Abstract
Wheat chromosome 5A plays a key role in cold acclimation and frost tolerance. The major frost tolerance gene Fr-A1 (formerly Fr1) and two loci that regulate the transcription of cold-regulated genes (Cor) have been mapped before on the long arm of this chromosome. In this study we report the existence of a new locus for frost tolerance designated Fr-A2. This new locus was mapped on the long arm of T. monococcum chromosome 5A, 40 cM from the centromere and 30 cM proximal to the major frost tolerance locus Fr-A1. We found also, that frost tolerant and frost susceptible T. monococcum parental lines differed in the transcription level of the cold induced gene Cor14b when plants were grown at 15°C. Transcription levels of this gene were measured in each of the recombinant inbreed lines and mapped as a QTL that perfectly overlapped the QTL for frost survival at the Fr-A2 locus. This result suggested that frost tolerance in this cross was mediated by a differential regulation of the expression of the Cor genes. In our previous study in hexaploid wheat we showed that Cor14b was regulated by two loci located on chromosome 5A, one in the same chromosome region as the T. monococcum Fr-A2 locus and the other one closely linked to Fr-A1. Since Cbf transcriptional activators in Arabidopsis regulate Cor genes and are involved in frost tolerance, we decided to map the cold regulated Cbf-like barley gene Cbf3 in the T. monococcum map. This gene was mapped on the peak of the Fr-A2 QTL for frost tolerance. This result suggest that the observed differential regulation of Cor14b at the Fr-A2 locus was originated by allelic variation at the XCbf3 locus, and that this transcriptional activator might be a candidate gene for the Fr-A2 frost tolerance locus on wheat chromosome 5A.
Keywords: frost tolerance, Cor genes, Cbf transcription factors, cold acclimation, wheat
INTRODUCTION
Freezing temperatures limit the geographical distribution of wheat (T. aestivum L.) and often cause severe losses in agricultural productivity. Therefore, increasing frost tolerance has been a major objective for most breeding programs in regions subject to severe winters. Frost tolerant wheat varieties show an increase in freezing tolerance after exposure to non-freezing low temperatures, a phenomenon known as cold acclimation (Sakai and Larcher 1985). During cold acclimation, winter cereals adjust their metabolism to low temperatures and protect critical cell structures against the effect of freezing temperatures.
The genetic control of frost tolerance in wheat is complex and at least 10 of the 21 pairs of chromosomes are involved in the regulatory gene network. However, the major genes affecting winter hardiness have been mapped on the long arms of homeologous groups 5 (Roberts 1990; Sutka 1994; Sutka and Snape 1989; Veisz and Sutka 1993). The major frost tolerance locus, Fr-A1 (formerly Fr1), was mapped on the long arm of chromosome 5A, 2-cM proximal to the vernalization gene Vrn-A1 (Galiba et al. 1995). Physical mapping using Chinese Spring deletion lines confirmed that Vrn-A1 and Fr-A1 were different genes and that Fr-A1 was located proximal to Vrn-A1 (Galiba et al. 1995). An additional QTL for frost tolerance designated QFr.jic-5D, was mapped on a colinear region of chromosome 5D, 10-cM proximal to Vrn-D1 (Snape et al. 1997).
In our previous work, we mapped two loci controlling the expression of Cold regulated gene Cor14b on the long arm of chromosome 5A (Vágújfalvi et al. 2000). One of these genes was closely linked to Fr-A1 whereas the other one was mapped linked to Xpsr911, an RFLP marker located approximately 35 cM proximal to Fr-A1 (Gale et al. 1995). The Cor genes are strictly regulated by low temperatures and are probably involved in the acquisition of frost tolerance (for a review see Cattivelli et al. 2002).
The well-studied Arabidopsis Cor15a gene encodes a 15-kDa protein that is targeted to the stromal compartment of the chloroplast. The over-expression of this gene in transgenic plants increased the frost tolerance not only of the chloroplasts but also of the protoplast isolated from leaves of the non-acclimated plants (Artus et al. 1996).
In Arabidopsis the Cor genes are regulated by three members of the CBF gene family of transcriptional activators that are organized in a tandem array on chromosome 4 (for a review see Thomashow et al. 2001). The CBF1, CBF2 and CBF3 proteins are 84% identical suggesting a relatively recent duplication (Medina et al. 1999). Genes similar to the Arabidopsis Cbf genes are present in the Triticeae EST databases. One of them, barley Cbf3, has been recently mapped on the long arm of chromosome 5H but in a region that has not been associated with frost tolerance in barley or wheat (Choi et al. 2002). Beside transcriptionally regulated Cor genes, as those described above, other Cor sequences are controlled at post-transcriptional level (Dunn et al. 1994; Phillips et al. 1997). These Cor genes may represent cold-response component not regulated by cold induced transcription factors.
In the present work we report the mapping of a new QTL for frost tolerance in diploid wheat T. monococcum and its linkage with the Cbf3 gene. We also show that variation at this locus is associated with differences in the threshold induction temperature of the cold regulated gene Cor14b, but does not affect expression of the post-transcriptionally regulated gene Ao86. The interaction between this new frost tolerance locus and growth habit is discussed.
