Abstract
We identified the lactic acid bacteria within rye sourdoughs and starters from four bakeries with different propagation parameters and tracked their dynamics for between 5–28 months after renewal. Evaluation of bacterial communities was performed using plating, denaturing gradient gel electrophoresis, and pyrosequencing of 16S rRNA gene amplicons. Lactobacillus amylovorus and Lactobacillus frumenti or Lactobacillus helveticus, Lactobacillus pontis and Lactobacillus panis prevailed in sourdoughs propagated at higher temperature, while ambient temperature combined with a short fermentation cycle selected for Lactobacillus sanfranciscensis, Lactobacillus pontis, and Lactobacillus zymae or Lactobacillus helveticus, Lactobacillus pontis and Lactobacillus zymae. The ratio of species in bakeries employing room-temperature propagation displayed a seasonal dependence. Introduction of different and controlled propagation parameters at one bakery (higher fermentation temperature, reduced inoculum size, and extended fermentation time) resulted in stabilization of the microbial community with an increased proportion of L. helveticus and L. pontis. Despite these new propagation parameters no new species were detected.
Introduction
Sourdough is a mixture of flour and water that is fermented with lactic acid bacteria (LAB) and yeasts. Traditional sourdoughs are propagated by backslopping over many decades by mixing a portion of mature sourdough with fresh flour and water and fermenting this into a new batch of sourdough [1, 2]. In mature sourdough, both homo- and hetero-fermentative species of LAB are prevalent and the community is typically dominated by members from the genus Lactobacillus [3].
Over 50 different species of LAB have been isolated from sourdoughs of different origin [4]. Despite this large number of identified species, mature sourdoughs typically contain only two or three dominant species. Lactobacillus brevis, Lactobacillus fermentum, Lactobacillus plantarum, Lactobacillus sanfranciscensis, and Lactobacillus acidophilus [1, 5–6] species are most often encountered in rye sourdoughs, which are used to make rye bread, a staple of the Nordic diet.
Sourdoughs can be classified into three types according to the technology used for their production [1, 7–8]. Type I sourdoughs are produced using a traditional method based on daily renewal. Type II sourdoughs used in large-scale production are semi-fluid and have good handling properties. Long term continuous propagations are common in type II sourdough processes. Type III sourdoughs are generally initiated by starter cultures and are dried before use. These sourdoughs are commonly used as taste and aroma enhancers.
A sourdough cycle can be started by either the spontaneous fermentation of flour, inoculation with mature sourdough, or with a starter culture [2]. The baking industry currently tends to begin sourdough fermentation with defined commercial starter cultures with specific properties [1]. Unfortunately, those strains may not adapt to the sourdough propagation conditions in the bakery and are often not competitive enough in the long term to fight off LAB that enter the process from either raw materials or the bakery environment. Therefore, to maintain a desirable microbial community, the sourdough cycle is frequently restarted [9–10]. The microbial composition of sourdoughs is affected by the process technology and applied conditions: fermentation temperature and time, inoculum size, water content (characterized by dough yield), production environment, and type of flour [1, 11–13].
Information regarding the composition and stability of sourdoughs used in industrial-scale production is limited. The traditions of sourdough preparation and bread making are region-dependent, which influences the sensory characteristics chosen for industrially prepared breads [14]. The aims of this work are i) to compare LAB communities in sourdoughs that originate from bakeries that apply different sourdough propagation parameters and ii) to evaluate the stability of these microbial communities and their influence on the chemical characteristics of the sourdough over many months of daily renewal. Both culture dependent and culture independent methods were used to characterize the microbial communities within the sourdough samples.
Materials and Methods
Sourdough samples from bakeries
The sourdoughs studied originate from four bakeries that use flour of the same origin (rye flour type 1370, extraction rate 85%, Tartu Mill AS, Estonia) and are referred to as Abakery, Bbakery, Cbakery, and Dbakery. The bakeries use different sourdough propagation parameters (presented in Table 1); two sourdoughs are type II (Abakery and Bbakery) and two are type I (Cbakery and Dbakery). In Abakery and Bbakery the sourdough fermentation temperature was precisely controlled while Cbakery and Dbakery propagated sourdoughs at room temperature which fluctuated seasonally. The temperature of each Dbakery sourdough sample is provided in Table 1.
Table 1. Sourdough propagation parameters (fermentation temperature and time, inoculum size, dough yield), starter used to initiate fermentation, and sampling schedule in four Estonian bakeries (Abakery, Bbakery, Cbakery and Dbakery).
