Abstract
Tea plant breeding is a topic of great economic importance. However, disease remains a major cause of yield and quality losses. In this study, an anthracnose-resistant cultivar, ZC108, was developed. An infection assay revealed different responses to Colletotrichum sp. infection between ZC108 and its parent cultivar LJ43. ZC108 had greater resistance than LJ43 to Colletotrichum camelliae. Additionally, ZC108 exhibited earlier sprouting in the spring, as well as different leaf shape and plant architecture. Microarray data revealed that the genes that are differentially expressed between LJ43 and ZC108 mapped to secondary metabolism-related pathways, including phenylpropanoid biosynthesis, phenylalanine metabolism, and flavonoid biosynthesis pathways. In addition, genes involved in plant hormone biosynthesis and signaling as well as plant-pathogen interaction pathways were also changed. Quantitative real-time PCR was used to examine the expression of 27 selected genes in infected and uninfected tea plant leaves. Genes encoding a MADS-box transcription factor, NBS-LRR disease-resistance protein, and phenylpropanoid metabolism pathway components (CAD, CCR, POD, beta-glucosidase, ALDH and PAL) were among those differentially expressed in ZC108.
Introduction
Tea is a widely consumed non-alcoholic beverage and the tea plant (Camellia sinensis (L.) O. Kuntze) is a woody crop of worldwide economic importance [1]. The tea plant is susceptible to many bacterial, fungal and viral diseases. A wide variety of tea plant germplasms exist worldwide [2, 3], and breeding continues to produce new superior cultivars with more beneficial elements (e.g. free amino acid content, lower caffeine content and higher tolerance to abiotic or biotic stress).
The yield and quality of the tea plant can be severely affected by biotic stressors [4, 5]. Anthracnose disease (AD), which is caused by Colletotrichum species (sp.), causes severe plant damage and loss of crop yield and quality. AD is an economically devastating disease prevalent under warm and humid conditions [6, 7]. As the disease progresses, circular spots appear on the leaves, enlarge, become sunken, and then produce lesions with round edges [8]. AD can cause plant wilt, shoot death, and fragility. Unfortunately, many tea plant cultivars are highly susceptible to AD [7]. Developing resistant cultivars may be the most efficient and economical strategy to reduce the threat of the disease.
Understanding tea plant defense mechanisms is important for breeding resistant cultivars. However, little is known about the molecular mechanisms regulating the defense response in tea plants. Plants have evolved multiple defense signaling pathways to respond to environmental conditions and pathogen attack [9]. Secondary metabolites (classified as alkaloids, phenolics and terpenoids) are important in plant protection to attenuate the effects of abiotic and biotic stress. Phenylpropanoid compounds are precursors to a wide range of phenolic compounds (e.g. flavonoids, isoflavonoids, anthocyanins, plant hormones, phytoalexins, and lignins) that play a vital role in host defense [10–12]. Phenylpropanoid pathway-associated genes comprise a large family involved in plant responses to various stressors. For example, induction of the phenylpropanoid pathway increases mango resistance to Ceratocystis fimbriata infection [13]. Furthermore, induction of four enzymes in the phenylpropanoid pathway is important for resistance to ascochyta blight disease in the chickpea [14]. Phenylalanine ammonia-lyase (PAL) is the first enzyme of phenylpropanoid metabolism. PAL controls a key branch point in the pathway of flavonoid biosynthesis, which generates antimicrobial phytoalexins and plays important roles in the response to biotic and abiotic stress [15]. Therefore, early or high PAL activity in response to infection is considered an indicator of pathogen resistance. For example, in the pepper plant, PAL1 enzymatic activity in the phenylpropanoid pathway acts as a positive regulator of salicylic acid (SA)-dependent defense signaling to combat microbial pathogens [16]. Furthermore, functional analysis of tobacco PAL showed that PAL-suppressed transgenic tobacco has undeveloped systemic acquired resistance and reduced SA levels in leaves [17].
Lignin is a major end-product of phenylpropanoid metabolism. Down-regulating lignin pathway genes alters metabolic flux and affects the biosynthesis of other secondary compounds [18]. Cinnamoyl-CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) catalyze the first and last steps in the lignin biosynthetic pathway, respectively [19]. Down-regulating CCR affects the composition of soluble phenolics by increasing various ferulate derivatives in Arabidopsis, poplar, and tobacco [20–23]. A similar pattern was observed in CAD-downregulated tobacco [24]. Reducing lignin content may trigger signaling pathways that activate stress genes [25]. Thus, lignin downregulation can alter phenylpropanoid metabolism, thereby affecting the biosynthesis of secondary compounds that play important roles in plant-environment interactions [18].
Many studies have investigated the relationship between AD and plants. In an AD-resistant muscadine cultivar, the chalcone synthase (CHS) gene in the phenylpropanoid pathway, which condenses three malony-CoA molecules with cinnamoyl-CoA to produce chalcone, may contribute to resistance [26]. Louime and colleagues observed differences in gene transcription between AD-susceptible and -tolerant grapevine cultivars, and identified many genes (CHS, stilbene synthase, polygalacturonase-inhibiting protein, chitinase and lipid transfer protein) that are only expressed in tolerant cultivars in response to anthracnose pathogen infection [26]. Vasanthaiah et al. demonstrated that chitinase and stilbene synthase genes were more rapidly expressed in tolerant cultivars after inoculation with an anthracnose pathogen [27]. In addition to PAL and CHS, other enzymes (e.g. chalcone isomerase (CHI) and dihydroflavonol 4-reductase (DFR)), which make up the flavonoid biosynthesis pathway, were also differentially expressed after inoculation in resistant and susceptible grapevines [8].
In this study, we characterized the anthracnose-resistant tea plant cultivar Zhongcha 108 (ZC108). ZC108 was produced by irradiating the offspring of Longjing 43 (LJ43) [28]. These two cultivars demonstrated different levels of resistance to Colletotrichum camelliae infection. Additionally, the two cultivars had many different characteristics, including bud sprouting time, leaf shape, and plant architecture. A 4 × 44 K custom microarray analysis revealed 2,453 differentially expressed genes between ZC108 and LJ43. These include genes involved in transcriptional regulation, phenylpropanoid metabolism, flavonoid biosynthesis, plant hormone signal transduction, and plant-pathogen interaction. Furthermore, several genes that were differentially regulated post-infection between ZC108 and LJ43 were identified by qRT-PCR.
Material and Methods
Plant material and sample preparation
Two tea plant (Camellia sinensis (L.) O. Kuntze) cultivars, LJ43 and ZC108, were used in this study. Plants were grown in the field at the Tea Research Institute of the Chinese Academy of Agricultural Sciences (TRI, CAAS, N 30°10', E 120°5'), Hangzhou, China. ZC108 was selected from the offspring of LJ43 cuttings under Co60γ-ray radiation [28]. The cutting shoots of LJ43 were irradiated by Co60γ-ray using 9.5 Gy irradiation dose in 1986, then the mutant offsprings (M1) were cuttage propagated for single plant selection. After 24 years breeding procedure, one new line was selected out and it performed early sprouting time, anti-anthracnose disease and high quality for processing green tea compared to LJ43. Then the line was named as ZC108 and registered as a new cultivar in China in 2010. And the parentage between LJ43 and ZC108 was identified by SSR markers and microarray method [29, 30].
