Significance
The mitotic checkpoint system has important roles to ensure accurate segregation of chromosomes in mitosis. This system regulates the activity of the ubiquitin ligase Anaphase-Promoting Complex/Cyclosome (APC/C) by the formation of inhibitors including the Mitotic Checkpoint Complex (MCC). The mode of the assembly of MCC is not sufficiently understood, and it is also not known whether checkpoint complexes different from MCC also inhibit the APC/C. We find that complexes lacking Mad2, a protein component of MCC, have greatly reduced APC/C inhibitory action. On the other hand, a previously unknown species of MCC that contains an additional molecule of Cdc20 is a strong inhibitor of APC/C. These results reveal important molecular mechanisms in the action of the mitotic checkpoint system.
Keywords: cell cycle, ubiquitin, protein degradation
Abstract
The mitotic (or spindle assembly) checkpoint system prevents premature separation of sister chromatids in mitosis and thus ensures the fidelity of chromosome segregation. Kinetochores that are not attached properly to the mitotic spindle produce an inhibitory signal that prevents progression into anaphase. The checkpoint system acts on the Anaphase-Promoting Complex/Cyclosome (APC/C) ubiquitin ligase, which targets for degradation inhibitors of anaphase initiation. APC/C is inhibited by the Mitotic Checkpoint Complex (MCC), which assembles when the checkpoint is activated. MCC is composed of the checkpoint proteins BubR1, Bub3, and Mad2, associated with the APC/C coactivator Cdc20. The intermediary processes in the assembly of MCC are not sufficiently understood. It is also not clear whether or not some subcomplexes of MCC inhibit the APC/C and whether Mad2 is required only for MCC assembly and not for its action on the APC/C. We used purified subcomplexes of mitotic checkpoint proteins to examine these problems. Our results do not support a model in which Mad2 catalytically generates a Mad2-free APC/C inhibitor. We also found that the release of Mad2 from MCC caused a marked (although not complete) decrease in inhibitory action, suggesting a role of Mad2 in MCC for APC/C inhibition. A previously unknown species of MCC, which consists of Mad2, BubR1, and two molecules of Cdc20, contributes to the inhibition of APC/C by the mitotic checkpoint system.
The mitotic (or spindle assembly) checkpoint system is a surveillance mechanism that monitors correct attachment of chromosomes to the mitotic spindle and thus ensures the fidelity of chromosome segregation (1–4). Kinetochores that are not attached, or are incorrectly attached to the spindle, generate an inhibitory signal that prevents progression into anaphase by inhibiting the Anaphase-Promoting Complex/Cyclosome (APC/C) ubiquitin ligase. APC/C ubiquitylates securin and cyclin, the degradation of which is necessary for chromosome segregation and for exit from mitosis. When the checkpoint is activated, a Mitotic Checkpoint Complex (MCC), which inhibits the APC/C, is assembled. MCC is composed of the checkpoint proteins BubR1, Bub3, and Mad2, associated with the APC/C coactivator Cdc20 (5). When the checkpoint is extinguished, MCC is disassembled (6–8). Disassembly of MCC is initiated by the release of Mad2, a process carried out by the joint action of the TRIP13 AAA-ATPase and the Mad2-binding protein p31comet (9–12).
The intermediary processes in the assembly of MCC and the possible role of different subcomplexes of MCC in the inhibition of APC/C are not sufficiently understood. When the checkpoint is activated, an early event that takes place on the kinetochore is the conversion of Mad2 from an open (O-Mad2) to a closed (C-Mad2) conformation that forms a tight complex with Cdc20 (1, 3). It has been proposed that the C-Mad2-Cdc20 (MC) subcomplex associates with BubR1-Bub3 to form the MCC (2, 4), but this has not yet been tested by direct experimentation. The roles of Cdc20-BubR1 intermediary subcomplexes are more obscure. [Bub3 is constitutively associated with BubR1, but has no appreciable influence on MCC function or structure (13, 14). Therefore, Bub3 was omitted from the subcomplexes of BubR1 and the mitotic checkpoint complexes described in this paper.] Cdc20 may bind to two different sites on BubR1. When the mitotic checkpoint is active, Cdc20 associates with a binding site at the N-terminal region of BubR1 in a process that requires Mad2 and that leads to MCC assembly (13, 15). Cdc20 also binds tightly to a second site at an internal region of BubR1 (15, 16, 17). The latter process does not require Mad2 and is not dependent on the mitotic checkpoint. The binding of Cdc20 to the internal site has been reported to inhibit the APC/C in vitro by the sequestration of Cdc20 (16, 17), but the role of this process in the regulation of APC/C with bound Cdc20 (APC/CCdc20) is not clear. It is also not clear whether or not Mad2-containing MCC is the major checkpoint inhibitor of APC/C. Nilsson et al. (18) reported that, in checkpoint complexes from HeLa cell extracts, only a small fraction of Cdc20 is associated with Mad2 and suggested that the role of Mad2 may be limited to the loading of Cdc20 onto BubR1. Cleveland and coworkers (19) furthermore proposed that Mad2-free Cdc20-BubR1 is the main checkpoint inhibitor of APC/C. These authors also suggested that C-Mad2 catalytically amplifies the production of the Cdc20-BubR1 inhibitor. According to this hypothesis, following the loading of Cdc20 onto BubR1, Mad2 dissociates and then binds another molecule of Cdc20 to produce the Cdc20-BubR1 inhibitor (19).
In the present study, the roles of subcomplexes of mitotic checkpoint proteins in MCC assembly and in the regulation of APC/C activity were investigated by the use of purified subcomplexes. Our results do not support the catalytic model of C-Mad2 action. We furthermore find that the release of Mad2 from MCC decreases markedly, but not completely, its APC/C inhibitory action, suggesting that that the presence of Mad2 in MCC is important for the inhibition of APC/CCdc20. A previously unknown species of MCC, which consists of Mad2 and two molecules of Cdc20 bound to the two binding sites of BubR1, contributes to the inhibition of APC/CCdc20.
Results
Isolation of Recombinant Subcomplexes of MCC.
