Abstract
Schimke Immunoosseous Dysplasia (SIOD) is a rare, autosomal recessive disorder of childhood characterized by spondyloepiphyseal dysplasia, focal segmental glomerulosclerosis and renal failure, T-cell immunodeficiency, and cancer in certain instances. Approximately half of patients with SIOD are reported to have biallelic mutations in SMARCAL1 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin, subfamily a-like 1), which encodes a DNA translocase that localizes to sites of DNA replication and repairs damaged replication forks. We present a novel mutation (NM_014140.3:c.2070+2insT) that results in defective SMARCAL1 mRNA splicing in a child with SIOD. This mutation, within the donor site of intron 12 results in the skipping of exon 12, which encodes part of a critical hinge region connecting the two lobes of the ATPase domain. This mutation was not identified as deleterious by diagnostic SMARCAL1 sequencing, but discovered through next generation sequencing, confirmed by Sanger sequencing, and found to result in absent SMARCAL1 expression in patient-derived lymphoblasts. The splicing defect caused by this mutation supports the concept of exon definition. Furthermore, it illustrates the need to broaden the search for SMARCAL1 defects in patients with SIOD lacking coding sequence mutations.
Keywords: SMARCAL1, DNA replication, RNA splicing, DNA repair, cancer
Introduction
Schimke Immunoosseous Dysplasia (SIOD) is a rare, autosomal recessive disorder of childhood characterized by spondyloepiphyseal dysplasia, focal segmental glomerulosclerosis and renal failure, T-cell immunodeficiency, and cancer in certain instances [Boerkoel et al., 2002; Carroll et al., 2013; Ehrich et al., 1990; Hunter et al., 2010; Spranger et al., 1991]. Approximately half of patients with SIOD are reported to have biallelic mutations in SMARCAL1 (SWI/SNF-related matrix-associated actin-dependent regulator of chromatin, subfamily a-like 1), which encodes the DNA translocase SMARCAL1 [Boerkoel et al., 2002; Clewing et al., 2007].
This protein localizes to stalled replication forks [Bansbach et al., 2009; Ciccia et al., 2009; Yuan et al., 2009; Yusufzai et al., 2009] where it catalyzes remodeling of damaged forks in a reaction regulated by the single-strand DNA binding protein Replication protein A (RPA) [Betous et al., 2013; Betous et al., 2012]. This remodeling is required to repair damaged forks and SIOD-associated SMARCAL1 mutations inactivate this function. Thus, SMARCAL1 is a genome maintenance protein and SIOD is a genome maintenance disorder [Bansbach et al., 2009; Bansbach et al., 2010].
A diversity of SMARCAL1 variants have been linked to SIOD [Clewing et al., 2007]. Published exon variants span the entirety of the gene while splice site variants involve the donor splice site consensus sequences of introns 4, 5, and 6. Accurate splicing requires a combination of canonical donor and acceptor splice site consensus sequences, branch points, and enhancers and silencers of splicing both within and outside of exons [De Conti et al., 2012]. Here we describe the clinical and molecular characterization of a patient with SIOD and an unusual SMARCAL1 splice-site mutation.
Materials and Methods
Transformation of patient B lymphocytes
Lymphocyte isolation and immortalization using Epstein Barr virus was completed by the University of North Carolina Lineberger Cancer Center Tissue Culture Facility.
Isolation of patient DNA and next generation sequencing
Genomic DNA (gDNA) was extracted using standard proteinase K digestion and phenol extraction. The gDNA was isolated from both untransformed and EBV-transformed patient cells. The untransformed gDNA was submitted for next generation sequencing, while transformed cells were used for all other experiments. The patient sample was analyzed with informed consent on an IRB-approved protocol.