MATERIALS AND METHODS
Plant material and frost tolerance tests
Analysis of frost tolerance and of Cor14b expression were performed in a single seed descent population derived from the F2 population used to construct a T. monococcum map that included 335 RFLP loci (Dubcovsky et al. 1996). These 74 RILs were derived from the cross between T. monococcum ssp. monococcum DV92 (spring and susceptible to frost) and T. monococcum ssp. aegilopoides G3116 (winter and tolerant to frost). DNA was extracted from F5 plants as described before (Dvorak et al. 1988) and frost tolerance and Cor14b expression studies were performed on the F5 derived F6 plants.
For the Cor14b expression studies plants were grown in modified Hoagland solution (Nagy and Galiba 1995) for two weeks with 16 hours illumination (260 μmol/m2s) at different temperatures: 5, 10, 15, 20 and 25°C. Plants were then kept at − 80°C for Northern analysis.
For the frost tolerance tests, seeds were potted in wooden boxes in a randomized block design arrangement. Seedlings were grown in phytotronic chambers at 15/10°C (day/night), 75% relative humidity, and the light intensity was 260 μmol/m2s. The hardening started when the temperature was reduced to 10/5°C for 2 weeks, than to 5/0°C for another 2 weeks, and to +2/−2°C for one week. Then the temperature was gradually lowered to the freezing temperature, −13°C and it was maintained for a day. After freezing the temperature was gradually increased to 17/16°C. At this temperature the leaves were cut several cm above the soil. Frost tolerance was estimated as the assessment of the re-growth of the plants scored on a scale running from 0 (death) to 5 (undamaged). Twenty five individual F6 plants were evaluated per each of 51 RILs in the first experiment in 1998 and 35 individual plants per each of 58 RILs were evaluated in the second experiment in 1999.
Northern and Southern analyses
Frozen shoots from each of the RILs were ground in liquid N2 and total RNA was extracted using Trizol reagent (GibcoBRL Cat. 15596-026). Twenty micrograms of total RNA was separated on denaturing formaldehyde-agarose gel, and than blotted to Hybond membranes. Radioactively labeled Cor14b and Ao86 barley cDNA probes (Cattivelli and Bartels 1990) were prepared by the random primer method (Feinberg and Vogelstein 1983) and hybridization was carried out overnight. The barley Cor gene Ao86, a member of the best characterized post transcriptionally regulated Cor gene family Blt14 (Grossi et al. 1998), was chosen to represent a sequence that is putatively not regulated by cold induced transcription factors. Filters were washed three times at 65°C in 2xSSC 0.1% SSD solution, then exposed to Kodak Scientific film. To adjust the Cor14b hybridization signal for differences in RNA loading, filters were hybridized with a barley DNA probe coding for the protein 12 of the ribosomal large subunit (RPL12) whose expression was prove to be unaffected by low temperature (Baldi et al. 2001). Developed films were scanned and the expression level of Cor14b and ribosomal probes in each line were quantified using Bio-Rad Molecular Analyst software version 1.5. Southern blots and hybridizations were performed as described before (Dubcovsky et al. 1994).
Mapping
The first frost tolerance QTL scan of the complete genome was performed with the marker information inferred from the homozygous lines available from the F2 generation (Dubcovsky et al. 1996). Marker information was then completed for the regions where significant QTLs for frost tolerance were detected. Ten markers previously mapped on chromosome 5A on the T. monococcum F2 population were remapped in the 74 F5 RILs and the QTL analyses was repeated with the complete mapping data matrix. Sources of these probes were described before (Dubcovsky et al. 1996). In addition, probes for RFLP loci Xpsr426, Xpsr2021 (Gale et al. 1995), Xmwg2062 (Graner et al. 1991), Xbcd926 (Anderson et al. 1992) and microsatellite markers Xgwm639 and Xgwm186 (Röder et al. 1998) were added to the map to facilitate comparisons with previous maps that include QTLs for frost tolerance (Snape et al. 2001) or genes that control the expression of Cor14b genes (Vágújfalvi et al. 2000). The probe from the barley Cbf3 gene was kindly provided by Jeff Skinner in the Chen and Hayes lab (Oregon State University). This probe was cloned from barley variety Dicktoo and is allelic to the Cbf3 gene from barley variety Morex (accession number AF298231). Maps were constructed with the software Mapmaker/EXP 3.0 and MapmakerQTL (Lander et al. 1987) using the Kosambi function (Kosambi 1943). QTL confidence intervals were calculated based on a difference of 1 LOD score with the peak of the QTL.
Nomenclature
The frost tolerance gene on the long arm of chromosome 5A has been historically referred as Fr-1 (McIntosh et al. 1998) whereas the QTL for frost tolerance mapped on chromosome 5D has been referred to as Fr-2 (Snape et al. 1997) or QFr.jic-5D (Snape et al. 2001). The presence of more that one frost tolerance locus in the long arm of chromosome 5A (results from this study) and of genes with similar effects on chromosomes 5B and 5D would make the use of the sequential number gene nomenclature complicated. Therefore, we propose to use a homoeologous system of nomenclature similar to the one currently in use for the vernalization genes in wheat (McIntosh et al. 1998). Hereafter, we will refer to Fr1 as Fr-A1, to the new locus described in this study as Fr-A2, and to the locus on 5DL as QFr.jic-5D until the orthology with Fr-A1 or Fr-A2 is more clearly demonstrated.
For the mapped CBF locus we use the XCbf3 name as published before by Choi et al (2002). However, this not necessarily indicates orthology between barley clone AF298231 and Arabidopsis Cbf3 gene. The predicted proteins from the three closely related Arabidopsis Cbf genes are more similar to each other than to the protein predicted from the barley Cbf3 sequence (data not shown).