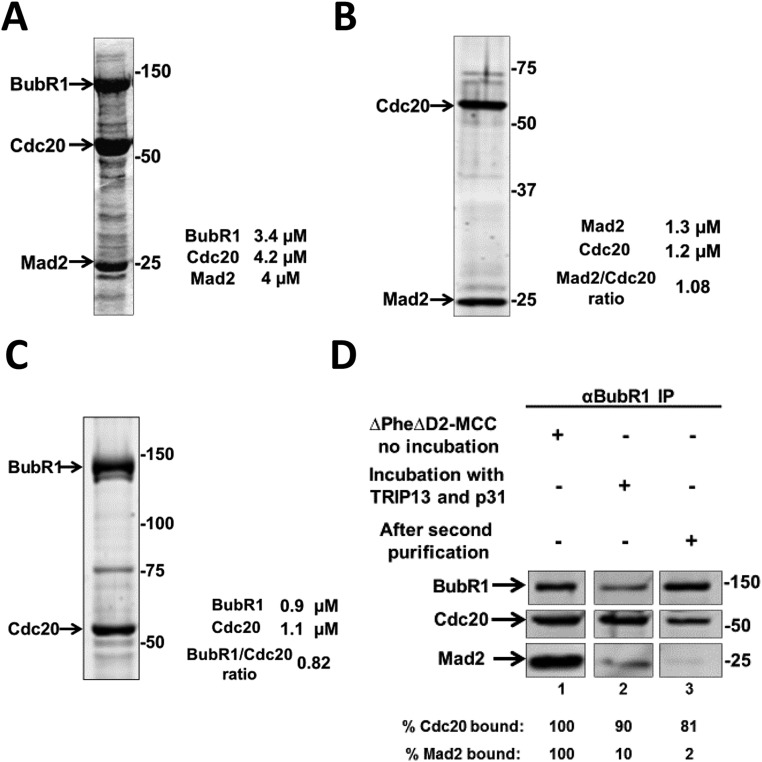
To study the role of different subcomplexes of mitotic checkpoint proteins in MCC assembly and in APC/C inhibition, the subcomplexes were prepared from recombinant checkpoint proteins. The subcomplex of Mad2 with Cdc20 (MC) was formed by coexpression in insect cells of Flag-Mad2 and his6-Cdc20, followed by two steps of affinity purification (SI Materials and Methods). This preparation of the Mad2-Cdc20 subcomplex had close to equimolar proportions of Mad2 and Cdc20 (Fig. S1B).
Fig. S1.
Isolation of recombinant MCC and its subcomplexes. (A) Formation of recombinant MCC in baculovirus-infected insect cells. Recombinant MCC was generated and purified as described in SI Materials and Methods. The preparation was subjected to SDS/PAGE and Coomassie Brilliant Blue staining, followed by quantification at 680 nm with LiCOR Odyssey scanner. Numbers on the right indicate the migration position of marker proteins (kDa). (B) Isolation of the recombinant Mad2-Cdc20 subcomplex. The recombinant Mad2-Cdc20 subcomplex was expressed and purified as described in SI Materials and Methods. The recombinant subcomplex was subjected to SDS/PAGE and quantified as described in SI Materials and Methods. Numbers on the right indicate the electrophoretic migration position of marker proteins (kDa). (C) Isolation of the recombinant subcomplex of Cdc20 bound to site 2 of BubR1 (BC-2). The recombinant Strep-WT-BubR1-Flag-Cdc20 subcomplex was expressed and purified as described in SI Materials and Methods. The recombinant subcomplex was subjected to SDS/PAGE, Krypton protein staining, and quantification as described in SI Materials and Methods. Numbers on the right side indicate the migration position of the marker proteins (kDa). (D) Formation of recombinant subcomplex of Cdc20 bound to site 1 of BubR1 (BC-1). ∆PheΔD2-MCC was incubated with recombinant TRIP13 and p31comet as described in SI Materials and Methods. Subsequently, the mixture was further purified on Strep-Tactin resin for removal of unbound proteins and reagents. Samples of MCC before incubation (lane 1), after incubation with TRIP13 and p31comet (lane 2), and the preparation of BC-1 after the second purification on Strep-Tactin (lane 3) were subjected to immunoprecipitation with anti-BubR1 and were immunoblotted for the indicated proteins. Results were expressed as the percentage of Cdc20 and of Mad2 bound to BubR1 relative to their maximal binding in MCC.
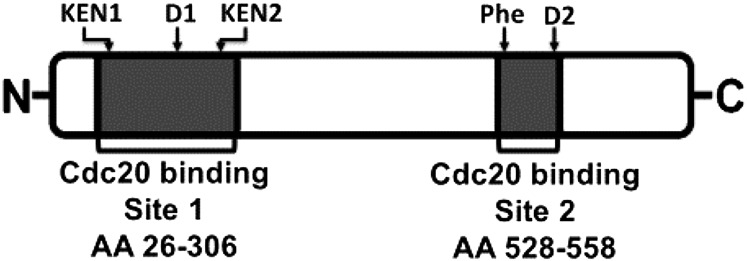
Based on previous information, two different types of BubR1-Cdc20 subcomplexes in which Cdc20 binds to different sites of BubR1 could be expected. BubR1 has a Cdc20-binding site in its N-terminal region (Fig. 1, “site 1”), which is involved in its Mad2-dependent assembly into MCC (13, 14, 15). In the absence of Mad2, Cdc20 binds strongly to a second site on BubR1, which is in its middle region (Fig. 1, “site 2”) (15, 16, 17). A subcomplex of Cdc20 bound to site 2 of BubR1 (“BC-2”) was formed by coexpression in insect cells of BubR1 and Cdc20 without Mad2. Following purification, the preparation of BC-2 had close to equimolar amounts of BubR1 and Cdc20 (Fig. S1C).
Fig. 1.
Schematic drawing of Cdc20-binding sites in human BubR1. Cdc20-binding site 1 is at the N-terminal region of BubR1- and Cdc20-binding site 2 is in the middle region of BubR1. The conserved functional motifs KEN1, KEN2, D1 box, D2 box, and Phe box are indicated on the top. The Cdc20-binding sites and their amino acid (AA) positions are shown below.