For leaf measurement, mature leaves of 5-year-old LJ43 and ZC108 were randomly collected from more than 20 tea plants. The width and length of the leaves was measured. Sprouting time of the first new shoot (one leaf and one bud) was observed in spring of 2013 and 2014. For microarray analysis, the fresh shoots (two leaves and one bud) were sampled from the 5-year-old plants grown in the field [30]. Because the AD always occur at mature leaves, so for infection assay, the third healthy mature leaf from the top of each cultivar was collected from 5-year-old tea plants for in vitro inoculation. For qRT-PCR analysis, 3-year-old plants were grown in the green house for 1 month before in vivo inoculation, after inoculation, the mature leaves of LJ43 and ZC108 at 0, 24 and 72 h post infection were sampled. Three independent biological replicates were performed.
Pathogen inoculations
Two strains of Colletotrichum camelliae were used in this study. They were isolated from diseased tea plants in Shaanxi (N 107°67', E 32°96') and Zhejiang (N 30°10', E 120°5'), China, and are indicated as Colletotrichum camelliae-1 and -2, respectively. The two strains were isolated from private land and did not require permits. Colletotrichum camelliae-1 and -2 were cultured on potato dextrose agar at 25°C under 12h light/12h dark for 7 days. Conidia were harvested and suspended in sterile water, and conidia concentrations were adjusted to 1×105 ml-1.
For pathogenicity tests, LJ43 and ZC108 were inoculated with Colletotrichum camelliae. Leaves were surface-sterilized with 75% ethanol and sterile distilled water. The leaves were wounded with a sterile needle, and 20μl conidial suspension were applied to the wound. Inoculated leaves were placed in wet cotton wool to maintain sample moisture and incubated at 30°C. Inoculation experiments were conducted independently and performed in triplicate.
Microarray data analysis
For the microarray assay, total RNA was purified using the RNeasy Mini Kit (QIAGEN, Hilden, Germany). The custom microarray used in this study was developed by our laboratory as previously described [30]. All microarray data were deposited into the Gene Expression Omnibus (GEO) database under Accession Number GSE52255 (Release date 2015-1-1).
Raw data were obtained and analyzed as previously described [31]. Differential gene expression was defined as >2-fold change (FC) with P< 0.05. To identify biological pathways, genes were annotated with the corresponding enzyme commission (EC) numbers from BLASTX alignments against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [32, 33]. Pathways were selected using Fisher’s exact test (P< 0.05, false discovery rate (FDR) < 0.05) [34]. Gene ontology (GO) terms were assigned based on the best BLASTx hit from the NR database using Blast2GO software [35].
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted from ZC108 and LJ43 leaf samples (0.5-1g) at 0, 24 and 72 hours post inoculation with Colletotrichum camelliae, as previously described [36]. RNA samples (5 μg) were treated with RNase-free DNase I (Invitrogen, Carlsbad, CA, USA) to remove residual genomic DNA, and then used for cDNA synthesis with SuperScript® III Reverse Transcriptase (Invitrogen). Reaction mixtures were diluted 1:40 with distilled water and used as templates for qRT-PCR. Primer information is listed in S1 Table. Quantitative assays were performed in triplicate on each cDNA sample, and transcript levels were calculated relative to the level of polypyrimidine tract-binding protein (PTB) using the 2-ΔΔCt formula [37, 38]. All data are presented as mean ± SD (n = 3).
Hierarchical clustering analysis of the expression data was carried out using Cluster 3.0. QRT-PCR data of LJ43 and ZC108 were normalized to LJ43 0h time point. Microarray data of ZC108 were normalized to microarray data of LJ43. The color scale represents log2 expression values and the expression levels presented in heatmap were log2-based.
Statistical analysis of the data
Data are expressed as the mean±SD from three independent biological replicates. Significance was determined via one-way analysis of variance followed by the post-hoc least significant difference (LSD) t-test (P < 0.05).
Results
Phenotypic performance of ZC108
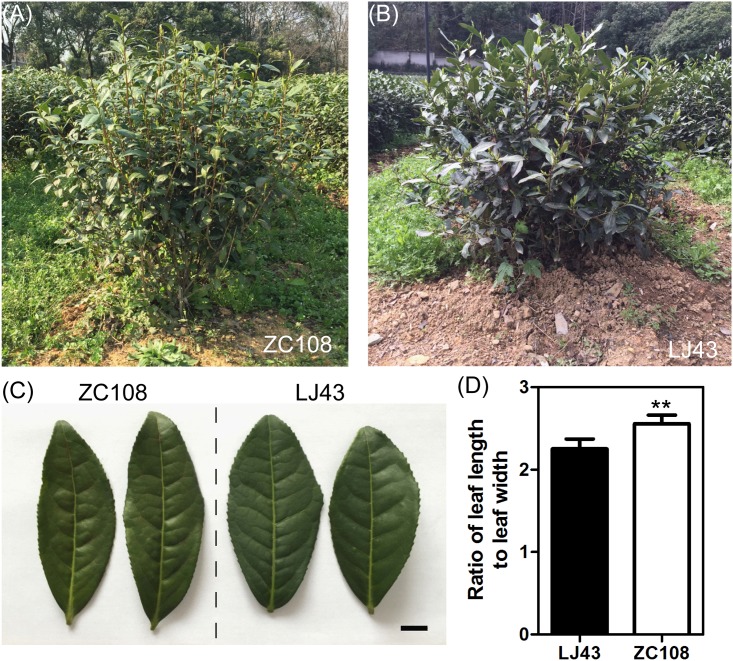
ZC108, the radiation-induced mutant of LJ43, showed many phenotypic differences compared to LJ43. One difference is bud sprouting time. Early sprouting in the spring is an attractive characteristic to tea growers because it confers higher economic value. Therefore, one goal of tea plant breeding is developing cultivars with earlier sprouting time. ZC108 sprouted earlier than LJ43 in the spring (Table 1). ZC108 and LJ43 also had different plant architecture (Fig 1A and 1B). Under normal conditions, the branch angle of ZC108 was smaller than that of LJ43 (Fig 1A and 1B). Shoot branching controls the yield of tea plant; therefore, ZC108 had higher yield than LJ43 [28]. Additionally, ZC108 leaves had a higher length to width ratio than LJ43 leaves (Fig 1C and 1D).
Table 1. Sprouting date of ZC108 and LJ43 cultivated in two areas in spring of 2013 and 2014.
| Date of one leaf and one bud (Area 1) | Date of one leaf and one bud (Area 2) | |||
|---|---|---|---|---|
| Cultivars | 2013 | 2014 | 2013 | 2014 |
| ZC108 | 23/3 | 29/3 | 2/4 | 1/4 |
| LJ43 | 2/4 | 31/3 | 6/4 | 3/4 |
Fig 1. Growth performance and leaf morphology of LJ43 and ZC108 grown in the field.
(A, B) Growth performance of ZC108 (A) and LJ43 (B) in the field in spring. (C) Mature leaf morphology of LJ43 and ZC108, bar = 1 cm. (D) Length:width ratio in mature leaves of LJ43 and ZC108. Data shown as the mean ±SD (n = 35). **P < 0.01 vs LJ43.