Paired-end (100 bp) sequencing of short-term cultured patient peripheral blood lymphocyte gDNA was performed on an Illumina HiSeq2000. A total of 509 million pair-end 100 base pair long reads were sequenced, and 97% of all reads were properly mapped. Thorough quality control was conducted based on multi-stage strategies using QC3 [Guo et al., 2014]. Alignment against the human hg19 reference genome was conducted using the Burrows-Wheeler Alignment tool (BWA) [Li and Durbin 2009]. Local realignment, recalibration and SNP/Indel analyses were carried out using Genome Analysis Tool Kit (GATK) [DePristo et al., 2011].
Immunoblotting and splicing analyses
Patient lymphoblasts and control normal lymphoblastoid cells (GM00130C, Coriell Institute) were lysed in 0.5% NP40 buffer containing 150mM NaCl, 1mM EDTA, 1mM DTT and protease inhibitors. Expression of SMARCAL1 was checked in both cell types by separating lysate on an 8% polyacrylamide gel and blotting with antibodies raised to the C-terminus and N-terminus of SMARCAL1 [Bansbach et al., 2009].
The RNA was isolated from normal lymphoblasts and patient-derived B lymphoblasts using the Qiagen RNeasy kit.
Results
Patient Characteristics
The patient was a Caucasian female of English, German, and French ancestry who came to our attention at 6 years of age due to persistent proteinuria. She was noted at the time to be short of stature (height < 3rd percentile). Her medical history was significant for disseminated Herpes Zoster. A renal biopsy revealed focal segmental glomerulosclerosis, skeletal survey showed spondyloepiphyseal dysplasia, and an immunologic work-up revealed significantly diminished CD4 and CD8 T cells. A clinical diagnosis of SIOD was made. A detailed family history revealed no additional cases of short stature or renal disease. Parents were reportedly nonconsanguineous and the patient had one full sibling who was healthy and normal in stature, and two maternal half-siblings who were healthy and normal in stature.
Identifying nucleotide variants in SMARCAL1 in patient-derived genomic DNA
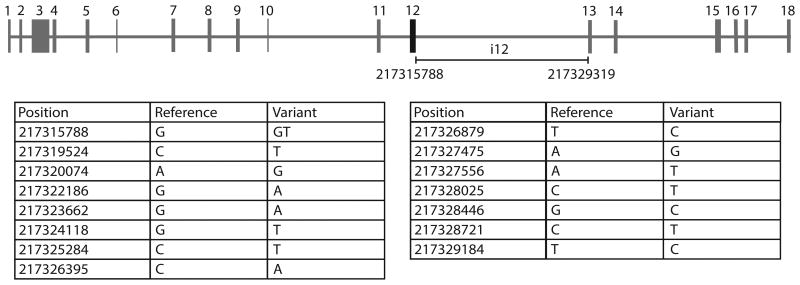
Diagnostic sequencing of SMARCAL1 exons and exon/intron boundaries in the patient failed to reveal a deleterious mutation. Mutations in SMARCAL1 are not apparent in approximately half of patients with SIOD tested by this method. Thus, we hypothesized that this patient's disease was due to another gene mutation, an unidentified mutation in a SMARCAL1 regulatory sequence, or possibly epigenetic silencing of SMARCAL1 expression. Genomic DNA from the patient-derived lymphocytes (prior to immortalization) was isolated and submitted for next generation sequencing. The gene is located on chromosome 2q35 spanning positions 217277473 to 217347774 of the hg19 reference genome, giving it a length of 70,302 nucleotides. It is comprised of 18 exons with exon 1 and exon 2 being untranslated (Figure 1). While no exonic variants were found in SMARCAL1, a number of intronic variants were identified, including a variant within the donor splice site consensus sequence of intron 12 (i12), resulting in a thymine insertion between positions 217315788 and 217315789 of the hg19 reference genome (NM_014140.3:c.2070+2insT or AC098820.3:g.115688insT). This mutation changes the exon 12/intron 12 boundary from ACT/GTG to ACT/GTTG. This was the only splice site variant identified in the entire gene, but one of 15 variants identified in intron 12 (Figure 1).