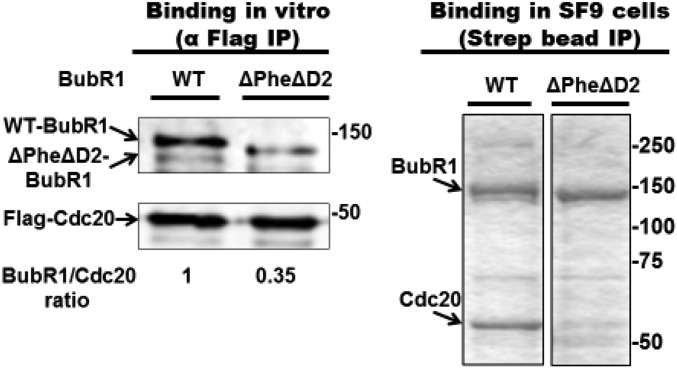
We used recombinant MCC (SI Materials and Methods and Fig. S1A) to produce a recombinant subcomplex in which Cdc20 is bound to site 1 in BubR1 (“BC-1”). We reasoned that because Cdc20 in MCC is bound to site 1 (14), following the release of Mad2 from MCC, residual Cdc20 attached to BubR1 remains bound to site 1. To minimize possible rebinding of Cdc20 to site 2 in BubR1, we took advantage of the information that binding of Cdc20 to site 2 requires an invariant Phe residue (“Phe box”) and a destruction box (“D2 box”) (17) (Fig. 1). Indeed, we have confirmed the observation (17) that the binding of Cdc20 to site 2 was markedly reduced in a variant of BubR1 that contained mutations in the Phe- and D2 boxes, both in vitro (Fig. S2, Left) and in proteins coexpressed in insect cells (Fig. S2, Right). To produce BC-1, we have therefore assembled recombinant MCC containing Δ-Phe, Δ-D2 box mutant of BubR1 and then subjected it to the joint action of TRIP13 and p31comet to liberate Mad2 (9). As expected, the amount of Mad2 bound to BubR1 was markedly reduced following this treatment (Fig. S1D, lane 2 vs. lane 1). Subsequently, BC-1 was further purified (SI Materials and Methods) to remove reagents used for its formation (Fig. S1D, lane 3).
Fig. S2.
Reduced binding of Cdc20 to site 2 of ΔPheΔD2 mutant BubR1. (Left) A total of 500 nM of either WT- or ΔPhe ΔD2-BubR1 was incubated with 300 nM of Flag-Cdc20 for 2 h at 23 °C, followed by immunoprecipitation on anti-Flag beads for 1 h at 4 °C. Subsequently, the beads were washed and subjected to immunoblotting for the indicated proteins. (Right) Insect cells were coinfected with baculoviruses expressing Flag-Cdc20 and either WT- or ΔPhe ΔD2-Strep-BubR1. Complexes of BubR1 were subjected to purification on Strep-Tactin resin as described in SI Materials and Methods, followed by SDS/PAGE and Coomassie staining. Equal amounts of BubR1 were loaded. Numbers on the right indicate the position of the marker proteins (kDa).
Mad2-Cdc20 Subcomplex Is a Preferred Precursor for MCC Formation.
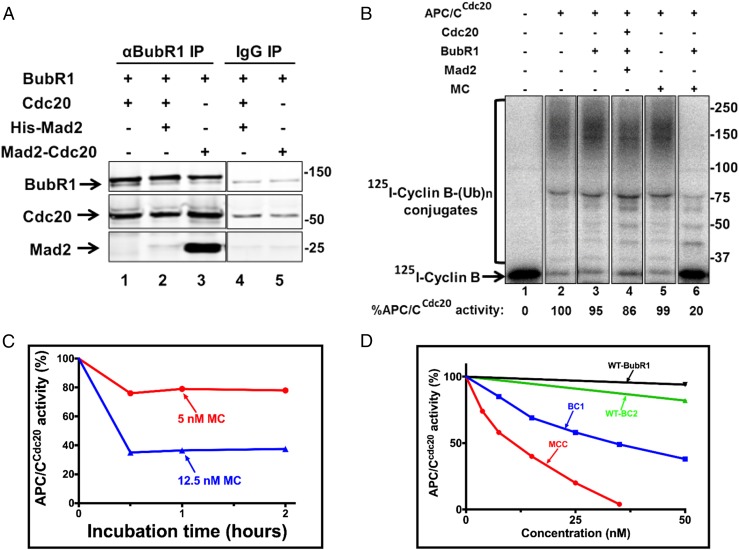
We first used our preparation of recombinant purified Mad2-Cdc20 to examine its putative role in MCC formation. A considerable body of evidence indicates that, when the mitotic checkpoint is active, the Mad2-Cdc20 subcomplex is formed at kinetochores not attached to the mitotic spindle (1–4). It has been assumed that Mad2-Cdc20 then assembles into MCC (2, 4), but, to our knowledge, this assumption has not been tested by direct experimentation. We used our preparation of recombinant Mad2-Cdc20 (MC) to examine its role in MCC assembly by a direct biochemical procedure. In the experiment shown in Fig. 2A, we compared the efficiency of the binding to BubR1 of the MC subcomplex with that of free Mad2 and Cdc20 by incubating equal concentrations of components followed by immunoprecipitation with anti-BubR1. Whereas there was marked binding to BubR1 of Mad2 originating from MC, indicating the formation of MCC (Fig. 2A, lane 3), incubation with free Mad2 and Cdc20 resulted in much less binding of Mad2 to BubR1 following (lane 2). Cdc20 from either source was bound to BubR1, but this was not surprising because free Cdc20 is expected to bind to site 2 of BubR1. We concluded that MC is a preferred intermediate for MCC formation. To further examine whether indeed functional MCC is formed by the assembly of the MC subcomplex with BubR1, we tested the effects of these incubations on APC/CCdc20 activity (Fig. 2B). The activity of APC/CCdc20 was not affected by BubR1 or MC when supplied by themselves (Fig. 2B, lanes 3 and 5, respectively), but was strongly inhibited following incubation with a combination of BubR1 and MC that led to MCC assembly (Fig. 2B, lane 6). By contrast, products of incubation of free Mad2, Cdc20 and BubR1 inhibited APC/CCdc20 activity only slightly (Fig. 2B, lane 4). These results indicate that the Mad2-Cdc20 subcomplex is indeed a preferred precursor for the assembly of MCC that is functional in APC/C inhibition.
Fig. 2.
