexpression of DKK1, Dab2 mutants had significantly higher cardiomyocyte numbers compared with DKK1− Dab2 mutants (Dab2 DKK−, 124.5 ± 12.5; Dab2 DKK+, 191.3 ± 13.5; P ≤ 0.01) (Fig. 4L). These data indicate that Dab2 plays a critical role during heart development, in part by negatively regulating the WNT/β-catenin signaling pathway, and that Dab2 deficiency can be rescued by overexpression of a distinct inhibitor of the WNT/β-catenin pathway (Fig. 4M).
Discussion
The combination of LFQ proteomics and cross-species validation can be used to reveal previously unidentified regulators of cardiac development that are relevant both in vitro and in vivo. WNT/β-catenin inhibitors have long been studied for their roles in modulating cell fate decisions during development and also as possible therapeutic options for the treatment of cancer (6, 8, 20). In this study, we have identified DAB2 as a critical WNT/β-catenin signaling inhibitor that is also essential for cardiomyocyte development. Although we have focused on the expression and function of DAB2 in the context of WNT/β-catenin signaling, this dataset also could be useful in determining other processes during heart development, such as metabolic transitions, cell–cell communications, and other signaling pathways.
In the presence of WNTs, the Frizzled (FZD) receptor and low-density lipoprotein receptor-related protein (LRP5/6) form a complex. WNT-mediated activation of FZD-LRP destabilizes a destruction complex, resulting in an accumulation of β-catenin and increased WNT signaling. By blocking the recruitment of β-catenin to the FZD-LRP complex, DAB2 promotes stabilization of the destruction complex and thereby inhibits WNT signaling (17). We found that the Dab2 mutation increases WNT/β-catenin signaling and diminishes cardiac development in vivo. By overexpressing an upstream WNT/β-catenin inhibitor (DKK1) in our Dab2 mutants, cardiomyocyte numbers and cardiac defects were partially rescued.
Here we link Dab2 to WNT/β-catenin signaling as a critical aspect of cardiac development; however, we note that the WNT/β-catenin signaling pathway also has known roles in controlling cellular proliferation and the survival of CPCs. Thus, our findings also may be attributed to the prosurvival or proliferative aspects of the WNT/β-catenin signaling pathway, and not solely from the control of mesoderm or CPC specification. Our findings highlight a previously unidentified role for DAB2’s regulation of cardiac development through modulation of the WNT/β-catenin signaling pathway.
DAB2 is also involved in TGF-β signaling (15). Cross-talk between TGF-β and WNT/β-catenin signaling is a critical component during development (5, 21–23). During hESC-directed differentiation, activation of both TGF-β and canonical WNT signaling pathways promote direct differentiation of hESCs toward precardiac mesoderm (2, 8), whereas subsequent WNT inhibition specifies CPCs. In zebrafish, Dab2 regulates TGF-β signaling by increasing the endocytosis of BMP2 to regulate angiogenesis (15). Furthermore, Morris et al. (24) demonstrated that a murine homozygous null mutation of Dab2 is embryo-lethal with a phenotype that closely resembles those of TGF-β mutations. Thus, the role of DAB2 in the whole organism is broad. Given that the heart is the first vertebrate organ to form and function, and requires tight regulation of WNT/β-catenin signaling (6, 25), knowledge of which proteins promote or repress WNT/β-catenin signaling is crucial for identifying regulators of cardiac development. Our work demonstrates that DAB2 is dynamically expressed in human stem cells, and supports cardiomyocyte development in vivo in part by suppressing WNT/β-catenin signaling.
Materials and Methods
Cell Culture and Cardiac-Directed Differentiation.
Undifferentiated RUES2 hESCs (female; Rockefeller University, NIHhESC-09-0013) were maintained as described previously (1). The hESCs were seeded on Matrigel (BD Biosciences)-coated plates and maintained with mouse embryonic fibroblast-conditioned medium containing 5 ng/mL human basic fibroblast growth factor (Peprotech; 100–18B) until appropriate confluency was observed. Directed differentiation of hESCs toward cardiomyocytes was performed in high-density monolayers, using a combination of activin A, BMP4, and small molecule activation and repression of WNT/β-catenin signaling, as described previously (1) and illustrated in Fig. 1A.
qRT-PCR.