RESULTS
QTL analyses for frost tolerance
The molecular characterization of the 74 F5 RILs with 10 RFLP markers from the QTL regions showed 7.8 % residual heterozygosity, a slightly higher value than the theoretical 3% expected in F5. Heterozygous markers were scored as missing data for all the QTL analyses.
Winter T. monococcum accession G3116 was significantly more frost tolerant than spring accession DV92 (P<0.0001) after freezing at −13°C. Average frost tolerance, evaluated as the capability of survival on a 0 to 5 scale, was 1.9 for G3116 (95% confidence interval 1.7 to 2.1) and 0.2 for DV92 (95% confidence interval 0.0 to 0.4).
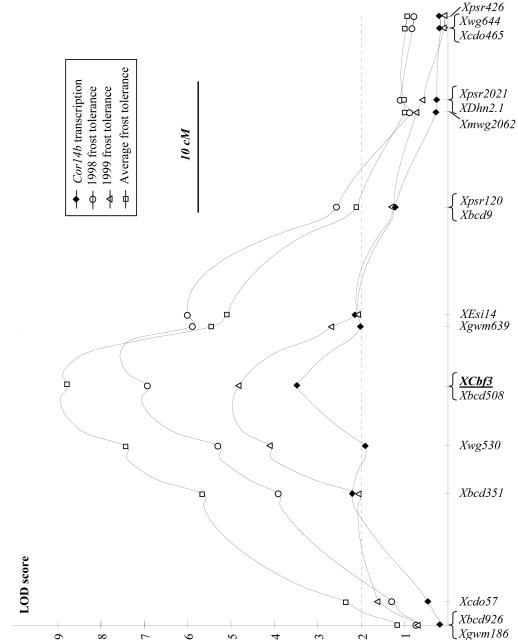
Segregation for frost tolerance scores was observed among the RILs in the 1998 (0-2.9) and 1999 (0-1.5) experiments. A QTL analysis of the seven T. monococcum chromosomes at a LOD threshold of three, showed only one QTL on the long arm of chromosome 5A for both experiments and for the average frost tolerance (Fig. 1). In the 1998 experiment, a QTL with a LOD score of 7.5 was mapped on a 7-cM confidence interval, which encompassed the Xbcd508 locus (Fig. 1). This QTL explained 48% of the variation in frost survival in this cross. A similar location was found for the QTL for frost tolerance mapped in the 1999 experiment (Fig. 1). This QTL with a LOD score of 5.0 explained 40% of the variation in frost tolerance and was mapped into a 9 cM confidence interval around locus Xbcd508. The combined data for both years showed a QTL with a LOD score of 8.9 with a confidence interval for the peak centered on Xbcd508 (7 cM confidence interval). This QTL explained 49% of the variation in frost tolerance in the DV92 x G3116 cross (Fig. 1).
Fig. 1.
QTL analysis for frost tolerance (○= 1998 experiment, □= 1999 experiment, and △= average from 1998 and 1999 experiments) and Cor14b transcript levels at 15°C (◆). Distances between markers in the X-axis are proportional to the distances in the RIL map.
QTL analyses for Cor14b transcription levels
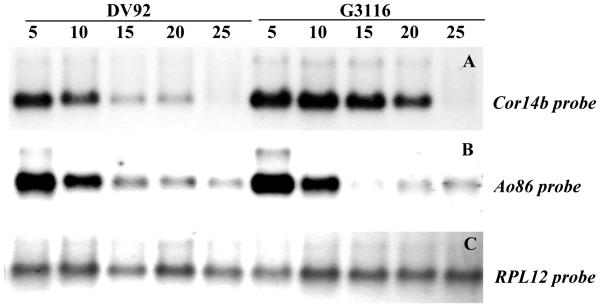
In our previous study in hexaploid wheat (Vágújfalvi et al. 2000) we found that that at 18/13°C (day/night temperatures) the Cor14b gene was transcribed in the frost tolerant genotypes, but not in the frost sensitive ones. All genotypes, independently from their frost tolerance, showed high levels of transcription at 2°C but non-detectable levels of transcripts at 25°C. To determine the optimum differential threshold temperature in T. monococcum, the level of transcription of Cor14b gene was determined in plants from the parental lines DV92 and G3116 grown at 5, 10, 15, 20 and 25°C. Northern blots of these samples showed that G3116 had a higher level of transcription of Cor14b than DV92 when plants were grown at 15 or 20°C. The largest difference between parental lines was observed at 15°C (Fig. 2A). When the same filter was hybridized with a probe from the cold regulated gene Ao86 (Grossi et al. 1998) similar expression profiles were observed from both genotypes (Fig. 2B) demonstrating that the cold induced expression of Cor14b and Ao86 is controlled by two different mechanisms.
Fig. 2.
Northern blot of G3116 and DV92 plants grown at different temperatures (°C) and hybridized with Cor14b (upper panel) Ao86 (middle panel) and with the ribosomal probe RPL12 (lower panel).