ZC108 is an anthracnose-resistant tea plant cultivar
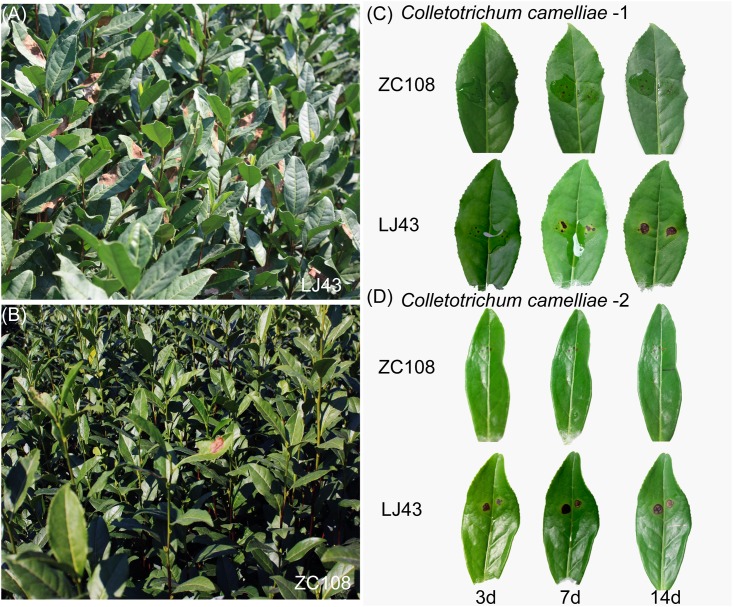
In the field, ZC108 exhibited higher resistance than LJ43 to AD. LJ43 plants showed disease symptoms on their leaves, but ZC108 plants appeared healthy (Fig 2A and 2B). To confirm the differential response to AD, leaves were inoculated in vitro. Both LJ43 and ZC108 were inoculated with two strains of Colletotrichum camelliae. Disease symptoms were observed on leaves 3, 7, and 14 days after inoculation. Anthracnose resistance was examined in the third mature leaf from the top of each cultivar. After inoculation, ZC108 symptoms appeared as tiny spots, and did not worsen over the time. However, LJ43 developed larger, sunken spots that produced lesions with round edges (Fig 2C and 2D). These results indicate that, compared to LJ43, ZC108 is more resistant to Colletotrichum camelliae infection.
Fig 2. Altered disease resistance of tea leaves to AD in ZC108.
(A, B) Leaf symptoms of LJ43 (A) and ZC108 (B) after suffering from AD in the field. (C, D) Differential disease resistance of tea leaves to Colletotrichum camelliae (1×105 ml-1) infection in LJ43 and ZC108. Two strains of Colletotrichum camelliae pathogens were used. Colletotrichum camelliae-1 and -2 were isolated from diseased tea plant leaves in Shaanxi and Zhejiang, respectively. Photos were taken on 3, 7 and 14 days post inoculation.
Microarray analysis
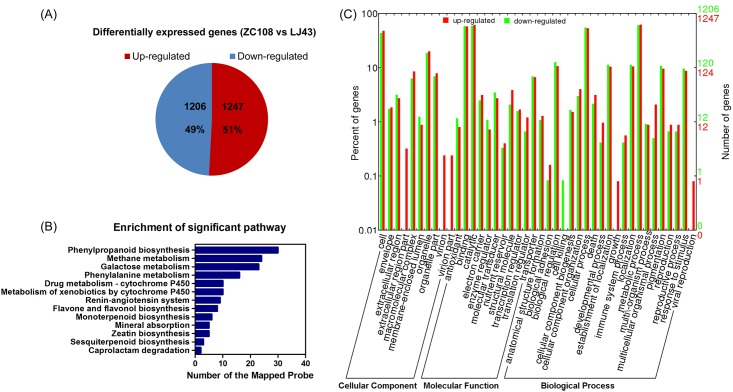
To investigate the mechanism underlying the phenotype of ZC108 plants, differences in gene expression between ZC108 and LJ43 were investigated using a Camellia sinensis custom microarray [30]. 2,453 genes were differentially expressed, 1,247 (51%) of which were up-regulated and 1,206 (49%) down-regulated in ZC108 (Fig 3A). Further analysis showed that genes mapped to 236 pathways (S2 Table). Notably, 13 pathways showed significant enrichment (P < 0.05, FDR < 0.05) (Fig 3B).
Fig 3. Differential expression analyses in ZC108 and LJ43.
(A) Number of significantly differentially expressed genes (P < 0.05, fold change (FC) > 2) in ZC108 compared with LJ43. (B) Significantly (P < 0.05, FDR < 0.05) enriched pathways (based on KEGG) among the 2,453 differentially expressed genes. (C) GO classification of the 2,453 differentially expressed genes.
The most significantly enriched pathway was phenylpropanoid biosynthesis, suggesting that genes related to phenylpropanoid metabolism may play important roles in the phenotype of ZC108 (Fig 3B). Genes involved in lignin biosynthesis (including PAL (EC:4.3.1.24), CAD (EC:1.1.1.195) and CCR (EC:1.2.1.44)), as well as peroxidase (EC:1.11.1.7, POD), beta-glucosidase (EC:3.2.1.21) and aldehyde dehydrogenase (EC:1.2.1.68, ALDH) genes, showed significant changes in ZC108. All ALDH and PAL probes were up-regulated, while most POD, all CCR, and all beta-glucosidase probes were down-regulated in ZC108 (Table 2). Notably, CAD, CCR, POD, beta-glucosidase, ALDH and PAL are important in the defense response and are related to plant pathogen resistance [39–45].
Table 2. Differentially expressed genes involved in phenylpropanoid biosynthesis (FC > 2, P < 0.05).
| Probe name | ZC108 vs LJ43 | Genes |
|---|---|---|
| Up-regulated | ||
| CUST_10940_PI428262022 | 137.69 | CAD |
| CUST_6622_PI428262022 | 90.23 | ALDH |
| CUST_33801_PI428262022 | 13.05 | CAD |
| CUST_506_PI428262022 | 6.89 | ALDH |
| CUST_3374_PI428262022 | 5.67 | PAL |
| CUST_21655_PI428262014 | 4.52 | PAL |
| CUST_35286_PI428262014 | 4.45 | ALDH |
| CUST_20888_PI428262022 | 3.47 | ALDH |
| CUST_25079_PI428262022 | 2.86 | POD |
| CUST_1515_PI428262022 | 2.75 | POD |
| CUST_11962_PI428262022 | 2.57 | caffeoyl-CoA O-methyltransferase (CCoAOMT) |
| CUST_20317_PI428262022 | 2.29 | PAL |
| CUST_1517_PI428262022 | 2.06 | POD |
| Down-regulated | ||
| CUST_42643_PI428262014 | -6.05 | POD |
| CUST_36442_PI428262014 | -5.36 | CAD |
| CUST_7937_PI428262014 | -5.24 | POD |
| CUST_2483_PI428262014 | -4.03 | CCR |
| CUST_33994_PI428262014 | -3.63 | POD |
| CUST_13516_PI428262014 | -3.06 | beta-glucosidase |
| CUST_41896_PI428262022 | -2.85 | POD |
| CUST_54961_PI428262014 | -2.77 | beta-glucosidase |
| CUST_6244_PI428262022 | -2.74 | POD |
| CUST_6222_PI428262014 | -2.73 | POD |
| CUST_10739_PI428262022 | -2.64 | CAD |
| CUST_371_PI428262014 | -2.53 | beta-glucosidase |
| CUST_50442_PI428262014 | -2.47 | POD |
| CUST_9847_PI428262014 | -2.27 | POD |
| CUST_16981_PI428262014 | -2.10 | 4-coumarate—CoA ligase |
| CUST_40717_PI428262022 | -2.09 | CCR |
| CUST_389_PI428262014 | -2.05 | beta-glucosidase |
In addition, genes related to plant hormone signaling (Table 3) and plant-pathogen interaction (Table 4) were affected. Among the differentially expressed hormone-related genes, the majority were auxin-related (Table 3). Differences in sprouting time and plant architecture between LJ43 and ZC108 may be due to these pathways. In addition, plant-pathogen interaction related genes were differentially expressed, including nucleotide-binding site (NBS) and leucine-rich repeat (LRR) resistance proteins and heat shock protein (HSP) [46–48], which may account for the different AD resistance of the two cultivars (Table 4).