In Cbakery and Dbakery the fermentation was carried out at room temperature (RT). ‘Months’ indicate time passed from the beginning of a new sourdough cycle (Abakery, Bbakery, and Cbakery) or from the collection of the first sourdough sample (Dbakery).
| Bakery | Abakery | Bbakery | Cbakery | Dbakery |
|---|---|---|---|---|
| Fermentation temperature | 32°C | 42–44°C | RT (up to 28°C) | RT (19–30°C) |
| Inoculum size | 10% | 6% | ~ 33% | ~ 33% |
| Fermentation time | 10 h | 16 h | ~ 4 h | ~ 4 h |
| Dough yield | 250 | 400 | ~ 200 | ~ 200 |
| Starter | Freeze-dried sourdough culture | Active sourdough starter | Commercial freeze-dried starter | Commercial freeze-dried starter |
| Analyzed samples | A0 –freeze-dried sourdough | Bs– 3 years propagated sourdough | C0 –freeze-dried commercial starter | D1 –approx. 30 years propagated sourdough (ambient temperature 19°C) |
| A1–1.2 months | B0 –fresh sourdough starter | C1–12 months | D2–3 months (30°C) | |
| A2–3.5 months | B1–0.25 months | C2–21 months | D3–5 months (28°C) | |
| A3–4.5 months | B2–1 month | C3–28 months | D4–19 months (23°C, before transfer to 30°C) | |
| A4–8.5 months | B3–2.5 months | D5–24 months (5 months after transfer to 30°C) | ||
| B4–6 months |
The sourdough process in Abakery was initiated with a freeze-dried sourdough made from a mature sourdough produced six years previously at the same bakery (sample A0). Bbakery sourdough had been successfully propagated for three years (sample Bs) and was then renewed from fresh cooled sourdough sourced from another bakery belonging to the same corporation (sample B0). Cbakery sourdough fermentation was initiated with a freeze-dried commercial starter (C0) one year before the first sourdough sample was collected. Dbakery sourdough was initiated in the 1980’s from a commercial starter. During the course of this study Dbakery adopted new propagation parameters with a well-controlled fermentation temperature in an attempt to improve both the stability of the sourdough and optimize the sourdough production cycle (Table 1). The cycle was extended from 4 h to 12 h and the fermentation temperature was increased to 30°C. In addition, the inoculum size was lowered from approximately 33% to 10%. Sourdough sample D4 was taken before adopting the new sourdough propagation cycle. Sample D5 was collected from sourdough that had been propagated for five months after the upgrade. In addition, 18 sourdough samples were collected between these two time points, however, only pH and DGGE analysis was performed.
Chemical analysis of sourdoughs
The pH and total titratable acidity (TTA) values of each sourdough sample were measured in triplicate. For each analysis 5 g of sourdough was homogenized with 45 ml of distilled water. The pH and TTA were measured with Food and Beverage Analyzer D22 (Mettler-Toledo International Inc., USA). TTA is given as ml of 0.1 N NaOH used to titrate 10 g of sourdough sample to pH 8.5.
Enumeration of lactic acid bacteria
The cell density of culturable LAB in each sourdough sample was determined by plate counting. 5 g of sourdough was mixed with 45 ml of sterile 0.85% NaCl solution. A series of decimal dilutions were plated on MRS agar (LabM, UK) in duplicate. In addition, sample C3 was plated on an mMRS agar (MRS with added 2% maltose; pH 5.6) and SDB agar (2% maltose; 0.03% Tween80; 0.6% trypticase; 1% yeast extract; pH 5.6) [15]. Plates from Abakery, Cbakery and Dbakery were incubated at 30°C, while those from Bbakery were incubated at 42°C; all under anaerobic conditions. A BD GasPak EZ System (Becton Dickinson Microbiology Systems, USA) was used to maintain an anaerobic environment.
DNA extraction from lactic acid bacteria isolates and sourdough samples
Selected colonies were checked for purity by streaking. The cultures were suspended in dH2O with a loop and subjected to DNA extraction using FTA membrane cards (Whatman Inc., USA) following the method provided by the manufacturer. Total DNA extraction from the sourdough samples was performed using 5 g of sourdough, which had been homogenized by vortexing with 45 ml of sterile physiological solution. This suspension was then centrifuged at 4°C for 5 minutes at 1000 × g. The supernatant was collected and centrifuged at 4°C for 15 minutes at 5000 × g. A GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich, Inc., USA) was used for DNA extraction from the obtained pellet in the case of Bbakery, Cbakery, and Dbakery according to the manufacturer’s instructions. A modified phenol-chloroform extraction was used for samples taken from Abakery [16].