The Mad2-Cdc20 subcomplex is a preferred precursor for MCC formation, but does not act catalytically to produce an inhibitor of APC/C activity. (A) The Mad2-Cdc20 subcomplex is a preferred substrate for MCC assembly. Reaction mixtures contained in the volume of 20 µL: 50 Mm Tris⋅HCl (pH 7.6), 100 mM NaCl, 5 mg/mL BSA, 1 mM DTT, and 10% (vol/vol) glycerol. The indicated additions were at 150 nM. Following incubation at 23 °C for 60 min, samples were subjected to immunoprecipitation with anti-BubR1 as described in Materials and Methods. Sham treatment was under similar conditions with nonimmune rabbit IgG. Samples of immunoprecipitates were subjected to immunoblotting for the indicated proteins as described in Materials and Methods. Numbers on the right side indicate the migration positions of marker proteins (kDa). (B) MCC assembled from the Mad2-Cdc20 subcomplex and BubR1 in vitro preferentially inhibits APC/CCdc20. APC/CCdc20 bound to anti-Cdc27 beads was prepared as described in Materials and Methods. A total of 100 nM of the indicated checkpoint proteins was incubated for 1 h at 23 °C, followed by the addition of APC/Cdc20 and further incubation with shaking (1,400 × g) for 1 h at 23 °C. Subsequently, beads were washed three times with Buffer A and were subjected to a cyclin-B ubiquitin ligation assay as described in Materials and Methods. Finally, samples were subjected to SDS/PAGE and radioautography as described in Materials and Methods. The results indicated at the bottom are expressed as the percentage of I125-cyclin B-(Ub)n conjugates formed relative to APC/C activity with 100 nM Cdc20. Numbers on the right indicate the migration position of marker proteins (kDa). MC, Mad2-Cdc20. (C) Limiting amounts of Mad2-Cdc20 do not act catalytically to stimulate the formation of an APC/C inhibitor. BubR1 and Cdc20 (300 nM, each) were incubated with the indicated concentrations of Mad-Cdc20 (MC) for 1 or 2 h. Subsequently, mixtures were added to immunopurified APC/CCdc20, and cyclin ubiquitylation activity was assayed as described in B. Results are expressed as the percentage of APC/CCdc20 activity without MC at time 0. (D) Comparison of the effectiveness of BC-1, BC-2, and MCC on the inhibition of APC/CCdc20 activity. Purified MCC and subcomplexes, at the concentrations indicated, were incubated with APC/CCd20, and samples were subjected to a cyclin-B ubiquitin ligation assay as described in B. Results are expressed as the percentage of cyclin–ubiquitin conjugates formed relative to APC/CCdc20 activity without additions.
Mad2-Cdc20 Subcomplex Does Not Act Catalytically to Produce an Inhibitor of APC/C.
We also could make use of the recombinant Mad2-Cdc20 subcomplex to examine the hypothesis of Cleveland and coworkers (19) according to which C-Mad2, or the C-Mad2-Cdc20 intermediate, acts catalytically to amplify the formation of BubR1-Cdc20. This hypothesis furthermore assumes that Mad2-free BubR1-Cdc20, rather than Mad2-containing MCC, is the major inhibitor of APC/CCdc20 (19). To test this hypothesis, we incubated low concentrations of MC with a large molar excess of BubR1 and of free Cdc20. Following incubation for different time periods, we examined the effects of incubation products on the activity of APC/CCdc20. If MC catalytically produced a BubR1-Cdc20 inhibitor of APC/C, it would be expected that, with longer incubation times of MC with excess BubR1 and Cdc20, more inhibitor would be produced. However, the degree of inhibition of APC/CCdc20 did not increase from 30 to 120 min of incubation (Fig. 2C). Instead, inhibition of APC/CCdc20 increased only when the concentration of MC was elevated, possibly reflecting the amounts of MCC produced. Thus, our results do not support the catalytic model of Cleveland and coworkers (19).
Comparison of the Effectiveness of BC-1, BC-2, and MCC on APC/C Inhibition.
We next used our preparations of purified recombinant subcomplexes to directly estimate their action on APC/CCdc20 activity. In the experiment shown in Fig. 2D, APC/CCdc20 was incubated with increasing concentrations of BC-1 or BC-2, and their effects on cyclin–ubiquitin ligation were monitored. For comparison, the effects of similar concentrations of MCC and of free BubR1 were also examined. Under our experimental conditions, free BubR1 had no appreciable effect on APC/CCdc20 activity, whereas BC-2 inhibited it only slightly. BC-1 did inhibit APC/CCdc20, but with less effectiveness than MCC. Thus, inhibition of 50% of APC/CCdc20 activity was obtained with ∼40 nM BC-1, compared with ∼10 nM of MCC. We conclude that, contrary to previous suggestions (18, 19), Mad2-containing MCC is a more effective inhibitor of APC/CCdc20 than are Mad2-free subcomplexes, although significant inhibition remains with a subcomplex in which Cdc20 is bound to site 1 of BubR1 (BC-1).
It is notable that we do not observe significant inhibition of APC/CCdc20 by free BubR1 (Fig. 2B), although BubR1 was reported as an APC/C inhibitor due to its association with Cdc20 (16, 17). This may be due to a difference in experimental conditions: in our procedure, Cdc20 is first bound to APC/C to form APC/CCdc20 (Materials and Methods), which may preclude the sequestration of Cdc20 by binding to site 2 of BubR1.
Mad2-Cdc20 Can Combine with BC-2 to Form MCC with an Additional Molecule of Cdc20.