Plants from 62 RILs were grown at 15°C, and the total RNA extracted from each line was blotted into a single membrane to facilitate comparisons. Hybridization of this membrane with Cor14b probe showed different levels of transcripts among the RILs. The same membrane was hybridized with a wheat ribosomal probe to normalize the hybridization signal of Cor14b for putative differences in total RNA loaded in the gel (Fig. 2C). The normalized Cor14b values were analyzed using Mapmaker QTL (Fig. 1). A significant QTL with a LOD score of 3.5 was mapped with a peak on RFLP marker Xbcd508. This locus explained 24% of the variation in intensity in the normalized Cor14b values in this segregating population. The QTL for differential transcription level of Cor14b overlapped with the QTL for frost survival (Fig. 1).
Mapping of the Cbf3 gene
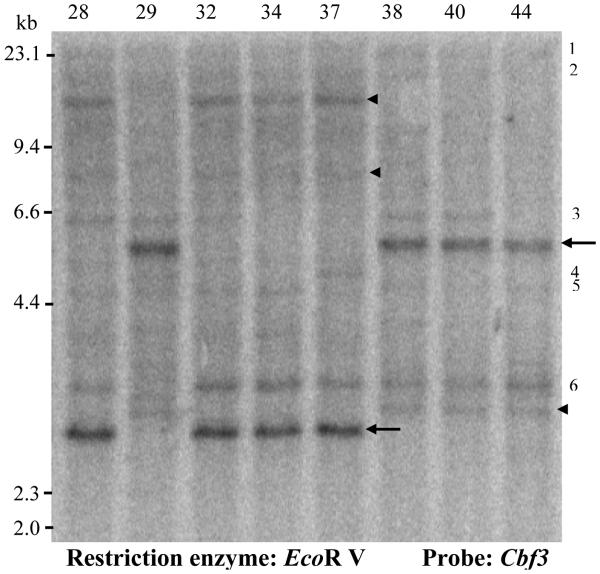
Barley clone Cbf3 hybridized with multiple RFLP fragments (Fig. 3) in T. monococcum, suggesting the presence of additional genes with at least partial sequence similarity to Cbf3. The RFLP fragment with stronger hybridization signal (Fig. 3, arrows) was mapped on the XCbf3 locus on the long arm of chromosome 5A completely linked to RFLP locus Xbcd508. This RFLP locus was located at the center of the confidence intervals for the QTLs for frost tolerance and for Cor14b transcription levels at 15°C (Fig. 1).
Fig. 3.
Southern blot of eight RILs from the DV92 × G3116 mapping population. DNAs were digested with restriction enzyme Eco RV and hybridized with the complete barley Cbf3 cDNA as a probe. Arrows indicate the RFLP fragments with strong hybridization signal mapped at the XCbf3 locus. Arrowheads indicate two additional RFLP fragments that cosegregate with the previous ones. Arrows and arrowheads on the left indicate G3116 alleles and on the right DV92 alleles. Numbers on the right of the figure indicate additional RFLP fragments with low hybridization signal that were not mapped.
Two additional restriction fragments with low hybridization signal (Fig. 3, arrowheads) cosegregate with the previous RFLP fragments, suggesting the possibility of multiple CBF-like genes at this locus. A similar result was found in barley (Choi et al. 2002). The other RFLP fragments with low hybridization signal were not polymorphic or did not cosegregate with the XCbf3 locus (e.g. RFLP indicated by numbers 3, 4, and 6 in the right side of Fig. 3).
Effect of vernalization requirement locus Vrn-A2 on frost tolerance
The 74 RILs were grown in the greenhouse without vernalization and growth habit was determined for each line. Growth habit showed complete linkage to RFLP marker Xbcd402 that was previously mapped completely linked at to the Vrn-A2 locus in the F2 population (Dubcovsky et al. 1998). This marker is not shown in Fig. 1 because it is 90 cM distal to the XCbf3 locus.
The Vrn-A2 locus showed only a marginal LOD score of two in the initial QTL analysis for frost tolerance, because of the simultaneous segregation of the Fr-A2 locus in this population. To increase the power to detect the effect of the Vrn-A2 locus on frost tolerance, a factorial analysis of variance was performed using the Fr-A2 and Vrn-A2 alleles as classification variables (heterozygous loci were coded as missing data). Significant differences in frost tolerance scores were detected for both Fr-A2 and Vrn-A2 in the 1998 and 1999 tests and in the combined results (Table 1). The two-gene model explained 58% of the variation in average frost tolerance scores in this population (R2=0.58). Plants carrying the G3116 allele for winter growth habit showed significantly higher frost tolerance scores (average 1.08) than plants carrying the DV92 alleles (average 0.65).
Table 1.
Factorial analysis of variance for frost tolerance scores and transcription levels of Cor14b at 15° C using Fr-A2 and Vrn-A2 as classification variables.
| Fr-A2 | Vrn-A2 | Interaction | |
|---|---|---|---|
| Frost tolerance1998 | P< 0.0001 | P= 0.0007 | P= 0.3605 |
| Frost tolerance1999 | P= 0.0001 | P= 0.0395 | P= 0.1625 |
| Frost tolerance average 2 years | P< 0.0001 | P= 0.0003 | P= 0.8589 |
| Cor14b transcription level | P< 0.0001 | P= 0.9466 | P= 0.8728 |
The levels of Cor14b transcripts at 15 °C showed significant differences when plants were grouped by the Fr-A2 alleles but no significant differences when they were grouped by the Vrn-A2 alleles.
The interactions between the two loci on frost tolerance were not significant in any of the analyses (Table 1). However, this result should be interpreted with caution because of the limited sample size of this study and the possibility that the developmental stage of the shoot apexes at the time of the frost tolerance test (not determined in this study) may influence the effect of this locus on frost tolerance.