Fingerprint typing of isolates with Rep-PCR
For each sourdough sample, 20 colonies were randomly picked from plates with suitable dilution (usually 20 to 200 colonies per plate) and analyzed by Rep-PCR. Rep-PCR with primer (GTG)5 (all primers in this work were obtained from Microsynth, Switzerland) was performed as described by Viiard [16] with slight modification: all PCR components were sourced from Solis BioDyne (Estonia). Share of each LAB fingerprint type within selected isolates was calculated as a percentage of the colonies analyzed.
Identification of lactic acid bacteria by 16 rRNA gene sequence analysis
One to two representatives of each fingerprint type group detected using Rep-PCR were selected for 16S rRNA gene analysis. 16S rRNA gene fragments were amplified using the universal primers 27f-YM [17] and 16R1522 [18] followed by column purification of the amplified fragment with a GeneJET PCR Purification Kit (Thermo Scientific Inc., USA) and sequenced in a commercial facility. The partial 16S rRNA gene sequences obtained (approximately 700 bp) were searched against GenBank database using the BLAST algorithm (Basic Local Alignment Search Tool, National Center for Biotechnology Information, USA) and the results were confirmed with the Greengenes 16S rRNA gene database (Lawrence Berkeley National Laboratory, USA).
Denaturing gradient gel electrophoresis analysis of microbial communities
Denaturing gradient gel electrophoresis (DGGE) analysis was performed to monitor the dynamics of microbial communities within sourdoughs. V3 region of the bacterial 16S rRNA genes was amplified using primers F357-GC and 518R as described by Gafan and Spratt [19] to obtain approx. 160 bp fragments. Polyacrylamide gel (8% acrylamide-N,N’-methylenebisacrylamide; 37.5:1) with a gradient from 35 to 70% urea and formamide (100% corresponding to 40% formamide and 7 M urea) was used. Electrophoresis was performed with an INGENY phorU (Ingeny BV International, The Netherlands) at a constant voltage of 70 V at 60°C for 17 h. For yeasts, the primers U1GC and U2 were used to amplify the 28S rRNA genes, as described by Meroth [20] to obtain approx. 300 bp amplicons. A gradient of 30–50% was used and electrophoresis was performed at a constant voltage of 130 V at 60°C for 4.5 h.
The gels were stained with ethidium bromide and digitized using an ImageQuant 400 system (GE Healthcare, USA). Bands of interest were excised and DNA within these bands was eluted by incubation in a TE buffer overnight at 4°C. The eluted fragments were reamplified using F357/518R or U1/U2 primer pairs for bacterial and yeast DNA, respectively, and sequenced in a commercial facility.
Pyrosequencing of bar-coded 16S rRNA gene amplicons
Universal primers 8F and 357R were used for PCR amplification of the V2–V3 hypervariable regions of 16S rRNA genes [21–22]. The amplicon mixtures were pyrosequenced using a 454 GS FLX+ System (Roche 454 Life Sciences, USA). The 454 pyrosequencing data was processed using MOTHUR v.1.32.1 [23] according to standard operating procedures [24]. Reads shorter than 150 bp or containing more than eight homopolymers were removed from the dataset. Sequences were aligned to the SILVA reference 16S rRNA database [25]. Operational Taxonomic Units (OTUs) were defined using an average neighbor clustering algorithm based on 97% sequence identity. Normalized OTU counts at 500 reads were calculated with the R (version 3.0.3) software package “vegan” version 2.0–10. In addition we calculated the rate of forming new OTUs when one sequence is added to the set of 500 sequences. The closest match to each OTU within the Greengenes 16S rRNA gene database (Lawrence Berkeley National Laboratory, USA) was identified using BLAST with a minimum of 97% similarity. The relative abundance of OTUs was calculated as the number of sequences for each OTU divided by the total number of bacterial sequences obtained for each sourdough sample. To estimate the beta-diversity, non-metric multidimensional scaling (NMDS) was conducted using Yue and Clayton distances [26] within MOTHUR and results were visualized using R software (version 3.0.3).
Nucleotide sequence accession numbers
Pyrosequencing data is available in the GenBank database under accession numbers KM972414—KM972548.