Table 3. Differentially expressed genes related to plant hormone biosynthesis and signaling pathways (FC > 2, P < 0.05).
| Probe name | ZC108 vs LJ43 | Definition |
|---|---|---|
| Auxin related | ||
| CUST_50500_PI428262014 | -6.16 | auxin influx carrier (AUX1 LAX family) |
| CUST_11577_PI428262014 | -4.74 | SAUR family protein |
| CUST_15249_PI428262022 | -3.08 | auxin responsive GH3 gene family |
| CUST_15250_PI428262014 | -2.53 | auxin responsive GH3 gene family |
| CUST_14183_PI428262014 | -2.46 | SAUR family protein |
| CUST_2448_PI428262014 | -2.30 | auxin-responsive protein IAA |
| CUST_20521_PI428262014 | -2.28 | auxin response factor |
| CUST_18314_PI428262022 | -2.12 | SAUR family protein |
| CUST_1953_PI428262022 | -2.11 | auxin-responsive protein IAA |
| CUST_16624_PI428262022 | 2.96 | SAUR family protein |
| CUST_12_PI428262022 | 2.09 | auxin response factor |
| CUST_10678_PI428262014 | 2.05 | auxin response factor |
| CUST_26069_PI428262014 | 2.05 | auxin-responsive protein IAA |
| ABA related | ||
| CUST_12149_PI428262014 | -25.2 | ABA receptor PYR/PYL family |
| CUST_20599_PI428262022 | -3.44 | brassinosteroid insensitive 1-associated receptor kinase 1 |
| CUST_7855_PI428262022 | -2.73 | ABA responsive element binding factor |
| CUST_10100_PI428262014 | -2.17 | ABA responsive element binding factor |
| Jasmonate related | ||
| CUST_8238_PI428262014 | -2.44 | jasmonate ZIM domain-containing protein |
| CUST_13372_PI428262014 | 4.26 | jasmonate ZIM domain-containing protein |
| CUST_3660_PI428262022 | 3.21 | jasmonate ZIM domain-containing protein |
| CUST_7185_PI428262022 | 2.93 | jasmonate ZIM domain-containing protein |
| Zeatin related | ||
| CUST_13969_PI428262022 | 13.38 | cis-zeatin O-glucosyltransferase |
| CUST_6601_PI428262014 | -12.43 | tRNA dimethylallyltransferase |
| CUST_17809_PI428262014 | -9.93 | cis-zeatin O-glucosyltransferase |
| CUST_19864_PI428262014 | -2.90 | cytokinin dehydrogenase |
| CUST_37226_PI428262014 | -2.68 | cytokinin dehydrogenase |
| Other hormones | ||
| CUST_548_PI428262014 | -2.44 | ethylene receptor |
| CUST_2735_PI428262014 | -2.13 | protein brassinosteroid insensitive 1 |
| CUST_809_PI428262022 | -2.11 | DELLA protein |
| Regulation | ||
| CUST_42121_PI428262022 | 7.32 | histidine-containing phosphotransfer peotein |
| CUST_3858_PI428262022 | 3.63 | serine/threonine-protein kinase SRK2 |
| CUST_48154_PI428262022 | 2.35 | transcription factor TGA |
| CUST_54248_PI428262022 | 2.35 | transcription factor MYC2 |
Table 4. Differentially expressed genes mapped to the plant-pathogen interaction pathway (KO04626) in KEGG (FC > 2, P < 0.05).
| Probe name | ZC108 vs LJ43 | Definition |
|---|---|---|
| Up-regulated | ||
| CUST_18562_PI428262022 | 14.79 | CC-NBS-LRR resistance protein |
| CUST_54537_PI428262014 | 9.15 | NBS-LRR resistance protein |
| CUST_50104_PI428262022 | 7.39 | CC-NBS-LRR resistance protein |
| CUST_13372_PI428262014 | 4.26 | jasmonate ZIM domain-containing protein |
| CUST_34812_PI428262022 | 4.01 | NBS-LRR resistance protein (RPS2) |
| CUST_53112_PI428262014 | 3.92 | calcium-dependent protein kinase |
| CUST_14941_PI428262022 | 3.57 | NBS-LRR resistance protein |
| CUST_47812_PI428262022 | 3.52 | cyclic nucleotide gated channel |
| CUST_15181_PI428262022 | 3.34 | NBS-LRR resistance protein (RPS2) |
| CUST_19468_PI428262022 | 3.24 | NBS-LRR disease resistance protein (RPM1) |
| CUST_3660_PI428262022 | 3.21 | jasmonate ZIM domain-containing protein |
| CUST_20165_PI428262022 | 2.98 | CC-NBS-LRR resistance protein |
| CUST_7185_PI428262022 | 2.93 | jasmonate ZIM domain-containing protein |
| CUST_19115_PI428262022 | 2.91 | transcription factor MYC2 |
| CUST_17805_PI428262022 | 2.4 | disease resistance protein RPM1 (NBS-LRR) |
| CUST_54248_PI428262022 | 2.35 | transcription factor MYC2 |
| CUST_56099_PI428262014 | 2.26 | glycerol kinase |
| CUST_20394_PI428262022 | 2.24 | NBS-LRR resistance protein |
| CUST_19763_PI428262014 | 2.18 | chitinase |
| CUST_14991_PI428262022 | 2.14 | molecular chaperone HtpG |
| CUST_20060_PI428262022 | 2.14 | respiratory burst oxidase |
| CUST_23265_PI428262014 | 2.11 | suppressor of G2 allele of SKP1 |
| CUST_7334_PI428262022 | 2.09 | cyclic nucleotide gated channel |
| CUST_7592_PI428262014 | 2.04 | suppressor of G2 allele of SKP1 |
| Down-regulated | ||
| CUST_18102_PI428262014 | -10.68 | heat shock protein 90kDa beta |
| CUST_36014_PI428262014 | -6.74 | NBS-LRR disease resistance protein (RPM1) |
| CUST_26291_PI428262014 | -4.09 | heat shock protein |
| CUST_9744_PI428262022 | -3.55 | mitogen-activated protein kinase kinase kinase 1 |
| CUST_52781_PI428262014 | -3.52 | cyclic nucleotide gated channel |
| CUST_40627_PI428262014 | -3.5 | cyclic nucleotide gated channel |
| CUST_20599_PI428262022 | -3.44 | brassinosteroid insensitive 1-associated receptor kinase 1 |
| CUST_51979_PI428262022 | -2.61 | cyclic nucleotide gated channel |
| CUST_8238_PI428262014 | -2.44 | jasmonate ZIM domain-containing protein |
| CUST_28392_PI428262014 | -2.30 | CC-NBS-LRR resistance protein |
| CUST_7836_PI428262014 | -2.27 | Calmodulin |
| CUST_19961_PI428262022 | -2.16 | Calmodulin |
| CUST_35507_PI428262022 | -2.08 | NBS-LRR resistance protein |
| CUST_49241_PI428262014 | -2.06 | CC-NBS-LRR resistance protein |
We performed a GO functional annotation of differentially-expressed genes. Genes were categorized as belonging to biological process (BP), molecular function (MF), and cellular component (CC) functional groups. Within each functional group, the largest categories were metabolic and cellular processes (BP), binding and catalytic activity (MF), and cell and organelle (CC) (Fig 3C). Microarray analysis also revealed that 7 probe sets corresponded to transcription factors (TFs) encoding AP2/ERF, MYB, ZF-HD, C2H2, MDAS, REM and bZIP family proteins (Table 5). Arabidopsis homologs of these TFs are involved in plant development and stress response.