DISCUSSION
Frost tolerance
Chromosome 5A from T. monococcum is colinear with chromosome 5A from wheat (Dubcovsky et al. 1996; Gale et al. 1995) facilitating the comparison of the results from this study with previous studies of frost tolerance in polyploid wheats. The major gene for frost tolerance Fr-A1 was mapped in hexaploid wheat on the long arm of chromosome 5A closely linked to RFLP marker Xwg644 (Galiba et al. 1995; Sutka et al. 1999). The same marker showed no association with frost tolerances in this T. monococcum mapping population indicating that the parental lines DV92 and G3116 did not differ at the Fr-A1 locus. This lack of segregation at the Fr-A1 locus increased the power of the QTL analysis to detect the new Fr-A2 locus.
The new frost tolerance locus Fr-A2 was mapped in diploid wheat 30-cM proximal to RFLP marker Xwg644, known to be tightly linked to Fr-A1 in polyploid wheat. Although no frost tolerance locus has been described before in the Fr-A2 region in polyploid wheats, it is possible that allelic variation at the Fr-A2 locus might have been obscured by simultaneous variation at a linked Fr-A1 locus. The presence of two loci for frost tolerance on chromosome 5A has been suggested more than 10 years ago based on inheritance studies using crosses between 'Winalta' and 'Winalta'-'Rescue'-5A chromosome substitution lines (Roberts 1990). The discovery of a locus controlling the expression of the Cor14b gene on the Fr-A2 region in T. aestivum chromosome 5A (Rcg1, for Regulator for cor14b gene) suggests that allelic variation at the Fr-A2 locus is also present in polyploid wheats (Vágújfalvi et al. 2000).
The Rcg1 locus was mapped linked to Xpsr911 on the long arm of chromosome 5A of polyploid wheat (Vágújfalvi et al. 2000). This RFLP marker was mapped 36 cM from the centromere and 35 cM proximal to Xpsr426, a marker tightly linked to Fr-A1 on chromosome 5A (Gale et al. 1995; Galiba et al. 1995). The Fr-A2 locus, which was also associated with the control of the expression of Cor14b (Fig.1), was mapped in this study completely linked to Xbcd508, 35 cM from the centromere and 35 cM proximal to Xpsr426 in T. monococcum (Dubcovsky et al. 1996). Furthermore, RFLP marker Xksu8 located 17 cM proximal to the Xpsr911 (Rcg1) locus on chromosome 5B (Gale et al. 1995) was mapped 18 cM proximal to the Xbcd508 locus on chromosome 5D (Gill et al. 1996). Similar distances to common reference markers suggest that the Rcg1 locus in T. aestivum and the Fr-A2 locus in T. monococcum are located in the same chromosome region.
Studies with chromosome substitution lines have shown that all three chromosomes of homoeologous group 5 carry major genes for frost tolerance (Sutka 2001). Since chromosomes 5A, 5B and 5D are colinear in the Fr1- Fr-2 region it is possible for each chromosome to carry different allelic variants for frost tolerance at each of the two loci. The complex segregation patterns resulting from these six genes would limit the success of breeding efforts to combine the best alleles for frost tolerance. The new molecular markers generated in this study for the Fr2 locus will facilitate the characterization and selection of optimum allele combinations for frost tolerance. RFLP markers mapped in the Fr-A2 region can be also used to develop isogenic lines for the 5B and 5D Fr2 region to test for the presence of allelic differences in Cor14b regulation or frost tolerance in these chromosomes.
The presence of two loci for frost tolerance in the same chromosome arm can also complicate the interpretation of orthologous relationships among frost tolerance loci on chromosomes from homoeologous group 5. For example, it was suggested that a QTL for frost tolerance designated QFr.jic-5D and mapped in the long arm of chromosome 5D was homeoallelic to Fr-A1 (Snape et al. 2001; Snape et al. 1997). However, the authors also pointed out that no clear segregational patterns were apparent in their mapping population, and that the peak of the QFr.jic-5D was mapped on a more proximal position (10 cM proximal to Vrn-D1) than Fr-A1 (2 cM proximal to Vrn-A1). This data should be reexamined for the possibility of simultaneous segregation at both Fr-D1 and Fr-D2 before deciding the allelic relationship between QFr.jic-5D and the two frost tolerance genes on chromosome 5A.
Effects of vernalization requirement on frost tolerance
The DV92 x G3116 mapping population segregates simultaneously for frost tolerance at the Fr-A2 locus and for vernalization requirement at the Vrn-A2 locus (Dubcovsky et al. 1998). The parental lines from this population have the same recessive vrn-A1 allele for winter growth habit and do not segregate at this locus, and the spring growth habit of DV92 is determined by the infrequent recessive vrn-A2 allele (Dubcovsky et al. 1998; Tranquilli and Dubcovsky 1999). The Vrn-A2 locus was mapped at the end of the long arm of chromosome 5A (in the region translocated from chromosome arm 4AL), approximately 60 cM distal to the Vrn-A1 locus and 90 cM distal to Fr-A2 (Dubcovsky et al. 1998). The independent segregation between Fr-A2 and Vrn-A2 in this mapping population provides a unique opportunity to study simultaneously the effects of frost tolerance and vernalization requirement on survival to freezing temperatures.