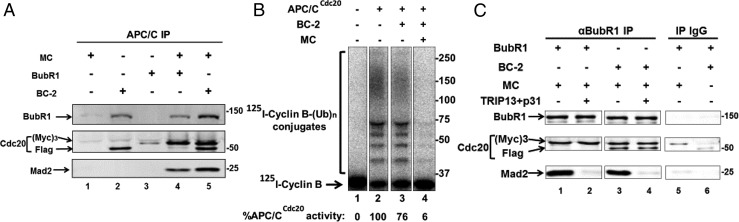
Because Cdc20 readily combines with BubR1 in vitro to form BC-2, it is possible that a similar process takes place in cells. This may sequester both Cdc20 and BubR1, which are present at roughly equal concentrations in mitotic cells (16). Therefore, the question arises whether BC-2 is a nonfunctional side product—or does it serve as an intermediate in the formation of an inhibitory complex? To address this problem, we examined whether BC-2 can combine with Mad2-Cdc20 to form an MCC-like complex. In the experiment shown in Fig. 3A, we incubated MC either with BC-2 or with free BubR1 and examined the formation of products bound to APC/C. To enable the identification of the source of products, we used MC that contained (Myc)3-Cdc20 and BC-2 that contained Flag-Cdc20. (Myc)3-Cdc20 migrates more slowly on SDS/PAGE than Flag-Cdc20. MC did not bind to APC/C (Fig. 3A, lane 1), whereas BC2 did bind (Fig. 3A, lane 2). As expected, following the incubation of MC with free BubR1, MCC was formed and was bound to APC/C (Fig. 3A, lane 4). Following incubation of MC with BC-2, an APC/C-bound product was formed that contained BubR1, Mad2, and both (Myc)3-Cdc20 and Flag-Cdc20 (Fig. 3A, lane 5). The results thus indicate that MC interacts with BC-2 to form a product that contains Cdc20 molecules from both MC and BC-2 and that this product binds to APC/C. We call this product “MCC-1–2” to indicate that it contains Cdc20 from two different sources. We suggest that Cdc20 originating from MC binds to site 1 of the BubR1 moiety of MCC-1–2 (Discussion), whereas Cdc20 that originates from BC-2 remains bound to site 2 of BubR1 in MCC-1–2. Thus, MCC-1–2 is different from the previously described species of MCC with two molecules of Cdc20 (20), in which the additional Cdc20 molecule is bound to the KEN2 box and the D1 box at the N-terminal region of BubR1.
Fig. 3.
Mad2-Cdc20 can combine with BC-2 to form MCC with an additional Cdc20 (MCC-1–2). (A) Formation of MCC-1–2 and its binding to APC/C. Incubation conditions were as described in Fig. 2A. The indicated subcomplexes and BubR1 were added at 150 nM. Reaction products were added to APC/C bound to anti-Cdc27 beads. Following further incubation for 60 min at 23 °C, proteins bound to APC/C were isolated by precipitation of anti-Cdc27 beads, followed by washing and APC/C elution with Cdc27 peptide as described in Materials and Methods. Subsequently, supernatants were immunoblotted for the indicated proteins. Numbers on the right indicate the electrophoretic migration of the marker proteins (kDa). (B) MCC-1–2 is an inhibitor of APC/CCdc20. BC-2 was incubated with or without MC as indicated. Incubation conditions were as described in Fig. 2A, and the final concentration of the specified subcomplexes was 100 nM. Subsequently, the products were added to APC/CCdc20, and following incubation their effects on cyclin B–ubiquitin ligation activity were determined as described in Fig. 2B. The results indicated at the bottom are expressed as the percentage of I125-cyclin B-(Ub)n conjugates formed relative to APC/CCdc20 without additions. Numbers on the right indicate the migration position of marker proteins (kDa). (C) MCC-1–2 is disassembled by the joint action of TRIP13 and p31comet. Different combinations of Mad2-(Myc)3-Cdc20 (MC) with either BubR1 or BubR1-Flag-Cdc20 (BC-2) were incubated at conditions described in Fig. 2A and then were precipitated with either anti-BubR1 or IgG beads, as indicated. Subsequently, beads were washed three times with Buffer A and then were incubated with TRIP13 and p31comet, where indicated, under conditions similar to those described in SI Materials and Methods for BC-1 formation. Sham treatment was under similar conditions but without TRIP13 and p31comet. Finally, the beads were washed, and samples were subjected to immunoblotting for the indicated proteins. Numbers on the right side indicate the migration position of marker proteins (kDa).
To examine whether MCC-1–2 is an inhibitor of APC/C, BC-2 was incubated with or without MC, incubation products were added to APC/CCdc20, and their effects on cyclin B-ubiquitin ligation activity were determined. As shown in Fig. 3B, BC-2 by itself inhibited APC/CCdc20 only slightly (lane 3), whereas, following incubation with MC, a strong inhibition of APC/CCdc20 was observed (Fig. 3B, lane 4). These results suggest that MCC-1–2, like MCC, is a strong inhibitor of APC/CCdc20.
If MCC-1–2 is a bona fide checkpoint inhibitor of APC/C, the question arises, how is it disassembled when the checkpoint signal is turned off? We examined the possibility that Mad2 is released from MCC-1–2 by a joint action of TRIP13 and p31comet, as is the case with MCC. In the experiment shown in Fig. 3C, MC was first incubated with BubR1 to form MCC (lane 1) or with BC-2 to form MCC-1–2 (lane 3). Subsequently, the samples were further incubated with TRIP13 and p31comet, and Mad2 associated with BubR1 was detected by immunoprecipitation with anti-BubR1. Treatment with TRIP13 and p31comet promoted the release of Mad2 from MCC-1–2 (lane 4) as it did from MCC (lane 2). These data indicate that MCC-1–2 may be disassembled by the joint action of TRIP13 and p31comet.
SI Materials and Methods
Recombinant MC was produced by coinfection of SF9 insect cells with baculoviruses expressing Myc3-His6-Cdc20 (generously provided by Jan-Michael Peters, Research Institute of Molecular Pathology (IMP), Vienna and described in ref. 28) with Flag-Mad2. The subcomplex was purified first with Ni-NTA-agarose (Qiagen) and then with anti-DYKDDDD (Flag) affinity resin (GeneScript), according to instructions of the manufacturers. A recombinant WT-BubR1-Cdc20 (BC-2) subcomplex was generated by coinfection of SF9 cells with baculoviruses expressing Flag-his6-Cdc20 and Streptavidin-Binding Peptide (SBP)-WT-BubR1. The complex was purified on Strep Tactin Agarose, High performance (GE Healthcare), followed by anti-DYKDDDD (Flag) affinity resin, according to the manufacturer’s instructions. Recombinant MCC was formed by coexpression in insect cells of Myc3-his6-Cdc20, SBP-ΔPhe-ΔD2-BubR1, and Flag-Mad2, followed by sequential purification on Strep Tactin Agarose and anti-Flag affinity resins. Baculoviruses expressing SBP-WT-BubR1 and the corresponding SBP-ΔPhe-ΔD2-BubR1 mutant were generously provided by Hongtao Yu, University of Texas Southwestern Medical Center, Dallas (described in ref. 17). Bub3 was not coexpressed in the formation of recombinant MCC or of its subcomplexes because it has no appreciable influence on MCC function or structure (13, 14). Protein concentrations and stoichiometry were estimated by SDS/PAGE separation followed by staining with Krypton Infrared Protein Stain (Pierce) in comparison with BSA standards, according to the instructions of the manufacturer, and detection with Li-COR Odyssey scanner. Preparations of recombinant MCC had close to equimolar concentrations of BubR1, Cdc20, and Mad2 (Fig. S1A).