Figure 1.
Multiple nucleotide variants were identified in intron 12 of SMARCAL1 by next generation sequencing. One of these was a splice variant (NM_014140.3:c.2070+2insT). While the functional consequences of the variants are of uncertain significance, none of the nucleotide changes create obvious cryptic splice sites.
SMARCAL1 is not expressed in patient-derived lymphoblasts
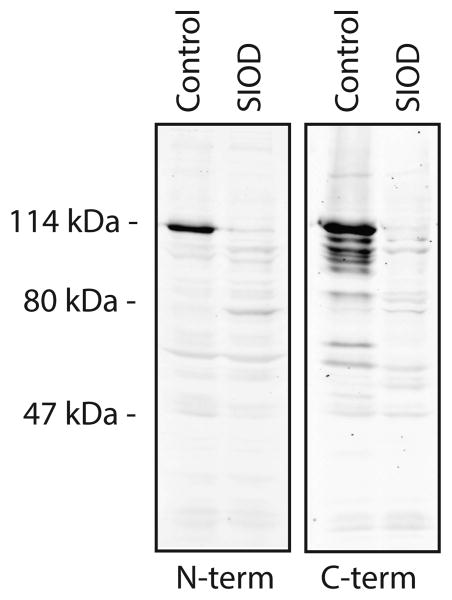
Since the minimal CT/GT exon/intron sequence is retained, it was not immediately clear whether the sequence variant would affect SMARCAL1 expression. Therefore, we looked for SMARCAL1 protein in immunoblots of whole cell lysates from control lymphoblasts and patient lymphoblasts using custom antibodies raised to the N-terminus and C-terminus of the protein (Figure 2). Full-length SMARCAL1 was not visible in whole cell lysate from patient-derived B lymphoblasts using either antibody (Figure 2). A faint, unique band that migrates just below 80 kDa in the patient-derived lysate was visible in immunoblots with the N-terminal antibody (Figure 2). However, this band was not observed with the C-terminal antibody.
Figure 2.
Patient-derived lymphoblasts do not express SMARCAL1. Lysates of normal and patient-derived lymphoblasts were separated by SDS-PAGE and immunoblotted with antibodies raised to the N-terminus or C-terminus of SMARCAL1.
Identifying a SMARCAL1 splicing defect by RT-PCR from patient-derived cDNA
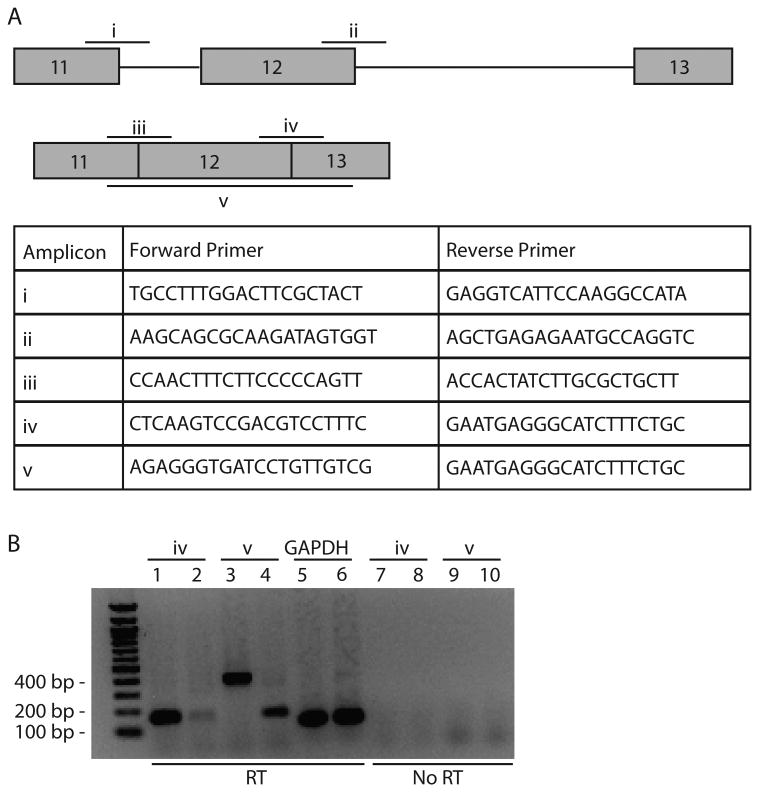
Based on the sequence alteration and the lack of SMARCAL1 expression, we tested if the donor site variant alters splicing of the mRNA transcript using RT-PCR. Primer pairs spanning the exon 11/intron 11 and exon 12/intron 12 transitions (i and ii in Figure 3A) did not produce RT-PCR products from either control or patient mRNA as would be expected (data not shown). The primer pair spanning the exon 12/exon 13 junction (iv in Figure 3A) yielded the expected product from control RNA (lane 1 in Figure 3B) but very little product from patient-derived RNA (lane 2 in Figure 3B).