The higher frost tolerance observed in winter RILs compared to the spring RILS, indicates that growth habit plays a significant role in the determination of frost tolerance. The delay in the differentiation of the vegetative apex into the more sensitive reproductive apex might be involved in the increased frost tolerance of the plants with winter growth habit. A delay in the transition from vegetative to reproductive stage in barley plants grown under short day conditions was shown to increase the expression of cold induced genes and the level of frost tolerance compared to plants grown under long day conditions (Fowler et al. 2001).
The significant effect of growth habit on frost tolerance, suggests that some caution is necessary in the interpretation of the magnitude of the effect of the Fr-A1 locus on frost tolerance in polyploid wheats. Because of its close linkage with Vrn-A1 (2cM), the effect of growth habit is difficult to separate from the effect of Fr-A1 in polyploid wheats segregating simultaneously at both loci. The T. monococcum mapping population used in this work may provide a better tool to study the interaction between growth habit and frost tolerance.
Cor14b transcript level
In our mapping population, the non-significant differences in Cor14b regulation between the spring and winter RILs indicates that the frost tolerance conferred by the allele for winter growth habit was, not mediated by differences on the threshold temperature of induction of the Cor genes. On the other hand, the overlap between the QTLs for frost tolerance and for Cor14b transcript levels suggests that frost tolerance conferred by the Fr-A2 locus might be mediated by the differential regulation of the Cor genes.
The Cor14b gene was mapped on chromosomes 2A but was regulated by two genes located on chromosome 5A and linked to the Fr-A1 and Fr-A2 loci in polyploid wheat (Vágújfalvi et al. 2000). Similarly, different expression levels of cold regulated gene wcs120 between frost resistant and susceptible wheat and rye cultivars were associated to variation at the Vrn-A1/ Fr-A1 region (Fowler et al. 1996). These observations suggest the possibility that the molecular basis for cold tolerance could be a regulatory gene able to control the simultaneous expression of many cold-related genes. After the discovery of the Arabidopsis Cbf transcription factors it has been suggested that the cereal loci controlling frost tolerance could represent Cbf cereal homologues but no direct evidence for this hypothesis was presented (Sarhan and Danyluk 1998).
Association of Cbf3 transcription factors with frost tolerance and differential Cor14b regulation
The Arabidopsis Cbf gene products bind to CRT (C-repeat)/DRE (dehydration responsive element) DNA regulatory sequences present in the promoters of many Cor genes, activating a regulon of genes involved in cold acclimation (Medina et al. 1999). The expression of the three Arabidopsis Cbf genes is regulated by cold but not by abscisic acid or dehydration, suggesting that they control the level of Cor gene expression and promote cold acclimation through an abscisic acid-independent pathway. It was also demonstrated that Cbf1 over expression in Arabidopsis induced coordinated Cor gene expression without a low-temperature stimulus (JagloOttosen et al. 1998). Constitutive expression of the cold-regulated Arabidopsis Cor15a gene increases both chloroplast and protoplast freezing tolerance (Artus et al. 1996).
Similar results to those reported in Arabidopsis have been found in different Triticeae species. The CRT/DRE core motif CCGAC recognized by the Cbf transcription factors is also present in the promoter sequences of cold-regulated genes from wheat (Ouellet et al. 1998) and barley (Dunn et al. 1998). Northern blot studies using a rye Cbf gene as a probe have shown that CBF-like transcripts accumulated rapidly (within 15-30 min) in both wheat and rye in response to low temperatures (Jaglo et al. 2001). More specific expression studies using quantitative PCR and Cbf3 specific primers have shown that the barley Cbf3 gene is transiently up-regulated by chilling treatment within 15 minutes of cold treatment (Choi et al. 2002). The cold induction of barley Cbf3 and the presence of CRT (C-repeat)/DRE (dehydration responsive element) DNA regulatory sequences in the promoter sequences of cold-regulated genes from wheat, make the Cbf3 gene an attractive candidate gene for frost tolerance. Although the promoter sequences of Cor14b have not yet been published, the genetic evidence reported in the present work suggest that the transcription of Cor14b but not of Ao86, could be under the control of CBF-like sequences. Notably, Ao86 is a member of the Blt14 gene family (Grossi et al. 1998), a class of sequences up-regulated in response to cold only through post-transcriptional mechanisms (Dunn et al. 1994; Phillips et al. 1997).
Choi at al (2002) mapped the Cbf3 gene in barley in a similar location to the one reported in this study, but as in previous studies they did not detect a frost tolerance effect associated with the Cbf3 region. Our study provides evidence for the first time for cosegregation of a Cbf gene, differences in the threshold temperatures of induction of Cor14b, and frost tolerance in wheat. However, the relationship between Cbf3 and frost tolerance needs further investigation because of the presence of multiple copies of Cbf like genes in cereals, some of them linked to Cbf3 (Fig. 3). Any of the Cbf like genes tightly linked to Cbf3 has a similar probability to be responsible for the observed differences in frost tolerance in this segregating population. We have recently initiated a systematic characterization of all the members of this family in T. monococcum to address these questions.
In summary, the Cbf3 transcription factor was mapped on the long arm of chromosome 5A in T. monococcum at the peak of the QTLs for frost tolerance and Cor14b differential regulation (Fig. 1). This association suggests the possibility that CBF-like transcriptional activators play a major role in the determination of frost tolerance in wheat. Demonstration of this relationship would be a fundamental step towards the biotechnological improvement of frost tolerance in wheat.