Table 5. Classification of differentially expressed genes encoding transcription factors.
| Probe | ZC108 vs LJ43 | Arabidopsis TF | Homologs | Target processes | Similarity | Matching Length (bp) | E value |
|---|---|---|---|---|---|---|---|
| CUST_3691_PI428262014 | -2.25 | AT1G15360 | AP2/ERF | Pathogenesis-related, wax biosynthesis, drought tolerance | 82.98 | 141 | 4.00E-30 |
| CUST_767_PI428262022 | 2.68 | AT3G01140 | MYB | Plant development, trichome branching | 80.44 | 501 | 5.00E-105 |
| CUST_4560_PI428262022 | 3.12 | AT2G02540 | ZF-HD | Expressed in vascular tissue | 82.72 | 162 | 3.00E-35 |
| CUST_8204_PI428262022 | 3.1 | AT5G16470 | C2H2 (MBS2) | Stress response, ROS signaling | 76.39 | 216 | 5.00E-27 |
| CUST_33289_PI428262022 | 2.42 | AT4G11880 | MADS (AGL14) | Agamous-like | 80.93 | 236 | 4.00E-41 |
| CUST_1834_PI428262022 | 2.42 | AT3G53310 | REM | Plant development | 100 | 28 | 3.00E-07 |
| CUST_4471_PI428262014 | -2.98 | AT5G06950 | bZIP (TGA2) | Plant pathogen interaction, regulate PR gene | 85 | 99 | 9.00E-16 |
Note: Genes from the tea plant showing similarity to Arabidopsis homologs higher than 75% with e value < 1e-5 are listed. The target process was described according to the TAIR database.
Validation of differentially expressed genes
To verify the microarray results, 27 genes from Tables 2–5 that were differentially expressed in ZC108 were analyzed using qRT-PCR. The microarray and qRT-PCR results are compared in Fig 4. Most of the selected genes analyzed by qRT-PCR at 0h showed an expression pattern consistent with the microarray data. About 6 genes including beta-glucosidase, C2H2, ERF, bZIP, ARP, CAD (CUST_36442) showed inconsistent regulation between qRT-PCR and microarray. This discrepancy may be attributed to several factors, including the different sensitivities of the analytical methods and the different samples used as templates in qRT-PCR and microarray.
Fig 4. Validation and expression analysis of 27 genes in response to Colletotrichum camelliae infection in leaves of LJ43 and ZC108.
Gene expression analysis were examined by qRT-PCR using cDNA from leaves of three-year-old LJ43 and ZC108 after inoculated by Colletotrichum camelliae for 0, 24 and 72 h. All of the qRT-PCR values were expressed relative to the expression level of LJ43-0h (control-set to 1.0). The data of ZC108 from microarray was expressed relative to the transcript abundance of LJ43 from microarray (control-set to 1.0). The color scale represents log2 expression values and the expression levels presented in heatmap were log2-based. All data are shown as the mean ± SD (n = 3).
To investigate molecular responses to pathogen infection in resistant ZC108 and wildtype LJ43 cultivars, leaves from both cultivars were infected with Colletotrichum camelliae and the expression of selected genes was measured. Expression of many of the selected genes changed in response to pathogen infection (Fig 4). Moreover, compared to LJ43, some genes also significantly changed in abundance at 24h and 72h post-infection in ZC108. These genes include a MADS-box transcription factor, 3 NBS-LRR family genes, plant hormone signal transduction-related genes (cytokinin dehydrogenase (CKX) and auxin-responsive protein IAA (AUX1)), and phenylpropanoid biosynthesis-related genes (CAD, CCR, POD, beta-glucosidase, ALDH and PAL) (Fig 4 and S1–S4 Figs). Notably, the expression of genes encoding defense enzymes (e.g. CAD, POD, PAL, ALDH and beta-glucosidase), the disease resistance protein NBS-LRR, and MADS-box transcription factor were significantly up-regulated in ZC108 after infection. Genes whose expression changed after infection in ZC108 may underlie tea plant responses against Colletotrichum camelliae infection.
Additionally, we found that the hormone signal transduction related genes ARF, SAUR, CKX, AUX1 and the transcription factor C2H2 showed significant differences in transcript abundance in ZC108 compared to LJ43 independent of pathogen infection condition (Fig 4, S1 and S3 Figs). These genes may therefore be responsible for the differential development and morphology between the two cultivars.
Discussion
Compared to its parent cultivar LJ43, the irradiated offspring cultivar ZC108 showed important phenotypic differences. These include the important agronomic characteristics plant architecture, bud sprouting time, and AD resistance. These changes were beneficial to the economic value of the cultivar [28]. In this study, we used microarrays to investigate which genes contribute to the phenotypic differences between LJ43 and ZC108. Our results provide insight into the molecular mechanisms underlying these changes, especially the response to Colletotrichum camelliae infection.
Early sprouting time in spring confers higher economic tea plant value in China. Therefore, producing cultivars with early sprouting time is an important goal of tea plant breeders. Plant hormone signaling is the most important of many factors influencing sprouting time. Many studies have shown that auxin (IAA), gibberellin (GA), abscisic acid (ABA) and cytokinins (e.g. zeatin) participate in sprouting progress [49, 50] and plant architecture [51–54]. In the tea plant, free GA and IAA increase before release from dormancy, suggesting they may promote sprouting [55, 56]. Moreover, we found that before tea plant bud sprouting, auxin and ABA content reach a peak and nadir, respectively (unpublished data). The microarray analysis results in this study show that most genes related to plant hormone biosynthesis and signaling pathways are down-regulated in ZC108 compared to LJ43 (Table 3). These results suggest that changes in biosynthesis and plant hormone signaling may underlie the differential sprouting time and plant architecture in ZC108.
Tea plant AD severely affects plant health and tea production [7]. Most green tea plant cultivars are susceptible to AD, and therefore breeding anthracnose resistant cultivars is of high importance. According to performance and pathogenicity tests, ZC108 is an anthracnose-resistant cultivar. To acquire further insight into the molecular mechanisms underlying anthracnose resistance in ZC108, differentially expressed genes related to plant-pathogen interaction were identified by microarray. Plant hormones not only regulate growth and development, but also interact with the immune system [9, 57]. Reports indicate that all plant hormones can induce plant defense responses and participate in systemic immunity [58]. Many of the differentially expressed genes identified between ZC108 and LJ43 are implicated in the biosynthesis of plant hormones, including auxin, ABA, ethylene, zeatin and jasmonic acid (Table 3). These genes may underlie the plant architecture, leaf shape, and disease resistance traits of ZC108 as well.
ESTs probe sets corresponded to abiotic stress-related transcription factors, encoding proteins in the AP2-EREBP, NAC, MYB, ZF-HD, C2H2, MDAS, REM and bZIP families. MADS-box transcription factor expression in response to the infection was different between ZC108 and LJ43 (Fig 4). After infection, a MADS gene defined by homology to AtAGL14 (AT4G11880) was down-regulated in LJ43 but up-regulated in ZC108, suggesting it might play an important role in regulating tea plant response to Colletotrichum camelliae. MADS is therefore a promising candidate gene for future studies of the molecular mechanisms underlying AD response.
Genes involved in phenylpropanoid biosynthesis were differentially expressed between ZC108 and LJ43 (Fig 3B). These differences may confer disease resistance to ZC108. Saravanakumar and colleagues have shown that accumulation of peroxidase and PAL increases in response to pathogen infection in the tea plant [4]. PAL is one of the most extensively studied enzymes in the plant biotic and abiotic stress response pathways [39]. Consistent with these previous findings, the present study demonstrated that PAL transcription increased after infection. All three PAL probes in our microarray were significantly up-regulated in ZC108 (by 2.29, 4.52, and 5.67-fold). Furthermore, PAL was up-regulated in ZC108 regardless of whether it was inoculated with Colletotrichum camelliae (Fig 4). These results suggest that PAL might be the key gene responsible for higher resistance to Colletotrichum camelliae in ZC108.