Figure 3.
Patient cells expressed an aberrant SMARCAL1 mRNA. (A) The primers for RT-PCR were designed to span the indicated exon-intron and exon-exon junctions. (B) The RNA was isolated from control and patient-derived lymphoblasts. The RT-PCR products were generated using primer pairs iv (lanes 1,2) and v (lanes 3,4). The RT-PCR reactions for GAPDH were included as positive controls (lanes 5,6). Reactions omitting the reverse-transcriptase step were included as negative controls (lanes 7-10).
The primer pair spanning the whole of exon 12 (v in Figure 3A) yielded different sized products from control and patient-derived RNA (lanes 3 and 4 in Figure 3B). The PCR product in lane 3, derived from control RNA, migrates at around 400 bp, which is the expected size if exon 12 is incorporated appropriately into mRNA. However, the product in lane 4, derived from patient RNA, migrates at just under 200 bp suggesting aberrant splicing. Again, we observed a difference in band intensity between control and patient conditions. Primers for GAPDH were included in this experiment as positive controls and yielded appropriately sized and abundant RT-PCR fragments in both conditions (lanes 5 and 6 in Figure 3B). Reactions with the reverse transcription step omitted were included as negative controls (lanes 7-10 in Figure 3B).
Confirming a SMARCAL1 splicing defect by Sanger sequencing
Sanger sequencing of the control PCR product in lane 3 of Figure 3B revealed the expected joining of exon 11 and exon 12 (Figure 4A). Sequencing of the patient-derived PCR product in lane 4 of Figure 3B confirmed an aberrant joining of exon 11 and exon 13 with no shift in the overall reading frame (Figure 4B).
Figure 4.
Sequencing of amplified cDNA from patient-lymphoblasts confirmed a SMARCAL1 splicing defect. (A and B) The PCR products from control (lane 3, Figure 3B) and patient (lane 4, Figure 3B) RT-PCR reactions were sequenced. (A) The control yielded the expected cDNA sequence containing exons 11, 12 and 13. (B) The patient cDNA sequence lacked exon 12 and showed aberrant joining of exons 11 and 13. The variant splice site in the patient sample is indicated (*).
Discussion
We present a child with SIOD in whom we identified a splice site variant affecting the donor splice site consensus sequence within intron 12 of SMARCAL1. This variant consists of a thymine insertion, which does not affect the -3, -2, or -1 positions of the intra-exonic donor consensus sequence, and does not change the canonical dinucleotide at the start of intron 12, but shifts the remainder of the consensus sequence in the 3′ direction by one position. That this insertion does not alter the canonical nucleotides at +1 and +2 may explain why the variant was not reported by diagnostic sequencing. Positions +3 to +6 of the donor consensus sequence can vary from intron to intron. The shifting of these nucleotides may not always result in a defect, especially when guanine and thymine are retained at +1 and +2 respectively.