Total RNA isolation was isolated using the Qiagen RNeasy Miniprep Kit, in accordance with the manufacturer’s protocol. First-strand cDNA from 500 ng of total RNA was synthesized using the SuperScript III Enzyme Kit (Invitrogen). qRT-PCR was performed using the Sensimix SYBR PCR Kit (Bioline) on a 7900HT Fast Real-Time PCR system (Applied Biosystems). Primers are listed in Table S1. For human in vitro studies, all transcripts were normalized to HPRT, and in vivo zebrafish studies were normalized to β-actin.
Table S1.
Primer sequences 5′ to 3′ for qRT-PCR and CRISPR/Cas9 mutagenesis
| Gene name | Forward primer | Reverse primer |
| Zebrafish qRT-PCR primers | ||
| β-actin | AAGCAGGAGTACGATGAGTC | TGGAGTCCTCAGATGCATTG |
| tcf7l1a | TGGCCTCTTCTTGACGTTCC | GGGTCCTTGGTATGCCAGTC |
| tcf3b | AACTCTTGACTCCCCTGGGC | CAAACATCGCACTGAAATCCAAC |
| wnt3 | GACCAGTGCTTGATAAAGCTAC | GTGCATGTTCTCCAATATGGTC |
| wnt8a | GTCACTCACGCAGCAATGAACC | GTTTTCGAAGTCCCCCATGCTG |
| wnt5a | GACATCAGTTAGGAGAGTCTTG | TAGAGCTGGCAGAGCTTCTTCT |
| wnt10a | ATTCACTCCAGGATGAGACTTCATA | GTTTCTGTTGTGGGCTTTGATTAG |
| dab2 | GCAAGCACAAGCAAAGAATATG | GCAAGCACAAGCAAAGAATATG |
| Human qRT-PCR primers | ||
| HPRT | TGACACTGGCAAAACAATGCA | GGTCCTTTTCACCAGCAAGCT |
| BryT | CAAATCCTCATCCTCAGTTTG | GTCAGAATAGGTTGGAGAATTG |
| SOX2 | GCCGAGTGGAAACTTTTGTCG | GGCAGCGTGTACTTATCCTTCT |
| GATA4 | ACACCCCAATCTCGATATGTTTG | GTTGCACAGATAGTGACCCGT |
| MYH6 | CAAGTTGGAAGACGAGTGCT | ATGGGCCTCTTGTAGAGCTT |
| DAB2 | GTAGAAACAAGTGCAACCAATGG | GCCTTTGAACCTTGCTAAGAGA |
| Primers for Dab2 CRISPR/Cas9 | ||
| sgRNA | TAGGTGATTGAGCATGAACATG | AAACCATGTTCATGCTCAATCA |
| dab2 “off” | GCAAGCACAAGCAAAGAATATG | GGTTATCAGTGACATCACGAGC |
| dab2 “on” | GCAAGCACAAGCAAAGAATATG | CACCACATGTTCATGCTCAATCAC |
Flow Cytometry.
Cardiomyocyte purity was obtained by flow cytometry using cTnT (Thermo Scientific; 1:100) antibody or isotype control. Cells were analyzed with a FACSCanto II flow cytometer using FACSDiva software (BD Biosciences). Raw FACS data were analyzed using FlowJo software (Tree Star).
Proteomics.