Notably, after inoculation, POD, ALDH, beta-glucosidase, and two CAD genes showed higher expression in ZC108 than LJ43 (Fig 4). CAD is a key enzyme in lignin biosynthesis. Deng et al. showed that one CAD, CsCAD3, was strongly up-regulated after E. oblique attack and mechanical damage, and three CsCADs responded to treatment with the defense-related hormones methyljasmonate (MeJA) and salicylic acid (SA) [40]. In the present study, the expression of two CADs was higher in ZC108 than LJ43 after Colletotrichum camelliae inoculation, suggesting that the CAD is important for ZC108 resistance (Fig 4). Our study indicates that beta-glucosidase may be involved in AD resistance, consistent with previous reports that it is important in the plant defense response [43]. In addition, VpALDH2B4 may be beneficial to the plant under conditions of abiotic stress. In our study, the expression of ALDHs were significantly up-regulated in both normal and Colletotrichum camelliae-infected ZC108 (Table 2 and Fig 4). This result agrees with previous reports and reveals that CsALDHs may be important for AD resistance. POD plays a key role in plant defense through lignin formation, cross-linking of cell wall components, phytoalexin synthesis, and metabolism of reactive oxygen species (ROS) reactive nitrogen species (RNS), and auxin [59]. In Arabidopsis, the French bean peroxidase type 1 (FBP1) knockdown line exhibits an impaired oxidative burst and is more susceptible than wild type to both fungal and bacterial pathogens [41]. Furthermore, the AtPOD33 knockdown line is more susceptible to Pseudomonas syringae than wild type plants [42]. In this study, several PODs were differentially expressed in ZC108. Most were down-regulated in the absence of pathogen. However, after Colletotrichum camelliae infection, POD was up-regulated in ZC108 compared to LJ43 (Fig 4). These results demonstrate that POD can be regulated in response to Colletotrichum camelliae infection in the tea plant. Beta-glucosidase is another important component of the defense response, and our study also indicated it may be involved in AD resistance [43]. Phenylpropanoid metabolism gives rise to numerous compounds, including lignin. Down-regulating lignin pathway genes can alter metabolic flux and differentially affect the biosynthesis of other secondary compounds involved in plant-pathogen interaction [18]. In this study, CCR was down-regulated and CADs were up-regulated in ZC108, indicating that the biosynthesis of lignin and secondary compounds may be altered in this cultivar. Together, these results demonstrate that the phenylpropanoid pathway plays a vital role in AD resistance in ZC108. Altered expression levels of genes involved in the phenylpropanoid pathway may underlie ZC108 resistance to Colletotrichum camelliae.
Several studies have reported that genes involved in flavonoid biosynthesis (CHS, DFR and CHI) are also involved in the response to AD [8, 26]. However, CHS, DFR and CHI transcription did not significantly change between ZC108 and LJ43, regardless of Colletotrichum camelliae inoculation (data not shown).
Levels of secondary metabolites, such as phenylpropanoid compounds, are controlled in response to environmental cues and play an important role in host defense [10, 60]. We have measured tea polyphenol (TP) content in leaves of ZC108 and LJ43, and found that the leaf TP content was increased in both ZC108 and LJ43 after inoculation, indicating that TP played an important role in resistance to AD in tea plant (S5 Fig). Compared to LJ43, the TP content in ZC108 was higher than LJ43 at the 24 h post inoculation, however, it was not significant (S5 Fig). Flavonoids, isoflavonoids, anthocyanins, phytoalexins and lignins belong to phenolic compounds. Although there was no significant difference between TP content in ZC108 and LJ43 after inoculation, it will be of great interest to examine other phenolic compounds, e.g. flavonoid, phytoalexins and lignin content as a possible mechanism for ZC108 pathogen resistance in future studies. Understanding defense mechanisms in anthracnose-resistant tea plants is important for plant breeding. Although differentially expressed genes in the transcriptomes of LJ43 and ZC108 were identified, we did not compare these transcriptomes after inoculation. This will be important to examine in the future to further elucidate the molecular mechanisms underlying AD resistance. ZC108 was selected from LJ43 cuttings that underwent Co60γ-ray radiation. Genes in ZC108 which were mutated to substantially change plant architecture and biochemical composition are still unknown. In future studies, identification of the mutated genes will contribute to our understanding of the molecular mechanisms underlying plant architecture and facilitate breeding elite varieties.
Tea plant breeding is very difficult due to very long procudure, very low breeding efficiency and natural mutant rate. So find a higher effective breeding method is very important for tea plant improvement. Radiaction mutation breeding is an effective breeding method in plant, especially some perennial species, such as tea plant. Radiaction is not only make some functional gene mutation, but also make epigenetic variations at DNA level [61, 62]. So in the following studies, we’ll conduct some projects on DNA epigenetic effects and functional genomics to illustrate the mechanisms of the phenotypic variation between ZC108 and LJ43 based on the whole genome sequnces of tea plant be sequenced. And this will be very useful for establishment a effective radiation mutant breeding method in tea plant.
In summary, we identified ZC108 as an anthracnose-resistant tea plant cultivar. We compared plant morphology, biochemistry, and transcriptomes of ZC108 and LJ43 and identified many differences. The levels of genes involved in plant hormone biosynthesis and signaling, plant-pathogen interaction, and secondary metabolic pathways (including phenylpropanoid biosynthesis), were changed in ZC108. We selected genes to examine in infected and uninfected tea plant leaves and confirmed that several genes may contribute to anthracnose disease tolerance in ZC108. These include MADS-box transcription factor and genes involved in phenylpropanoid metabolism (CAD, CCR, POD, beta-glucosidase, ALDH and PAL). Additional studies to measure phenylpropanoid compound content and transcriptomes after inoculation will expand our understanding of the mechanisms of AD resistance.
Supporting Information
Gene expression analysis were examined by qRT-PCR using cDNA from leaves of three-year-old LJ43 and ZC108 after inoculated by Colletotrichum camelliae for 0, 24 and 72 h. All of the qRT-PCR values were expressed relative to the expression level of LJ43-0h (control-set to 1.0). The data of ZC108 from microarray was expressed relative to the transcript abundance of LJ43 from microarray (control-set to 1.0). Data shown as the mean ±SD (n = 3).
(TIF)
(TIF)
(TIF)
(TIF)
Three-year-old plants were grown in the green house for 1 month before in vivo inoculation. The first to third mature leaves from the top of LJ43 and ZC108 without inoculation and 24 h post inoculation were sampled. Data shown as the mean ±SD (n = 3).
(TIF)
(DOCX)
(XLS)
Acknowledgments
This manuscript has been thoroughly edited by a native English speaker from an editing company. Editing Certificate will be provided upon request.