ACKNOWLEDGMENTS
J. Dubcovsky acknowledge financial support from the United States National Science Foundation (grant NSF-MTA-OTKA 0095051) and Department of Agriculture, National Research Initiative (grant 2000-1678); G. Galiba thanks the financial support of OTKA T34277, OMFB-02579/2000, NKFP4/038, Marton-3,2, by the CNR-MTA Hungarian Italian cooperation program (Project number: 15/MTA 401), and by the MTA-OTKA-NSF cooperation program (Project number: MTA 1/2001/A.; and L. Cattivelli the support of GENFUN-MIUR and MiToS-MiPAF. A. Vágújfalvi thanks for the János Bolyai Research Grant . The authors also express their gratitude to Tony Chen and Pat Hayes for supplying the Dicktoo CBF3 barley clone and to L. Stheli for excellent technical assistance.
REFERENCES
- Anderson JA, Ogihara Y, Sorrells ME, Tanksley SD. Development of a chromosomal arm map for wheat based on RFLP markers. Theoretical and Applied Genetics. 1992;83:1035–1043. doi: 10.1007/BF00232969. [DOI] [PubMed] [Google Scholar]
- Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin CT, Thomashow MF. Constitutive expression of the cold-regulated Arabidopsis thaliana Cor15a gene affects both chloroplast and protoplast freezing tolerance. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13404–13409. doi: 10.1073/pnas.93.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P, Vale G, Mazzucotelli E, Govoni C, Faccioli P, Stanca AM, Cattivelli L. The transcripts of several components of the protein synthesis machinery are cold-regulated in a chloroplast-dependent manner in barley and wheat. Journal of Plant Physiology. 2001;158:1541–1546. [Google Scholar]
- Cattivelli L, Baldi P, Crosatti C, Di Fonzo N, Faccioli P, Grossi M, Mastrangelo AM, Pecchioni N, Stanca AM. Chromosome regions and stress-related sequences involved in resistance to abiotic stress in Triticeae. Plant Molecular Biology. 2002;48:649–665. doi: 10.1023/a:1014824404623. [DOI] [PubMed] [Google Scholar]
- Cattivelli L, Bartels D. Molecular cloning and characterization of cold-regulated genes in barley. Plant Physiology. 1990;93:1504–1510. doi: 10.1104/pp.93.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D-W, Rodriguez EM, Close TJ. Barley Cbf3 gene identification , expression pattern, and map location. Plant Physiology. 2002;129:1–7. doi: 10.1104/pp.003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Galvez AF, Dvorak J. Comparison of the genetic organization of the early salt stress response gene system in salt-tolerant Lophopyrum elongatum and salt-sensitive wheat. Theoretical and Applied Genetics. 1994;87:957–964. doi: 10.1007/BF00225790. [DOI] [PubMed] [Google Scholar]
- Dubcovsky J, Lijavetzky D, Appendino L, Tranquilli G. Comparative RFLP mapping of Triticum monococcum genes controlling vernalization requirement. Theoretical and Applied Genetics. 1998;97:968–975. [Google Scholar]
- Dubcovsky J, Luo M-C, Zhong G-Y, Bransteiter R, Desai A, Kilian A, Kleinhofs A, Dvorak J. Genetic map of diploid wheat, Triticum monococcum L., and its comparison with maps of Hordeum vulgare L. Genetics. 1996;143:983–999. doi: 10.1093/genetics/143.2.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn MA, Goddard NJ, Zhang L, Pearce RS, Hughes MA. Low-temperature-responsive barley genes have different control mechanisms. Plant Molecular Biology. 1994;24:879–888. doi: 10.1007/BF00014442. [DOI] [PubMed] [Google Scholar]
- Dunn MA, White AJ, Vural S, Hughes MA. Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.) Plant Molecular Biology. 1998;38:551–564. doi: 10.1023/a:1006098132352. [DOI] [PubMed] [Google Scholar]
- Dvorak J, McGuire PE, Cassidy B. Apparent sources of the A genomes of wheats inferred from the polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome. 1988;30:680–689. [Google Scholar]
- Feinberg AP, Vogelstein P. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Analytical Biochemistry. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fowler DB, Breton G, Limin AE, Mahfoozi S, Sarhan F. Photoperiod and temperature interactions regulate low-temperature-induced gene expression in barley. Plant Physiology. 2001;127:1676–1681. [PMC free article] [PubMed] [Google Scholar]
- Fowler DB, Chauvin LP, Limin AE, Sarhan F. The regulatory role of vernalization in the expression of low-temperature-induced genes in wheat and rye. Theoretical and Applied Genetics. 1996;93:554–559. doi: 10.1007/BF00417947. [DOI] [PubMed] [Google Scholar]
- Gale MD, Atkinson MD, Chinoy CN, Harcourt RL, Jia J, Li QY, Devos KM. Genetic maps of hexaploid wheat. In: Li ZS, Xin ZY, editors. Proceedings 8th International Wheat Genetic Symposium. China Agricultural Scientech Press; Beijin: 1995. pp. 29–40. [Google Scholar]
- Galiba G, Quarrie SA, Sutka J, Morgounov A. RFLP mapping of the vernalization (Vrn1 ) and frost resistance (Fr1 ) genes on chromosome 5A of wheat. Theoretical and Applied Genetics. 1995;90:1174–1179. doi: 10.1007/BF00222940. [DOI] [PubMed] [Google Scholar]