Cells were analyzed as described previously (10). In brief, cell pellets were lysed in 1 M urea and 50 mM ammonium bicarbonate (pH 7.8) and heated to 50 °C for 20 min. Normalized quantities of protein were reduced, alkylated, and digested overnight with trypsin. Three biological replicates per time point were run in technical triplicate. Peptides were measured by nano LC-MS/MS on a Thermo Scientific Orbitrap Fusion mass spectrometer. Peptides were separated in a 180-min gradient [1–45% (vol/vol) acetonitrile] at 250 nL/min. The Orbitrap was operated in data-dependent mode with a 60,000 resolution, 400–1,600 m/z full scan, top speed of 3 s, and 1.8-m/z isolation window. Identification and LFQ of peptides was done with MaxQuant 1.5 (26) using a 1% false discovery rate (FDR) against the UniProt human proteome dataset. To identify proteins that were significantly different between conditions, Perseus 1.4.1.3 was used to perform permutation-based t tests (FDR, 5%) and hierarchical clustering analysis. PCA was conducted in MetaboAnalyst 3.0, and the data were filtered using mean intensity values and Pareto-scaled (27). GO terms linked to proteins enriched in hESCs, CPCs, or cardiomyocytes were analyzed in g:Profiler using the CoCoA algorithm (28). Mean protein abundance was quantified by LFQ, and samples were normalized to hESCs to compare protein expression patterns during the time course of cardiomyocyte differentiation.
Zebrafish Strains and Husbandry.
WT (AB; Zebrafish International Resource Center), cmlc2:DsRed [Tg(cmlc2:DsRed-nuc] (29), Siam:mCherry [Tg(7xTCF-Xla.Siam:nlsmCherry)] (30), Tg(hsDKK1:GFP), and Tg(flk1:GFP) zebrafish strains were used in these experiments. All fish were maintained using standard procedures in accordance with the Institutional Animal Care and Use Committee-approved protocols. Heat shocking of heterozygous hsDKK1:GFP fish was conducted as described previously (31). Blinded scoring of DKK1 overexpression was done as described in Results with genetic DKK1− internal heat-shocked controls. Zebrafish biological replicates are considered to represent individual spawning pairs and experiments conducted on separate days unless stated otherwise.
Analysis of Zebrafish WNT/β-Catenin Reporter Expression.
Homozygous Siam:mCherry and flk1:GFP lines were crossed to obtain a heterozygous line (7xTCF-Xla.Siam:nlsmCherry+/−;flk1:GFP+/−). Fish were injected with either control (dab2 sgRNA) or Dab2 (dab2 sgRNA/Cas9) at the one-cell stage as described previously. At 24 hpf, fish were fixed in 4% (vol/vol) paraformaldehyde (PFA) and mounted in Vectashield containing DAPI (Vector Laboratories) for confocal microscopy. Whole-mount anterior and posterior images (z-series) were obtained, and total fluorescence intensity was quantified using ImageJ software. Confocal images were obtained using a Nikon A1R confocal mounted on a Nikon TiE inverted microscope.
Cardiomyocyte Nuclei Quantification.
Heterozygous cmlc2:DsRed-nuc and hsDKK1:GFP+/−;cmlc2:DsRed-nuc+/− control (dab2 sgRNA) and Dab2 (dab2 sgRNA/Cas9)-injected fish were collected at 48 hpf and fixed in 4% (vol/vol) PFA overnight, then subjected to immunhistochemistry as described previously (32). Rabbit anti-DsRed (1:200; AnaSpec) and mouse anti-activated leukocyte cell adhesion molecule (ALCAM; 1:50) primary antibodies were used, followed by staining with Alexa Fluor 596 anti-rabbit and Alexa Fluor 633 anti-mouse secondary antibodies at 1:100. Blocking and dilutions were done in PBS with 0.03% Triton and 4% (wt/vol) BSA. The ALCAM antibody was used to decipher the zebrafish heart ventricle from atrium owing to preferential staining of the ventricle. Hearts were imaged (z-series) by confocal microscopy, and cardiomyocytes were counted in a blinded fashion.
Statistics.
Single variable analyses between two samples were performed using Student’s t test. Univariate and multivariable assays were analyzed by one- or two-way ANOVA.
CRISPR-Cas9 genome editing in zebrafish embryos is described in SI Methods.
