Abstract
We previously demonstrated the feasibility of using silicon double-sided strip detectors (DSSDs) for SPECT imaging of the activity distribution of iodine-125 using a 300-micrometer thick detector. Based on this experience, we now have developed fully customized silicon DSSDs and associated readout electronics with the intent of developing a multi-pinhole SPECT system. Each DSSD has a 60.4 mm × 60.4 mm active area and is 1 mm thick. The strip pitch is 59 micrometers, and the readout of the 1024 strips on each side gives rise to a detector with over one million pixels. Combining four high-resolution DSSDs into a SPECT system offers an unprecedented space-bandwidth product for the imaging of single-photon emitters. The system consists of two camera heads with two silicon detectors stacked one behind the other in each head. The collimator has a focused pinhole system with cylindrical-shaped pinholes that are laser-drilled in a 250 μm tungsten plate. The unique ability to collect projection data at two magnifications simultaneously allows for multiplexed data at high resolution to be combined with lower magnification data with little or no multiplexing. With the current multi-pinhole collimator design, our SPECT system will be capable of offering high spatial resolution, sensitivity and angular sampling for small field-of-view applications, such as molecular imaging of the mouse brain.
I. Introduction
This work presents the design characteristics of a dual-headed small-animal SPECT based on silicon double-sided strip detectors. The high intrinsic detector resolution (~ 60 μm) [1] is combined with a multi-pinhole collimator system for task-specific applications in molecular imaging that require both high resolution and sensitivity in a small field of view such as mouse brain. We are particularly interested in detecting and visualizing micro-structures such as Amyloid beta plaques [2] that aggregate in hippocampus and frontal cortex during the progress of Alzheimer’s disease. There are a number of 125I-based tracers available that can bind to these plaques, and their low-energy radiation can be detected with silicon-based detectors.
II. Silispect, Dual-headed Spect With Silicon Dssds
Fig. 1 shows the dual-headed stationary SiliSPECT scanner with two silicon DSSDs stacked in each camera head. The silicon DSSDs have an active area of 60 mm × 60 mm and a thickness of 1mm.
Fig. 1.
A photograph of SiliSPECT, a dual-headed SPECT with two silicon DSSDs stacked in each camera head.
The stacked detector design compensates the modest detection efficiency of silicon (39% at around 30 keV). The front detectors contribute to 64% of the detected events and the back detectors to 36 %. This configuration enables the front detectors to acquire projections at a different magnification than the back detectors. We intend to use this type of acquisition, known as synthetic collimation [3], for 3D image reconstruction using a limited number of projection angles. Fig. 2 shows an example of projection images on the front/back detectors at two orthogonal camera views with a pair of brachytherapy seeds acquired using a multi-pinhole collimator.
Fig. 2.
The stacked acquisition geometry (left) and the projection images (right) on stacked detectors at two orthogonal camera views. The magnification on the front detectors (upper images) is 0.6 and the back detectors (lower images) 1.6. The phantom consists of a pair of brachytherapy seeds with a total activity of 63 μCi. The acquisition time was 87 minutes.
We used an off-line algorithm to sort coincident events between independently triggered P and N strips. These strips are orthogonal on the two detector sides, so each pair of coincident strips determines the coordinates of the photon interaction in silicon. The magnitude of photon scatter in the front detectors is not significant enough to degrade the back detector projection images. This fact was verified by comparing the projection profiles on the back detectors with and without the presence of the front detectors with respect to their full width at half maxima and the signal-to-background ratios [1].