- Gill KS, Gill BS, Endo TR, Boyko EV. Identification and high-density mapping of gene-rich regions in chromosome group 5 of wheat. Genetics. 1996;143:1001–1012. doi: 10.1093/genetics/143.2.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graner A, Jahoor A, Schondelmeier J, Siedler H, Pillen K, Fischbeck G, Wenzel G, Herrman RG. Construction of an RFLP map of barley. Theoretical and Applied Genetics. 1991;83:250–256. doi: 10.1007/BF00226259. [DOI] [PubMed] [Google Scholar]
- Grossi M, Giorni E, Rizza F, Stanca AM, Cattivelli L. Wild and cultivated barleys show differences in the expression pattern of a cold-regulated gene family under different light and temperature conditions. Plant Molecular Biology. 1998;38:1061–1069. doi: 10.1023/a:1006079916917. [DOI] [PubMed] [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiology. 2001;127:910–917. [PMC free article] [PubMed] [Google Scholar]
- JagloOttosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann. Eugen. 1943;12:172–175. [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly M, Lincoln SE, Newburg L. MAPMAKER: An integrated computer package for construction of primary linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- McIntosh RA, Hart GE, Devos KM, Gale MD, Rogers WJ. Proc. 9th Vol. 5. Int. Wheat Genetics Symp.; Saskatchewan, Canada: 1998. Catalogue of Gene Symbols for Wheat; pp. 1–235. [Google Scholar]
- Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiology. 1999;119:463–469. doi: 10.1104/pp.119.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Galiba G. Drought and salt tolerance are not necessarily linked: A study on wheat varieties differing in drought tolerance under consecutive water and salinity stresses. Journal of Plant Physiology. 1995;145:168–174. [Google Scholar]
- Ouellet F, Vazquez-Tello A, Sarhan F. The wheat wcs120 promoter is cold-inducible in both monocotyledonous and dicotyledonous species. FEBS Letters. 1998;423:324–328. doi: 10.1016/s0014-5793(98)00116-1. [DOI] [PubMed] [Google Scholar]
- Phillips JR, Dunn MA, Hughes MA. mRNA stability and localisation of the low-temperature-responsive barley gene family blt14. Plant Molecular Biology. 1997;33:1013–1023. doi: 10.1023/a:1005717613224. [DOI] [PubMed] [Google Scholar]
- Roberts DWA. Identification of loci on chromosome-5A of wheat involved in control of cold hardiness, vernalization, leaf length, rosette growth habit, and height of hardened plants. Genome. 1990;33:247–259. [Google Scholar]
- Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier M, Leroy P, Ganal MW. A microsatellites map of wheat. Genetics. 1998;149:2007–2023. doi: 10.1093/genetics/149.4.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai JW, Larcher V. Responses and adaptations to freezing stress. Springer-Verlag, Berlin, Heidelberg; New York: 1985. Frost survival of plants. [Google Scholar]
- Sarhan F, Danyluk J. Engineering cold-tolerant crops - throwing the master switch. Trends in Plant Science. 1998;3:289–290. [Google Scholar]
- Snape JW, Sarma R, Quarrie SA, Fish L, Galiba G, Sutka J. Mapping genes for flowering time and frost tolerance in cereals using precise genetic stocks. Euphytica. 2001;120:309–315. [Google Scholar]
- Snape JW, Semikhodskii A, Fish L, Sarma RN, Quarrie SA, Galiba G, Sutka J. Mapping frost tolerance loci in wheat and comparative mapping with other cereals. Acta Agronomica Hungarica. 1997;45:268–270. [Google Scholar]
- Sutka J. Genetic control of frost tolerance in wheat (Triticum aestivum L. Euphytica. 1994;77:277–282. [Google Scholar]
- Sutka J. Genes for frost resistance in wheat. Euphytica. 2001;119:167–172. [Google Scholar]
- Sutka J, Galiba G, Vagujfalvi A, Gill BS, Snape JW. Physical mapping of the Vrn-A1 and Fr1 genes on chromosome 5A of wheat using deletion lines. Theoretical and Applied Genetics. 1999;99:199–202. [Google Scholar]
- Sutka J, Snape JW. Location of a gene for frost resistance on chromosome 5A of wheat. Euphytica. 1989;42:41–44. [Google Scholar]
- Thomashow MF, Gilmour SJ, Stockinger EJ, Jaglo-Ottosen KR, Zarka DG. Role of the Arabidopsis CBF transcriptional activators in cold acclimation. Physiologia Plantarum. 2001;112:171–175. [Google Scholar]
- Tranquilli GE, Dubcovsky J. Epistatic interactions between vernalization genes Vrn-Am1 and Vrn-Am2 in diploid wheat. Journal of Heredity. 1999;91:304–306. doi: 10.1093/jhered/91.4.304. [DOI] [PubMed] [Google Scholar]
- Vágújfalvi A, Crosatti C, Galiba G, Dubcovsky J, Cattivelli L. Two loci on wheat chromosome 5A regulate the differential cold-dependent expression of the cor14b gene in frost tolerant and sensitive genotypes. Molecular and General Genetics. 2000;263:194–200. doi: 10.1007/s004380051160. [DOI] [PubMed] [Google Scholar]
- Veisz O, Sutka J. Ditelosomic analysis of frost resistance in wheat (Cv Chinese Spring) Cereal Research Communications. 1993;21:263–267. [Google Scholar]