Results
Chemical analysis and LAB enumeration of rye sourdoughs
The chemical properties of Abakery, Bbakery and Cbakery sourdoughs were rather stable throughout the study (Table 2). The average pH value of the sourdough throughout the study was 3.64 ± 0.08 in Abakery, 3.56 ± 0.08 in Bbakery and 4.10 ± 0.09 in Cbakery. Bbakery sourdough was characterized by a high TTA in all samples except the initiating starter dough B0. Significant seasonal fluctuations in pH occurred in the sourdough from Dbakery, where the average pH value of the sourdough was 4.06 ± 0.18. Sample D1 with a pH value of 4.28 was taken in February, when temperature of the sourdough was 19°C. During warmer months June (D2) and August (D3) the pH values decreased to 3.96 and 3.86, respectively.
Table 2. Mean values ± standard deviation of pH, total titratable acidity (TTA) and cell density of presumptive lactic acid bacteria (LAB) of rye sourdoughs from four Estonian bakeries (Abakery, Bbakery, Cbakery and Dbakery).
Samples are coded according to the description reported in Table 1.
| Sample | pH | TTA | LAB |
|---|---|---|---|
| (ml 0.1 N NaOH / 10 g) | (log CFU g-1) | ||
| A0 | NA* | NA | 7.08 ± 0.11 |
| A1 | 3.67 ± 0.06 | 22.30 ± 0.56 | 8.82 ± 0.07 |
| A2 | 3.71 ± 0.01 | 18.89 ± 0.05 | 9.04 ± 0.06 |
| A3 | 3.63 ± 0.04 | 21.30 ± 0.41 | 8.84 ± 0.07 |
| A4 | 3.53 ± 0.02 | 21.84 ± 0.30 | 9.08 ± 0.03 |
| Bs | 3.60 ± 0.05 | 31.33 ± 0.13 | 8.63 ± 0.16 |
| B0 | 3.40 ± 0.12 | 21.60 ± 0.58 | 8.56 ± 0.14 |
| B1 | 3.63 ± 0.08 | 30.23 ± 0.02 | 8.11 ± 0.06 |
| B2 | 3.57 ± 0.06 | 31.38 ± 1.27 | 8.94 ± 0.08 |
| B3 | 3.57 ± 0.05 | 34.51 ± 1.28 | 8.93 ± 0.24 |
| B4 | 3.58 ± 0.06 | 33.18 ± 1.09 | 8.85 ± 0.05 |
| C0 | NA | NA | 6.95 ± 0.04 |
| C1 | 4.11 ± 0.09 | 16.50 ± 1.11 | 6.56 ± 0.07 |
| C2 | 4.00 ± 0.07 | 18.20 ± 0.03 | 6.64 ± 0.01 |
| C3 | 4.18 ± 0.11 | 17.10 ± 0.58 | 8.28 ± 0.03 |
| D1 | 4.28 ± 0.06 | 16.94 ± 1.08 | 8.00 ± 0.03 |
| D2 | 3.96 ± 0.16 | 18.30 ± 1.06 | 8.80 ± 0.08 |
| D3 | 3.86 ± 0.06 | 23.19 ± 1.10 | 9.05 ± 0.12 |
| D4 | 4.12 ± 0.11 | 17.85 ± 0.03 | 8.01 ± 0.11 |
| D5 | 3.78 ± 0.07 | 18.98 ± 0.03 | 8.31 ± 0.08 |
* NA–not acquired
The cell density of LAB was high and stable in Abakery (on average 8.95 ± 0.13 log CFU g-1) and Bbakery (on average 8.67 ± 0.32 log CFU g-1), with the exception of sample B1, which exhibited a lower cell density (Table 2). In case of Cbakery unusually low values of LAB cell density (order of magnitude: 6 log CFU g-1) were obtained for samples C1 and C2 (Table 2). Two fold higher cell density was found for sample C3, but the related plates were incubated for additional 24 h compared to the samples C1 and C2. In Dbakery the LAB cell density (on average 8.47 ± 0.54 CFU g-1) depended on the ambient temperature in the bakery and was higher during summer (samples D2 and D3).