SI Materials and Methods
The single guide RNA (sgRNA) was designed using the online sgRNA generator CHOPCHOP (33). Primers were annealed and ligated into a BsaI-digested pDR274 vector containing a T7 promoter upstream of the partial guide sequence (34). Following sequencing (Operon) to confirm sgRNA insertion, RNA was synthesized by in vitro transcription (MEGAscript T7 Transcription Kit, AM1334; Life Technologies). Cas9 (pT3TS-nCas9n; Addgene) RNA was synthesized as described previously (35). In brief, Xbal-digested pTSTS-nCas9n plasmid was in vitro-transcribed using the MEGAscript T3 Transcription Kit (AM1338; Life Technologies). sgRNA and cas9 RNA were cleaned using the MEGAclear Transcription Clean-Up Kit (AM1908; Life Technologies), and band size was confirmed by gel electrophoresis and quantified with a Nano Drop 1000 spectrophotometer (Thermo Scientific). RNA (sgRNA, 50 ng/µL; Cas9 RNA, 300 ng/µL; ∼2 nL) was injected into zebrafish embryos at the one-cell stage as described previously (35).
Validation of targeting was conducted by isolating genomic DNA from uninjected and Dab2-injected zebrafish at 24 hpf, as described previously (16) and shown in Fig. 4. In brief, qRT-PCR was performed comparing uninjected and Dab2 genomic DNA with “OFF” primers flanking the protospacer adjacent motif (PAM) site and “ON” primers, of which the 3′ end of the reverse primer overlapped the PAM site. Primer efficiency was calculated using the following equation: (mutant primer ON/mutant primer OFF)/(WT primer ON/WT primer OFF) (16). Further validation of the presence of mosaic mutagenesis was conducted by subjecting “OFF” PCR products to a T7 endonuclease assay, followed by sequencing (Operon). Sequencing indicated that the Dab2 mutants analyzed in this study had both insertions and deletions downstream of the PAM site. Primer sequences are presented in Table S1.
Supplementary Material
Acknowledgments
We thank Stanley Kim for zebrafish husbandry and Kaytlyn Gerbin for assistance with principal component analysis. P.H. is funded through the National Institutes of Health’s Experimental Pathology of Cardiovascular Disease Training Grant T32 HL007312. This work was supported in part by the University of Washington's Proteomics Resource (Grant UWPR95794); National Institutes of Health Grants U01 HL100405 (to C.E.M. and R.T.M.), P01 GM081619 (to C.E.M. and R.T.M.), R01 HL084642 (to C.E.M.), and P01 HL094374 (to C.E.M.); and an award from the Fondation Leducq Transatlantic Network of Excellence (to C.E.M.). R.T.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: Proteins identified by mass spectrometry in the datasets have been deposited in the UniProt database (www.uniprot.org).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523930113/-/DCSupplemental.
References
- 1.Palpant NJ, Hofsteen P, Pabon L, Reinecke H, Murry CE. Cardiac development in zebrafish and human embryonic stem cells is inhibited by exposure to tobacco cigarettes and e-cigarettes. PLoS One. 2015;10(5):e0126259. doi: 10.1371/journal.pone.0126259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paige SL, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151(1):221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palpant NJ, et al. Inhibition of β-catenin signaling respecifies anterior-like endothelium into beating human cardiomyocytes. Development. 2015;142(18):3198–3209. doi: 10.1242/dev.117010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gessert S, Kühl M. The multiple phases and faces of Wnt signaling during cardiac differentiation and development. Circ Res. 2010;107(2):186–199. doi: 10.1161/CIRCRESAHA.110.221531. [DOI] [PubMed] [Google Scholar]
- 5.Paige SL, et al. Endogenous Wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One. 2010;5(6):e11134. doi: 10.1371/journal.pone.0011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ueno S, et al. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci USA. 2007;104(23):9685–9690. doi: 10.1073/pnas.0702859104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naito AT, et al. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci USA. 2006;103(52):19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palpant NJ, et al. Transmembrane protein 88: A Wnt regulatory protein that specifies cardiomyocyte development. Development. 2013;140(18):3799–3808. doi: 10.1242/dev.094789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luber CA, et al. Quantitative proteomics reveals subset-specific viral recognition in dendritic cells. Immunity. 2010;32(2):279–289. doi: 10.1016/j.immuni.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Clarke RL, Robitaille AM, Moon RT, Keller G. A quantitative proteomic analysis of hemogenic endothelium reveals differential regulation of hematopoiesis by SOX17. Stem Cell Rep. 2015;5(2):291–304. doi: 10.1016/j.stemcr.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howe K, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stainier DY, Fishman MC. The zebrafish as a model system to study cardiovascular development. Trends Cardiovasc Med. 1994;4(5):207–212. doi: 10.1016/1050-1738(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 13.Thisse C, Thisse B. 2005. High-throughput expression analysis of ZFModels Consortium clones. Available at zfin.org/ZDB-PUB-051025-1. Accessed September 7, 2015.