III. Multi-pinhole Collimator For Silispect
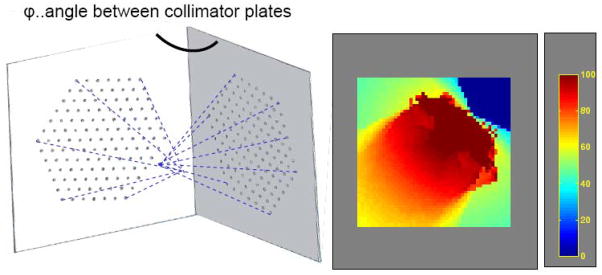
The experimental projection data were acquired using a multi-pinhole collimator system with two collimator plates each made of a 250 μm thick tungsten layer. 127 pinholes were laser-drilled through each collimator plate resulting in a total number of 254 pinholes of 250 μm diameter. The pinhole pitch is 2.5 mm. The pinholes are cylindrical in shape and are all tilted toward a common focal point at a distance of 30 mm away from the collimator surface. In a dual-headed camera the angle between the two collimator plates can be adjusted in a way that an overlap region with relatively high sampling completeness [4] is created (Fig. 3).
Fig. 3.
Schematic of focused pinhole systems on each collimator plate and the overlap region created with two collimators (left). A single axial slice of a 3D sampling map (Metzler et al. 2002).
Cylindrical pinholes can be considered as knife-edge pinholes with minimum (almost zero) opening angle. This geometry not only makes the penetrative component of the collimator sensitivity insignificant [5] but also causes a steeper fall off of the geometric sensitivity across the field of view (Fig. 4). Particularly at small source-collimator distances (such as 20–25 mm in our application) the geometric sensitivity profile becomes a function of the pinhole opening angle and collimator thickness (Fig. 5). At larger source-collimator distances and pinhole opening angles the sensitivity profile follows again the conventional formulation: cos3(θ).d3/16h2.
Fig. 4.
Left: The collimator system of SiliSPECT is characterized by small pinhole opening angle and short source-collimator normal distances. Under these geometric conditions, the geometric sensitivity profile (right) becomes a function of pinhole opening angle and collimator thickness, and can be made very narrow. At larger source-collimator distances and pinhole opening angle the geometric sensitivity fall-off follows the conventional formulation (dashed line).
Fig. 5.
Left: At small pinhole opening angles the geometric sensitivity profile can be varied with the collimator thickness, whereas with larger pinhole angles (right) the collimator thickness does not impact the geometric sensitivity, which follows the conventional term (cos3(θ).d3/16h2).
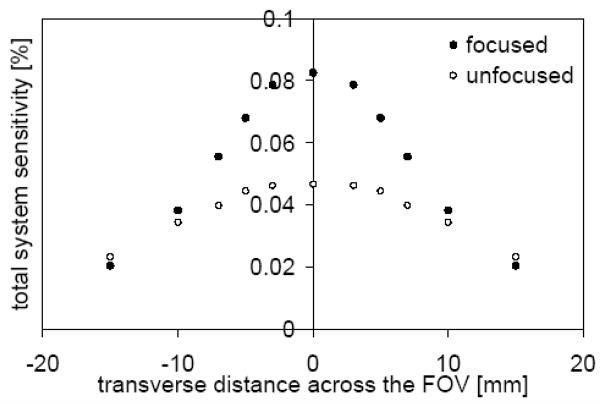
Our main goal in utilizing a focused pinhole system is achieving high sensitivity in a small region and being able to reduce the system sensitivity for photons emitted from background regions. Fig. 6 shows an enhancement of the total system sensitivity by almost a factor of two at the center of the FOV compared to a pinhole system that uses the same pinhole pattern but with non-tilted pinholes.
Fig. 6.
The total system sensitivity (including a dual-headed camera with stacked, one mm thick silicon detectors) for a pair of collimators (each 127 pinholes) and a focused pinhole system compared to an unfocused pinhole system.
IV. Sumary And Discussion
We have highlighted the design characteristics of SiliSPECT for some task-specific molecular imaging applications that require good sensitivity/resolution to resolve microstructures in very small fields such as the whole mouse brain or even a small region within the brain. An example of such a task-specific application is the visualization of Amyloid beta plaques in hippocampus and frontal cortex. Fig. 7 shows our initial attempts of reconstructing microstructures (diameter: 60 – 250 μm) using a simulation study with stacked detectors in a dual-headed camera and a pair of 127 focused multi-pinhole collimators. A total of 5,000,000 projection counts were generated, and 25 ML-EM iterations were used to reconstruct the 3D image. The image field is 0.2 mm × 0.2 mm × 0.128 mm.