Each position of the donor splice site consensus sequence is potentially important for proper exon definition and splicing. While variants at the +1, +2, and +5 positions are most commonly linked to human disease, variants at any position can be deleterious [Buratti et al., 2007]. We hypothesize that the shift in the donor splice site consensus sequence brought about by this mutation explains the patient's apparent lack of SMARCAL1 expression and clinical phenotype. However, our sequencing revealed additional variants within intron 12. We cannot exclude the possibility that one or more contribute to altered splicing in this case.
The sequencing data did not indicate any evidence for copy number variations within the SMARCAL1 gene region, suggesting the mutation is homozygous. However, we cannot exclude other possibilities in the absence of additional family data.
The variant in the patient is the only one described to affect splicing of an exon that encodes for part of the critical SNF2 motor domain of SMARCAL1. Deletion of exon 12 results in loss of amino acids 618-690, which comprises the hinge domain between the two lobes of the ATPase domain. Skipping exon 12 does not alter the reading frame, so no premature stop codon is created. Exactly why the faulty protein is not expressed in the present patient is unclear, but it may be unstable due to improper protein folding.
Our findings illustrate the phenomenon of exon definition, which in its original conception argued that exons are recognized and defined as units during early assembly by binding of factors to the 3′ end of the intron, followed by a search for the downstream 5′ splice site [Robberson et al., 1990]. In the present patient, an insertion affecting the donor splice site consensus sequence traversing the exon 12/intron 12 transition affects exon 12 definition. The exons prior and subsequent to exon 12 are unaffected.
It is widely cited in the SIOD literature that nearly half of patients with SIOD have no detectable SMARCAL1 mutation [Clewing et al., 2007]. However, this conclusion is based on limited sequencing of the coding sequence and intron/exon boundaries. As our analysis shows, not all relevant SMARCAL1 mutations are identified (or characterized as deleterious) by the current sequencing methodology. We propose that when encountering a patient with a clear SIOD phenotype and no SMARCAL1 mutation identified by conventional sequencing of exons and exon/intron boundaries, the most logical way to proceed is to first identify whether the patient expresses SMARCAL1 in peripheral white blood cells. With the current availability of antibodies to multiple SMARCAL1 domains, this step is feasible, quick, and relatively inexpensive. In the absence of protein expression, further sequencing of gDNA or RNA would be warranted.
Acknowledgments
The DNA extraction and facilitation of genome sequencing was performed by the Vanderbilt Innovative Translational Research Shared Resource supported by the Vanderbilt-Ingram Cancer Center, the TJ Martell Foundation, and the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation. This research was supported from grant R01CA102729 and R01CA160432 to D.C. and 5K12CA090625-14 to C.C. The authors declare no conflicts of interest.