- 14.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3(1):59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 15.Kim JD, et al. Context-dependent proangiogenic function of bone morphogenetic protein signaling is mediated by disabled homolog 2. Dev Cell. 2012;23(2):441–448. doi: 10.1016/j.devcel.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shah AN, Davey CF, Whitebirch AC, Miller AC, Moens CB. Rapid reverse genetic screening using CRISPR in zebrafish. Nat Methods. 2015;12(6):535–540. doi: 10.1038/nmeth.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, He X, Howe PH. Disabled-2 (Dab2) inhibits Wnt/β-catenin signalling by binding LRP6 and promoting its internalization through clathrin. EMBO J. 2012;31(10):2336–2349. doi: 10.1038/emboj.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y, Luo W, Howe PH. Dab2 stabilizes Axin and attenuates Wnt/beta-catenin signaling by preventing protein phosphatase 1 (PP1)–Axin interactions. Oncogene. 2009;28(33):2999–3007. doi: 10.1038/onc.2009.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James RG, et al. Bruton’s tyrosine kinase revealed as a negative regulator of Wnt/beta-catenin signaling. Sci Signal. 2009;2(72):ra25. doi: 10.1126/scisignal.2000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13(1):11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 21.Akhmetshina A, et al. Activation of canonical Wnt signalling is required for TGF-β mediated fibrosis. Nat Commun. 2012;3:735. doi: 10.1038/ncomms1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishita M, et al. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann’s organizer. Nature. 2000;403(6771):781–785. doi: 10.1038/35001602. [DOI] [PubMed] [Google Scholar]
- 23.Gadue P, Huber TL, Paddison PJ, Keller GM. Wnt and TGF-beta signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci USA. 2006;103(45):16806–16811. doi: 10.1073/pnas.0603916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris SM, Tallquist MD, Rock CO, Cooper JA. Dual roles for the Dab2 adaptor protein in embryonic development and kidney transport. EMBO J. 2002;21(7):1555–1564. doi: 10.1093/emboj/21.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dohn TE, Waxman JS. Distinct phases of Wnt/β-catenin signaling direct cardiomyocyte formation in zebrafish. Dev Biol. 2012;361(2):364–376. doi: 10.1016/j.ydbio.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies, and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 27.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: A Web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37(Web Server issue):W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler: A Web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35(Web Server issue):W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mably JD, Mohideen MA, Burns CG, Chen JN, Fishman MC. Heart of glass regulates the concentric growth of the heart in zebrafish. Curr Biol. 2003;13(24):2138–2147. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 30.Moro E, et al. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev Biol. 2012;366(2):327–340. doi: 10.1016/j.ydbio.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 31.Novikov N, Evans T. Tmem88a mediates GATA-dependent specification of cardiomyocyte progenitors by restricting WNT signaling. Development. 2013;140(18):3787–3798. doi: 10.1242/dev.093567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plavicki J, Hofsteen P, Peterson RE, Heideman W. Dioxin inhibits zebrafish epicardium and proepicardium development. Toxicol Sci. 2013;131(2):558–567. doi: 10.1093/toxsci/kfs301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: A CRISPR/Cas9 and TALEN Web tool for genome editing. Nucleic Acids Res. 2014;42(Web Server issue):W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 2013;110(34):13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.