His6-TRIP13 and p31comet were expressed in bacteria and were purified on Ni-NTA-agarose (Qiagen) as was previously described (7, 9). For the generation of the BC-1 subcomplex, MCC was subjected to disassembly by the joint action of TRIP13 and p31comet. The reaction mixture was contained in the volume of 150 μL: 1.5 μM recombinant MCC (containing the ΔPhe-ΔD2-BubR1 mutant), 25 mM Tris⋅HCl (pH 7.6), 5 mM MgCl2, 1 mM DTT, 20% (vol/vol) glycerol, 2 mg/mL BSA, 1 mM ATP, 10 mM phosphocreatine, 100 μg/mL creatine phosphokinase, 1.5 µM TRIP-13, and 2.5 µM p31comet. The reaction was carried out at 30 °C for 2 h as was previously described for the disassembly of endogenous MCC from checkpoint extracts (9). Then the incubation mixture was further purified on Strep-Tactin beads to remove incubation reagents. The maximal contamination of the final preparation of BC-1 by residual MCC was estimated at around 2–4% by comparison of the concentration of Mad2 with either Cdc20 or BubR1.
Discussion
In this study we investigated the roles of subcomplexes of mitotic checkpoint proteins in MCC assembly and in the regulation of APC/C activity by the use of purified subcomplexes. The preparation of specific subcomplexes of BubR1 with Cdc20 was hindered by the existence of two separate Cdc20-binding sites on BubR1 (Fig. 1). We have prepared a subcomplex of Cdc20 bound to site 1 of BubR1 (“BC1”) by releasing Mad2 from recombinant MCC by treatment with TRIP13 and p31comet (9) (Fig. S1D).
The intermediary steps in the assembly of MCC are not sufficiently understood. Because Cdc20 binds preferentially to C-Mad2 (21, 22), it appeared reasonable to assume that the formation of the C-Mad2-Cdc20 subcomplex follows the conformational transition of O-Mad2 to C-Mad2 at the kinetochore. Indeed, considerable levels of the C-Mad2-Cdc20 subcomplex were detected in checkpoint-arrested cells (23). Although the binding of C-Mad2 to Cdc20 may sequester Cdc20 from activating the APC/C (24), we observed that, unlike MCC, the Mad2-Cdc20 subcomplex did not inhibit preformed APC/CCdc20 (Fig. 2B). It has been proposed that the Mad2-Cdc20 subcomplex associates with BubR1-Bub3 to form the MCC (2, 4), but this suggestion has not been previously tested by direct biochemical methods. Using preparations of recombinant purified MC, we showed that it is indeed a preferred precursor for assembly with BubR1, compared with the lower extent of assembly from free Mad2 and Cdc20 (Fig. 2). It should be noted that other investigators observed more significant formation of MCC from free Mad2 in vitro (19, 25). The difference in results is possibly due to the higher extent of formation of C-Mad2 from O-Mad2 in vitro under the experimental conditions of these investigators.
Although Mad2 is undoubtedly required for MCC assembly, it remained questionable whether or not Mad2-containing MCC is the major checkpoint inhibitor of APC/C. Nilsson et al. (18) observed that, in checkpoint complexes of extracts from nocodazole-arrested HeLa cells, only a small fraction of Cdc20 was bound to Mad2, whereas most of it was associated with BubR1. Cleveland and coworkers furthermore proposed that Mad2-free Cdc20-BubR1 complex is the main checkpoint inhibitor of APC/C (19). These authors also suggested that the role of C-Mad2, or of the C-Mad2-Cdc20 intermediate, is to catalytically amplify the production of the putative Cdc20-BubR1 inhibitor of APC/C. The availability of purified subcomplexes of MCC in the present study allowed us to critically examine these suggestions. We found that incubation of low concentrations of the C-Mad2-Cdc20 intermediate with large molar excess of Cdc20 and BubR1 did not lead to a time-dependent progressive increase in the extent of the inhibition of APC/CCdc20 (Fig. 2C), as would be predicted by the catalytic model of Cleveland and coworkers (19). Furthermore, direct estimation of the effects of purified BubR1-Cdc20 subcomplexes on the activity of APC/CCdc20 showed that BC-2 inhibited APC/CCdc20 only slightly, whereas BC-1 did inhibit APC/CCdc20, although less effectively than MCC (Fig. 2D). We therefore concluded that the major checkpoint inhibitor of APC/C is the Mad2-containing mitotic checkpoint complex. It should be noted that, although the above-mentioned studies reported substoichiometric amounts of Mad2 (relative to Cdc20 and BubR1) in mitotic checkpoint complexes (18, 19), they did not actually estimate the action of such complexes on APC/C activity. It is thus possible that, in checkpoint-arrested living cells, there is a mixture of MCC, BC-1, and BC-2, of which only MCC powerfully inhibits the APC/C. The exact role of the Mad2 moiety in the inhibition of APC/CCdc20 remains to be elucidated. It is possible that Mad2 in MCC interacts with a part of the MCC-binding site of APC/C, but it is also possible that the role of the Mad2 moiety is more indirect. Because C-Mad2 in MCC interacts with both Cdc20 and BubR1 (14), it is possible that it promotes conformational alterations in Cdc20 and/or BubR1 that tighten their interaction with APC/C. The significant, although decreased, inhibition of APC/CCdc20 activity by BC-1 (Fig. 2D) suggests that some weak interactions of the two moieties of this subcomplex with APC/C take place in the absence of Mad2. These problems should be resolved by detailed structural analysis of the interaction of MCC with its binding site on APC/C.