Fig. 7.
Simulation study with SiliSPECT. The phantom projections were acquired at two different magnifications, M1 = 1, M2 = 1.5 (top). The phantom consists of microstructure activity hot spots of diameters between 60–250 μm (lower, left). A slice from simulated data reconstructed using the MLEM algorithm is shown in the lower right.
Our pinhole system is designed with respect to high-resolution silicon detectors and the potential imaging tasks. Based on the simulation studies, we showed that despite using silicon as detector material, which has low stopping power, relatively high system sensitivity is achievable with a focused pinhole geometry. Due to the narrow sensitivity profile, the high sensitivity and sampling completeness regions are limited to small objects. This condition could be considered as an advantage by reducing the impact of background radiation on the pinhole projections. The small pinhole pitch of 2.5 mm in our current collimators is expected to be sufficient for acquiring multi-pinhole projection images from mouse hippocampus with minimum projection overlap (multiplexing). Implementing the concept of synthetic collimation with high resolution and stacked detectors, the front detectors can acquire projections at magnification levels as small as 0.5. The back detectors can have higher magnification and possible multiplexing. Our initial simulation studies show feasible image reconstruction of microstructures under these conditions. In the future, we will extend our simulation studies to more complex and realistic microstructure phantoms with the presence of out-of-field background to study its impact on image reconstruction with synthetic collimation.
Acknowledgments
This work was supported by the NIH/NIBB grants R33 EB000776, P41 EB002035 and a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Contributor Information
Sepideh Shokouhi, Email: sepideh.shokouhi@vanderbilt.edu, Vanderbilt University, Nashville, TN, USA, telephone: 615-322-6214.
Mark A. Fritz, Vanderbilt University, Nashville, TN, USA, telephone: 615-322-6214
Benjamin S. McDonald, Vanderbilt University, Nashville, TN, USA, telephone: 615-322-6214
Heather L. Durko, University of Arizona, Center for Gamma-Ray Imaging and the College of Optical Sciences, Tucson, AZ, USA, telephone: 520-626-4256
Lars R. Furenlid, Email: furen@radiology.arizona.edu, University of Arizona, Center for Gamma-Ray Imaging and the College of Optical Sciences, Tucson, AZ, USA, telephone: 520-626-4256
Donald W. Wilson, Email: dwwilson@radiology.arizona.edu, University of Arizona, Tucson, AZ, USA, telephone: 520-626-9433
Todd E. Peterson, Email: todd.peterson@vanderbilt.edu, Vanderbilt University, Nashville, TN, USA, telephone: 615-322-2648
References
- 1.Shokouhi S, McDonald BS, Durko HL, Fritz M, Furenlid LR, Peterson TE. Performance characteristics of thick silicon double-sided strip detector. IEEE NSS-MIC Conference; 2007; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shokouhi S, Wilson WD, Pham W, Peterson TE. System evaluation for In Vivo imaging of Amyloid beta plaques in a mouse brain using statistical decision theory. IEEE NSS-MIC Conference; 2007. [Google Scholar]
- 3.Wilson DW, Barrett HH, Clarkson EW. Reconstruction of two- and three-dimensional images from synthetic-collimator data. IEEE Trans Med Imaging. 2000;19:412–22. doi: 10.1109/42.870252. [DOI] [PubMed] [Google Scholar]
- 4.Metzler SD, Bowsher JE, Jaszczak RJ. Geometrical similarities of the Orlov and Tuy sampling criteria and a numerical algorithm for assessing sampling completeness. IEEE Trans Med Imaging. 2003;50:1550–55. [Google Scholar]
- 5.Metzler SD, Bowsher JE, Smith MF, Jaszczak RJ. Analytic determination of pinhole collimator sensitivity with penetration. IEEE Trans Med Imaging. 2001;20:730–41. doi: 10.1109/42.938241. [DOI] [PubMed] [Google Scholar]