References
- Bansbach CE, Betous R, Lovejoy CA, Glick GG, Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23:2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansbach CE, Boerkoel CF, Cortez D. SMARCAL1 and replication stress: an explanation for SIOD? Nucleus. 2010;1:245–248. doi: 10.4161/nucl.1.3.11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Couch FB, Mason AC, Eichman BF, Manosas M, Cortez D. Substrate-selective repair and restart of replication forks by DNA translocases. Cell Rep. 2013;3:1958–1969. doi: 10.1016/j.celrep.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26:151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel CF, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, Andre JL, Bogdanovic R, Burguet A, Cockfield S, Cordeiro I, Frund S, Illies F, Joseph M, Kaitila I, Lama G, Loirat C, McLeod DR, Milford DV, Petty EM, Rodrigo F, Saraiva JM, Schmidt B, Smith GC, Spranger J, Stein A, Thiele H, Tizard J, Weksberg R, Lupski JR, Stockton DW. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet. 2002;30:215–220. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- Buratti E, Chivers M, Kralovicova J, Romano M, Baralle M, Krainer AR, Vorechovsky I. Aberrant 5′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2007;35:4250–4263. doi: 10.1093/nar/gkm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll C, Badu-Nkansah A, Hunley T, Baradaran-Heravi A, Cortez D, Frangoul H. Schimke Immunoosseous Dysplasia associated with undifferentiated carcinoma and a novel SMARCAL1 mutation in a child. Pediatr Blood Cancer. 2013;60:E88–90. doi: 10.1002/pbc.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009;23:2415–2425. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewing JM, Fryssira H, Goodman D, Smithson SF, Sloan EA, Lou S, Huang Y, Choi K, Lucke T, Alpay H, Andre JL, Asakura Y, Biebuyck-Gouge N, Bogdanovic R, Bonneau D, Cancrini C, Cochat P, Cockfield S, Collard L, Cordeiro I, Cormier-Daire V, Cransberg K, Cutka K, Deschenes G, Ehrich JH, Frund S, Georgaki H, Guillen-Navarro E, Hinkelmann B, Kanariou M, Kasap B, Kilic SS, Lama G, Lamfers P, Loirat C, Majore S, Milford D, Morin D, Ozdemir N, Pontz BF, Proesmans W, Psoni S, Reichenbach H, Reif S, Rusu C, Saraiva JM, Sakallioglu O, Schmidt B, Shoemaker L, Sigaudy S, Smith G, Sotsiou F, Stajic N, Stein A, Stray-Pedersen A, Taha D, Taque S, Tizard J, Tsimaratos M, Wong NA, Boerkoel CF. Schimke immunoosseous dysplasia: suggestions of genetic diversity. Hum Mutat. 2007;28:273–283. doi: 10.1002/humu.20432. [DOI] [PubMed] [Google Scholar]
- De Conti L, Skoko N, Buratti E, Baralle M. Complexities of 5'splice site definition: implications in clinical analyses. RNA Biol. 2012;9:911–923. doi: 10.4161/rna.20386. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich JH, Offner G, Schirg E, Hoyer PF, Helmchen U, Brodehl J. Association of spondylo-epiphyseal dysplasia with nephrotic syndrome. Pediatr Nephrol. 1990;4:117–121. doi: 10.1007/BF00858821. [DOI] [PubMed] [Google Scholar]
- Guo Y, Zhao S, Sheng Q, Ye F, Li J, Lehmann B, Pietenpol J, Samuels DC, Shyr Y. Multi-perspective quality control of Illumina exome sequencing data using QC3. Genomics. 2014;103:323–328. doi: 10.1016/j.ygeno.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter KB, Lucke T, Spranger J, Smithson SF, Alpay H, Andre JL, Asakura Y, Bogdanovic R, Bonneau D, Cairns R, Cransberg K, Frund S, Fryssira H, Goodman D, Helmke K, Hinkelmann B, Lama G, Lamfers P, Loirat C, Majore S, Mayfield C, Pontz BF, Rusu C, Saraiva JM, Schmidt B, Shoemaker L, Sigaudy S, Stajic N, Taha D, Boerkoel CF. Schimke immunoosseous dysplasia: defining skeletal features. Eur J Pediatr. 2010;169:801–811. doi: 10.1007/s00431-009-1115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberson BL, Cote GJ, Berget SM. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger J, Hinkel GK, Stoss H, Thoenes W, Wargowski D, Zepp F. Schimke immuno-osseous dysplasia: a newly recognized multisystem disease. J Pediatr. 1991;119:64–72. doi: 10.1016/s0022-3476(05)81040-6. [DOI] [PubMed] [Google Scholar]
- Yuan J, Ghosal G, Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009;23:2394–2399. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kong X, Yokomori K, Kadonaga JT. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 2009;23:2400–2404. doi: 10.1101/gad.1831509. [DOI] [PMC free article] [PubMed] [Google Scholar]