Although the interaction of Cdc20 with site 1 of BubR1 requires its binding to Mad2 and mitotic checkpoint signaling, the binding of Cdc20 to site 2 does not. Thus, BC-2 may accumulate in cells with the rise of Cdc20 levels at the G2/M phases of the cell cycle (see fig. 1 in ref. 23). The formation of BC-2 may sequester both Cdc20 and BubR1, which are present in approximately equal concentrations in mitotic cells (16). We therefore examined the possibility that BC-2 may interact with MC to form an MCC-like mitotic checkpoint complex. We indeed found that, under these conditions, a complex with an additional molecule of Cdc20, MCC-1–2, is formed, which contains Cdc20 from both MC and BC-2 (Fig. 3A). Because Cdc20 originating from MC binds to site 1 of BubR1 (15), whereas in BC-2 Cdc20 is bound to site 2 (16, 17), it appears reasonable to assume that MCC-1–2 contains Cdc20 molecules bound to both sites 1 and 2 of BubR1. It should be noted that MCC-1–2 is different from a complex of MCC with a second Cdc20, reported previously by Izawa and Pines (20). There, the additional Cdc20 molecule was bound to KEN2 box and D1 box at the N-terminal region of BubR1. We showed that MCC-1–2 inhibits APC/C activity (Fig. 3B); thus, it may serve as an additional mitotic checkpoint inhibitor complex to MCC. We also showed that Mad2 can be released from MCC-1–2 by the joint action of TRIP13 and p31comet (Fig. 3C), similar to their action on MCC (9). This process may terminate the action of MCC-1–2 when the mitotic checkpoint is inactivated. It is interesting to note that estimation of the molar ratios of checkpoint complexes from HeLa cell extracts indicated an ∼50% excess of Cdc20 over BubR1 (18) (Fig. S2). This excess may be explained by the presence of the dissociation product of MCC-1–2 in these complexes. Further investigation is required to examine the presence and abundance of MCC-1–2 in checkpoint-inhibited cells.
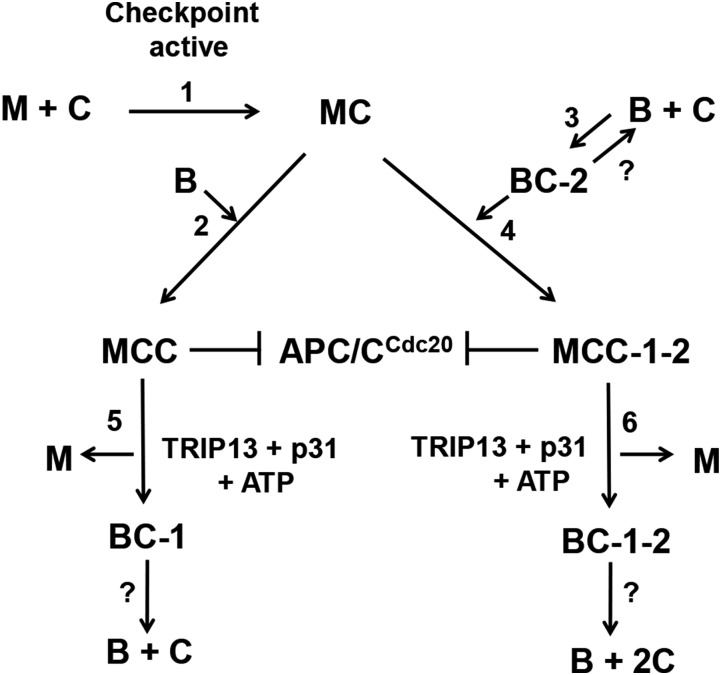
Fig. 4 summarizes our proposal for the roles of intermediary subcomplexes in the assembly of mitotic checkpoint complexes and in the inhibition of the ubiquitin ligase activity of APC/CCdc20. When the mitotic checkpoint is active, C-Mad2 is generated and associates with Cdc20 to form MC (step 1). MC binds to BubR1 to form MCC (step 2) in a process in which the Cdc20 moiety binds to site 1 of BubR1. Cdc20 may also bind to site 2 of BubR1 with the formation of BC-2 in a process independent of the mitotic checkpoint (step 3). MC may also associate with BC-2 to form MCC-1–2 (step 4). Both MCC and MCC-1–2 inhibit the APC/CCdc20. When the checkpoint is extinguished, Mad2 is released from both MCC and MCC-1–2 by the joint action of TRIP13 and p31comet (steps 5 and 6). This process is accompanied by the conversion of C-Mad2 to O-Mad2 (12). The conversion of MCC to BC-1 (step 5) decreases the inhibition of APC/C, but does not abolish it completely (Fig. 2D). For complete release of APC/CCdc20 from checkpoint inhibition, the further dissociation of BC-1 to free BubR1 and Cdc20 is required. The molecular mechanisms of the disassembly of BC-1, BC-2, and BC-1–2 remain unknown and require further investigation.
Fig. 4.
Proposed intermediary processes in the assembly and disassembly of mitotic checkpoint complexes (Discussion). Note that Bub3 is not depicted in this scheme, but it is constitutively bound to BubR1. M, O-Mad2; C, Cdc20; B, BubR1; MC, C-Mad2-Cdc20; BC-1, BubR1 with Cdc20 bound to site 1; BC-2, BubR1 with Cdc20 bound to site 2; BC-1, -2, BubR1 with Cdc20 molecules bound to sites 1 and 2; MCC, Mitotic Checkpoint Complex (with Cdc20 bound to site 1 of BubR1); MCC-1–2, Mitotic Checkpoint Complex with Cdc20 molecules bound to sites 1 and 2 of BubR1; APC/CCdc20, Anaphase-Promoting Complex/Cyclosome with bound Cdc20.
Materials and Methods
Immunoprecipitation and Immunoblotting.
Rabbit polyclonal antibodies directed against human proteins were used for immunoprecipitation. All antibodies were purified by affinity chromatography on their respective antigens: α-Cdc27, 17-amino acid C-terminal peptide of Cdc27; and α-BubR1, an his6-tagged 38- to 468-aa fragment of BubR1. For immunoprecipitation, all antibodies were bound to Affi-Prep Protein A beads (Bio-Rad) at a concentration of 0.5 mg/mL of packed beads. For sham immunoprecipitations, purified rabbit IgG (Pierce) was bound to the Protein A beads at a similar concentration. For immunoblotting of human proteins, the following mouse monoclonal antibodies were used: Cdc27 and BubR1 (BD Transduction Laboratories 3290559 and 612053, respectively); Cdc20 (Santa Cruz Biotechnology sc-13162); Mad2 (MBL Laboratories K0167); c-Myc (Sigma M5546); and Flag (Sigma F3165). Immunoblots were detected and quantified with fluorescently labeled secondary antibodies using an Odyssey (Li-Cor) scanner.
APC/C-Binding Assay.