Abbreviations
- AD
anthracnose disease
- ALDH
coniferyl-aldehyde dehydrogenase
- ARF
auxin response factor
- AUX/IAA
auxin-responsive protein IAA
- CAD
cinnamyl-alcohol dehydrogenase
- CCR
cinnamoyl-CoA reductase
- CHI
chalcone isomerase
- CHS
chalcone synthase
- CKX
cytokinin dehydrogenase
- DFR
dihydroflavonol 4-reductase
- EC
enzyme commission
- GO
gene ontology
- HSP
heat shock protein
- KEGG
kyoto encyclopedia of genes and genomes
- FC
fold change
- FDR
false discovery rate
- NBS-LRR
nucleotide-binding site (NBS) and leucine-reach repeats (LRR)
- PAL
phenylalanine ammonia-lyase
- PTB
polypyrimidine tract-binding protein
- POD
peroxidase
- TP
tea polyphenol
Data Availability
All relevant data are within the paper and its Supporting Information files. All microarray files are available from the Gene Expression Omnibus (GEO) database (accession number GSE52255).
Funding Statement
This work was supported by the Earmarked Fund for China Agriculture Research System (CARS-23), the Major Project for New Agricultural Varieties Breeding of Zhejiang Province (2012C2905-3) and the Chinese Academy of Agricultural Sciences through an Innovation Project for Agricultural Sciences and Technology (CAAS-ASTIP-2014-TRICAAS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wang X, Zhao Q, Ma C, Zhang Z, Cao H, Kong Y, et al. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC genomics. 2013;14(1):415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen J, Wang P, Xia Y, Xu M, Pei S. Genetic diversity and differentiation of Camellia sinensis L. (cultivated tea) and its wild relatives in Yunnan province of China, revealed by morphology, biochemistry and allozyme studies. Genet Resour Crop Evol. 2005;52(1):41–52. [Google Scholar]
- 3.Wachira F, Tanaka J, Takeda Y. Genetic variation and differentiation in tea (Camellia sinensis) germplasm revealed by RAPD and AFLP variation. Journal of Horticultural Science and Biotechnology. 2001;76:557–563. [Google Scholar]
- 4.Saravanakumar D, Vijayakumar C, Kumar N, Samiyappan R. PGPR-induced defense responses in the tea plant against blister blight disease. Crop Protection. 2007;26(4):556–565. [Google Scholar]
- 5.Gulati A, Ravindranath S, Chakrabarty D. Economic yield losses caused by Exobasidium vexans in tea plantations. Indian Phytopathol. 1993;46:155–159. [Google Scholar]
- 6.Prom L, Perumal R, Isakeit T, Radwan G, Rooney W, Magill C. The impact of weather conditions on response of Sorghum genotypes to anthracnose (Colletotrichum sublineola) infection. American Journal of Experimental Agriculture. 2015;6(4):242–250. [Google Scholar]
- 7.Yoshida K, Takeda Y. Evaluation of anthracnose resistance among tea genetic resources by wound-inoculation assay. Japan Agricultural Research Quarterly: JARQ. 2006;40(4):379–386. [Google Scholar]
- 8.Gao M, Wang Q, Wan R, Fei Z, Wang X. Identification of genes differentially expressed in grapevine associated with resistance to Elsinoe ampelina through suppressive subtraction hybridization. Plant Physiology and Biochemistry. 2012;58(0):253–268. [DOI] [PubMed] [Google Scholar]
- 9.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson R. Phenolic compounds and their role in disease resistance. Annu Rev Phytopathol. 1992;30:369–389. [Google Scholar]
- 11.Dixon R, Paiva N. Stress-induced phenylpropanoid metabolism. The Plant cell. 1995;7(7):1085–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.La Camera S, Gouzerh G, Dhondt S, Hoffmann L, Fritig B, Legrand M, et al. Metabolic reprogramming in plant innate immunity: the contributions of phenylpropanoid and oxylipin pathways. Immunological Reviews. 2004;198(1):267–284. [DOI] [PubMed] [Google Scholar]
- 13.Araujo L, Bispo WMS, Rios VS, Fernandes SA, Rodrigues FA. Induction of the phenylpropanoid pathway by acibenzolar-s-methyl and potassium phosphite increases Mango resistance to Ceratocystis fimbriata infection. Plant Disease. 2014;99(4):447–459. [DOI] [PubMed] [Google Scholar]
- 14.Kavousi H, Marashi H, Mozafari J, Bagheri A. Expression of phenylpropanoid pathway genes in Chickpea defense against race 3 of Ascochyta rabiei. Plant Pathology Journal. 2009;8(3):127–132. [Google Scholar]
- 15.Bowles D. Defense-related proteins in higher plants. Annu Rev Biochem. 1990;59:873–907. [DOI] [PubMed] [Google Scholar]
- 16.Kim D, Hwang B. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. Journal of experimental botany. 2014;65(9):2295–306. 10.1093/jxb/eru109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pallas J, Paiva N, Lamb C, Dixon R. Tobacco plants epigenetically suppressed in phenylalanine ammonia-lyase expression do not develop systemic acquired resistance in response to infection by tobacco mosaic virus. The Plant Journal. 1996;10:281–293. [Google Scholar]
- 18.Baxter H, Stewart C. Effects of altered lignin biosynthesis on phenylpropanoid metabolism and plant stress. Biofuels. 2013;4(6):635–650. [Google Scholar]
- 19.Lacombe E, Hawkins S, Doorsselaere J, Piquemal J, Goffner D, Poeydomenge O, et al. Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: cloning, expression and phylogenetic relationships. Plant Journal. 1997;11(3):429–441. [DOI] [PubMed] [Google Scholar]
- 20.Dauwe R, Morreel K, Goeminne G, Gielen B, Rohde A, Beeumen J, et al. Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. The Plant Journal. 2007;52:263–285. [DOI] [PubMed] [Google Scholar]
- 21.Leplé JC, Dauwe R, Morreel K, Storme V, Lapierre C, Pollet B, et al. Downregulation of cinnamoyl-coenzyme a reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. The Plant cell. 2007;19(11):3669–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mir Derikvand M, Sierra J, Ruel K, Pollet B, Do CT, Thévenin J, et al. Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta. 2008;227(5):943–956. [DOI] [PubMed] [Google Scholar]
- 23.Prashant S, Srilakshmi Sunita M, Pramod S, Gupta R, Anil Kumar S, Rao Karumanchi S, et al. Down-regulation of Leucaena leucocephala cinnamoyl CoA reductase (LlCCR) gene induces significant changes in phenotype, soluble phenolic pools and lignin in transgenic tobacco. Plant cell reports. 2011;30(12):2215–2231. 10.1007/s00299-011-1127-6 [DOI] [PubMed] [Google Scholar]
- 24.Sirisha VL, Prashant S, Ranadheer Kumar D, Pramod S, Jalaja N, Hima Kumari P, et al. Cloning, characterization and impact of up- and down-regulating subabul cinnamyl alcohol dehydrogenase (CAD) gene on plant growth and lignin profiles in transgenic tobacco. Plant growth regulation. 2012;66(3):239–253. [Google Scholar]
- 25.Fornalé S, Shi X, Chai C, Encina A, Irar S, Capellades M, et al. ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. The Plant Journal. 2010;64(4):633–644. 10.1111/j.1365-313X.2010.04363.x [DOI] [PubMed] [Google Scholar]
- 26.Louime C, Lu J, Onokpise O, Vasanthaiah HKN, Kambiranda D, Basha S, et al. Resistance to Elsinoë Ampelina and expression of related resistant genes in Vitis Rotundifolia Michx. grapes. Int J Mol Sci. 2011;12(6):3473–3488. 10.3390/ijms12063473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasanthaiah HN, Basha S, Katam R. Differential expression of chitinase and stilbene synthase genes in Florida hybrid bunch grapes to Elsinoë ampelina infection. Plant growth regulation. 2010;61(2):127–134. [Google Scholar]
- 28.Yang Y, Yang S, Yang Y, Zeng J. Investigation on the breeding of new tea cultivar, Zhongcha 108, with early-sprouting, superior-quality and suitable for manufacturing high-qulity green tea. China Tea. 2003;25(2):12–14. [Google Scholar]