Xenopus interphase egg extracts were prepared as described (26), and APC/C was immunoprecipitated from these extracts with anti-Cdc27 antibody bound to Affiprep Protein A (Bio-Rad) beads at an extract-to-bead ratio of 10:1 (vol/vol) for 2 h at 4 °C. The APC/C beads were washed twice with a buffer consisting of 50 mM Tris⋅HCl (pH 7.2), 20% (vol/vol) glycerol, 1 mg/mL BSA, 1 mM DTT, and 0.3 M NaCl and then twice with the same buffer without NaCl (Buffer A). APC/C beads (1 μL) were suspended in 20 μL of Buffer A and were then incubated with checkpoint proteins or complexes as specified in the figure legends for the indicated time and temperature with shaking (1,400 × g) by a Thermomix shaker (Eppendorf). Unbound proteins were removed by washing the beads three times with Buffer A containing 0.15 M NaCl and 0.1% Triton X-100 (“washing buffer”). Then, the APC/C complex was eluted from the beads by a peptide similar to the 17 C-terminal amino acids of Cdc27 (“Cdc27 peptide”). Elution was carried out with at 2 mg/mL of Cdc27 peptide in 20 µL of washing buffer for 2 h at 30 °C, and samples of the supernatants were analyzed by immunoblotting.
Assay of Inhibition of APC/CCdc20 Activity (Cyclin B-Ubiquitin Ligation Assay).
APC/C from interphase Xenopus extracts was immunopurified on anti-Cdc27 beads, as described above, and then was incubated with recombinant purified Cdc20 to prepare APC/Cdc20. For this purpose, 1 μL of APC/C beads suspended in 20 μL of Buffer A were mixed with 100 nM Cdc20 at 50 × g for 1 h at 4 °C and then washed three times with Buffer A. Samples of 1-μL beads containing APC/Cdc20 were incubated with checkpoint proteins or complexes under conditions specified in figure legends and then washed again three times with Buffer A. Finally, APC/C activity was assayed by the conversion of the 125I-cyclin B N-terminal fragment to higher molecular ubiquitylated derivatives, as described previously (27).
Acknowledgments
We thank Dr. Jan-Michael Peters for providing baculovirus for the expression of Myc3-His6-Cdc20; Dr. Hongtao Yu for providing baculoviruses expressing SBP-BubR1 (wild type and Δ-Phe Δ-D2 mutant); and Dr. Michael Fry for comments on the manuscript. This work was supported by grants from the Israel Science Foundation; the Israel Cancer Research Fund; and the Ricbac Foundation for Research in Cell Biology and Cancer. A part of this work was performed at the Marine Biological Laboratory (Woods Hole, MA).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1524551113/-/DCSupplemental.
References
- 1.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 2.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22(22):R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Jia L, Kim S, Yu H. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem Sci. 2013;38(6):302–311. doi: 10.1016/j.tibs.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Primorac I, Musacchio A. Panta rhei: The APC/C at steady state. J Cell Biol. 2013;201(2):177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154(5):925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miniowitz-Shemtov S, Teichner A, Sitry-Shevah D, Hershko A. ATP is required for the release of the anaphase-promoting complex/cyclosome from inhibition by the mitotic checkpoint. Proc Natl Acad Sci USA. 2010;107(12):5351–5356. doi: 10.1073/pnas.1001875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teichner A, et al. p31comet promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc Natl Acad Sci USA. 2011;108(8):3187–3192. doi: 10.1073/pnas.1100023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miniowitz-Shemtov S, et al. Role of phosphorylation of Cdc20 in p31(comet)-stimulated disassembly of the mitotic checkpoint complex. Proc Natl Acad Sci USA. 2012;109(21):8056–8060. doi: 10.1073/pnas.1204081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eytan E, et al. Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and p31(comet) Proc Natl Acad Sci USA. 2014;111(33):12019–12024. doi: 10.1073/pnas.1412901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, et al. Thyroid hormone receptor interacting protein 13 (TRIP13) AAA-ATPase is a novel mitotic checkpoint-silencing protein. J Biol Chem. 2014;289(34):23928–23937. doi: 10.1074/jbc.M114.585315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miniowitz-Shemtov S, Eytan E, Kaisari S, Sitry-Shevah D, Hershko A. Mode of interaction of TRIP13 AAA-ATPase with the Mad2-binding protein p31comet and with mitotic checkpoint complexes. Proc Natl Acad Sci USA. 2015;112(37):11536–11540. doi: 10.1073/pnas.1515358112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye Q, et al. 2015. TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching. eLife 2015(4):e07367.
- 13.Malureanu LA, et al. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev Cell. 2009;16(1):118–131. doi: 10.1016/j.devcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484(7393):208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 15.Davenport J, Harris LD, Goorha R. Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1. Exp Cell Res. 2006;312(10):1831–1842. doi: 10.1016/j.yexcr.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 16.Tang Z, Bharadwaj R, Li B, Yu H. Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1(2):227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 17.Diaz-Martinez LA, et al. The Cdc20-binding Phe box of the spindle checkpoint protein BubR1 maintains the mitotic checkpoint complex during mitosis. J Biol Chem. 2015;290(4):2431–2443. doi: 10.1074/jbc.M114.616490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10(12):1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han JS, et al. Catalytic assembly of the mitotic checkpoint inhibitor BubR1-Cdc20 by a Mad2-induced functional switch in Cdc20. Mol Cell. 2013;51(1):92–104. doi: 10.1016/j.molcel.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izawa D, Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2015;517(7536):631–634. doi: 10.1038/nature13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9(1):59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 22.Sironi L, et al. Crystal structure of the tetrameric Mad1-Mad2 core complex: Implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21(10):2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell. 2002;13(3):755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izawa D, Pines J. Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation. J Cell Biol. 2012;199(1):27–37. doi: 10.1083/jcb.201205170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lara-Gonzalez P, Scott MI, Diez M, Sen O, Taylor SS. BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. J Cell Sci. 2011;124(Pt 24):4332–4345. doi: 10.1242/jcs.094763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- 27.Braunstein I, Miniowitz S, Moshe Y, Hershko A. Inhibitory factors associated with anaphase-promoting complex/cylosome in mitotic checkpoint. Proc Natl Acad Sci USA. 2007;104(12):4870–4875. doi: 10.1073/pnas.0700523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uzunova K, et al. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol. 2012;19(11):1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]