- 29.Tan L, Peng M, Xu L, Wang L, Chen S, Zou Y, et al. Fingerprinting 128 Chinese clonal tea cultivars using SSR markers provides new insights into their pedigree relationships. Tree Genetics & Genomes. 2015;11(5):1–12. [Google Scholar]
- 30.Wang L, Wang X, Yue C, Cao H, Zhou Y, Yang Y. Development of a 44K custom olligo microarray using 454 pyrosequencing data for large-scale gene expression analysis of Camellia sinensis. Scientia Horticulturae. 2014;174:133–141. [Google Scholar]
- 31.Wang L, Yue C, Cao H, Zhou Y, Zeng J, Yang Y, et al. Biochemical and transcriptome analyses of a novel chlorophyll-deficient chlorina tea plant cultivar. BMC plant biology. 2014;14(1):352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic acids research. 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic acids research. 2014;42(D1):D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological). 1995;57(1):289–300. [Google Scholar]
- 35.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. [DOI] [PubMed] [Google Scholar]
- 36.Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- 37.Livaka KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 38.Hao X, Horvath D, Chao W, Yang Y, Wang X, Xiao B. Identification and evaluation of reliable reference genes for quantitative real-time PCR analysis in tea plant (Camellia sinensis (L). O. Kuntze). International journal of molecular sciences. 2014;15:22155–22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacDonald M, D'Cunha G. A modern view of phenylalanine ammonia lyase. Biochemistry and Cell Biology. 2007;85(3):273–282. [DOI] [PubMed] [Google Scholar]
- 40.Deng W, Zhang M, Wu J, Jiang Z, Tang L, Li Y, et al. Molecular cloning, functional analysis of three cinnamyl alcohol dehydrogenase (CAD) genes in the leaves of tea plant, Camellia sinensis. Journal of plant physiology. 2013;170(3):272–282. [DOI] [PubMed] [Google Scholar]
- 41.Bindschedler L, Dewdney J, Blee K, Stone J, Asai T, Plotnikov J, et al. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. The Plant Journal. 2006;47:851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daudi A, Cheng Z, O’Brien JA, Mammarella N, Khan S, Ausubel FM, et al. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. The Plant cell. 2012;24(1):275–287. 10.1105/tpc.111.093039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mattiacci L, Dicke M, Posthumus M. beta-glucosidase: an elicitor of herbivore-induced plant odor that attracts host-searching parasitic wasps. PNAS. 1995;92(6):2036–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lauvergeat V, Lacomme C, Lacombe E, Lasserre E, Roby D, Grima-Pettenati J. Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry. 2001;57(7):1187–1195. [DOI] [PubMed] [Google Scholar]
- 45.Wen Y, Wang X, Xiao S, Wang Y. Ectopic expression of VpALDH2B4, a novel aldehyde dehydrogenase gene from Chinese wild grapevine (Vitis pseudoreticulata), enhances resistance to mildew pathogens and salt stress in Arabidopsis. Planta. 2012;236(2):525–539. 10.1007/s00425-012-1624-z [DOI] [PubMed] [Google Scholar]
- 46.Pedras MS, Minic Z. Differential protein expression in response to the phytoalexin brassinin allows the identification of molecular targets in the phytopathogenic fungus Alternaria brassicicola. Mol Plant Pathol. 2012;13(5):483–493. 10.1111/j.1364-3703.2011.00765.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome biology. 2006;7(4):212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeYoung BJ, Innes RW. Plant NBS-LRR proteins in pathogen sensing and host defense. Nature immunology. 2006;7(12):1243–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powell LE. The hormonal control of bud and seed dormancy in woody plants In: Davies PJ, editor. Plant Hormones and their Role in Plant Growth and Development: Springer; Netherlands; 1987. p. pp 539–552. [Google Scholar]
- 50.Vanstraelen M, Benkova E. Hormonal interactions in the regulation of plant development. Annual review of cell and developmental biology. 2012;28:463–487. 10.1146/annurev-cellbio-101011-155741 [DOI] [PubMed] [Google Scholar]
- 51.Ongaro V, Leyser O. Hormonal control of shoot branching. Journal of experimental botany. 2008;59(1):67–74. [DOI] [PubMed] [Google Scholar]
- 52.Gallavotti A. The role of auxin in shaping shoot architecture. Journal of experimental botany. 2013;64(9):2593–2608. 10.1093/jxb/ert141 [DOI] [PubMed] [Google Scholar]
- 53.Finlayson SA, Krishnareddy SR, Kebrom TH, Casal JJ. Phytochrome regulation of branching in Arabidopsis. Plant physiology. 2010;152(4):1914–1927. 10.1104/pp.109.148833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teichmann T, Muhr M. Shaping plant architecture. Frontiers in plant science. 2015;6:233 10.3389/fpls.2015.00233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagar PK, Kumar A. Changes in endogenous gibberellin activity during winter dormancy in tea (Camellia sinensis (L.) O. Kuntze). Acta Physiologiae Plantarum. 2000;22(4):439–443. [Google Scholar]
- 56.Nagar PK, Sood S. Changes in endogenous auxins during winter dormancy in tea (Camellia sinensis L.) O. Kuntze. Acta Physiologiae Plantarum. 2006;28(2):165–169. [Google Scholar]
- 57.Spoel SH, Dong XN. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe. 2008;3(6):348–351. 10.1016/j.chom.2008.05.009 [DOI] [PubMed] [Google Scholar]
- 58.Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. Networking by small-molecule hormones in plant immunity. Nature chemical biology. 2009;5(5):308–316. 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- 59.Almagro L, Gómez Ros LV, Belchi-Navarro S, Bru R, Ros Barceló A, Pedreño MA. Class III peroxidases in plant defence reactions. Journal of experimental botany. 2009;60(2):377–390. 10.1093/jxb/ern277 [DOI] [PubMed] [Google Scholar]
- 60.Payyavula R, Navarre D, Kuhl J, Pantoja A, Pillai S. Differential effects of environment on potato phenylpropanoid and carotenoid expression. BMC plant biology. 2012;12(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H, Feng Q, Zhang M, Yang C, Sha W, Liu B. Alteration of DNA methylation level and pattern in sorghum (Sorghum bicolor L.) pure-lines and inter-line F1 hybrids following low-dose laser irradiation. Journal of photochemistry and photobiology B, Biology. 2010;99(3):150–153. [DOI] [PubMed] [Google Scholar]
- 62.Xu W, Wang T, Xu S, Xu S, Wu L, Wu Y, et al. Radiation-induced epigenetic bystander effects demonstrated in Arabidopsis thaliana. Radiation research. 2015;183(5):511–524. 10.1667/RR13909.1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene expression analysis were examined by qRT-PCR using cDNA from leaves of three-year-old LJ43 and ZC108 after inoculated by Colletotrichum camelliae for 0, 24 and 72 h. All of the qRT-PCR values were expressed relative to the expression level of LJ43-0h (control-set to 1.0). The data of ZC108 from microarray was expressed relative to the transcript abundance of LJ43 from microarray (control-set to 1.0). Data shown as the mean ±SD (n = 3).
(TIF)
(TIF)
(TIF)
(TIF)
Three-year-old plants were grown in the green house for 1 month before in vivo inoculation. The first to third mature leaves from the top of LJ43 and ZC108 without inoculation and 24 h post inoculation were sampled. Data shown as the mean ±SD (n = 3).
(TIF)
(DOCX)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All microarray files are available from the Gene Expression Omnibus (GEO) database (accession number GSE52255).