Analysis of the LAB community in rye sourdoughs
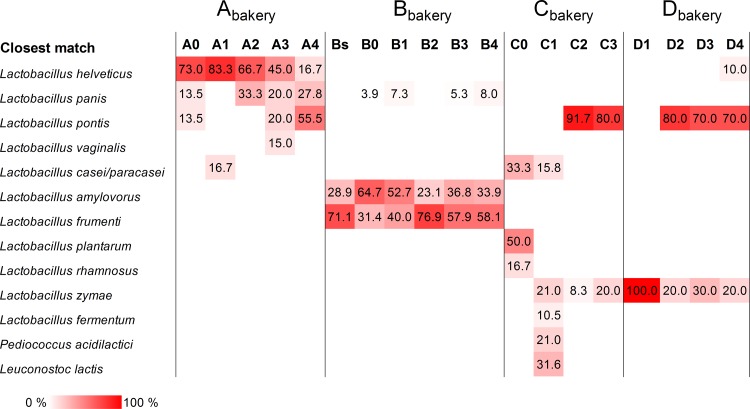
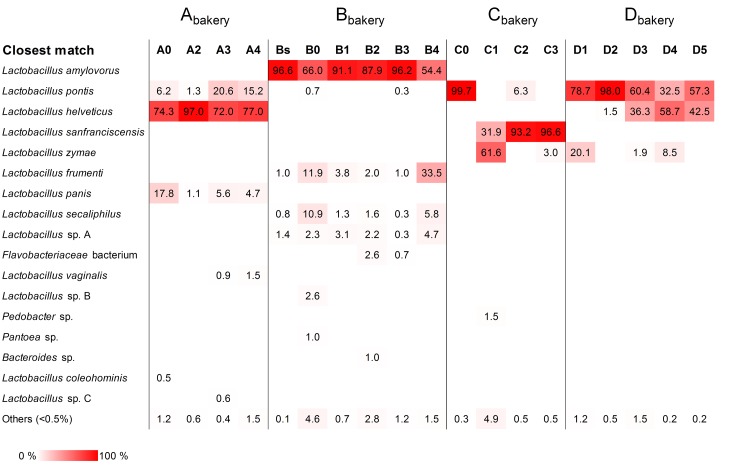
Sourdough samples from Abakery were monitored for over eight months after renewal from a freeze-dried starter (previously published by Viiard [16]). Based on the results from culture dependent analysis, the dominating LAB in the freeze-dried starter dough belonged to species Lactobacillus helveticus, Lactobacillus panis and Lactobacillus pontis (Table 3 and Fig 1). It was shown that during continuous propagation of sourdough in Abakery the proportion of L. helveticus colonies decreased, and that of L. panis and L. pontis increased. DGGE analysis, however, revealed L. helveticus as a prevalent species during over eight months of propagation (Table 3 and S1 Fig). The pyrosequencing analysis confirmed that the microbial community within the Abakery sourdough was remarkably stable (Table 3 and Fig 2). Data of sequences and OTUs from 16S rRNA pyrosequencing performed using DNA extracted from sourdoughs sampled at Abakery, Bbakery, Cbakery and Dbakery are shown in Table 4. The trimmed amplicon length of all sourdough samples was in the range 228–262 bp. The total number of sequences before processing (raw reads) was 42,388; on average 2231 sequences per sample were obtained. After data processing (reads) in total 34,906 sequences remained.
Table 3. Bacterial species/genera found in the rye sourdough samples from four Estonian bakeries (Abakery, Bbakery, Cbakery and Dbakery) through culture dependent analysis, DGGE or 16S pyrosequencing.
Presence (+) or absence (‒) of species is indicated for each sample in the following order: culture dependent analysis / DGGE / 16S pyrosequencing. N–not analyzed. Samples are coded according to the description reported in Table 1.
| Bacteroides sp. | Flavobacteriaceae bacterium | Lactobacillus amylovorus | Lactobacillus casei/paracasei | Lactobacillus coleohominis | Lactobacillus fermentum | Lactobacillus frumenti | Lactobacillus helveticus | ||
|---|---|---|---|---|---|---|---|---|---|
| Abakery | A0 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/+ | ‒/‒/‒ | ‒/‒/‒ | +/+/+ |
| A1 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | |
| A2 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | |
| A3 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | |
| A4 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | |
| Bbakery | Bs | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | ‒/‒/‒ |
| B0 | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | ‒/‒/‒ | |
| B1 | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | ‒/‒/‒ | |
| B2 | ‒/‒/+ | ‒/‒/+ | +/+/+ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | ‒/‒/‒ | |
| B3 | ‒/‒/‒ | ‒/‒/+ | +/+/+ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | ‒/‒/‒ | |
| B4 | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | ‒/‒/‒ | |
| Cbakery | C0 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ |
| C1 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/‒/‒ | ‒/‒/‒ | +/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | |
| C2 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | |
| C3 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | |
| Dbakery | D1 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/+/‒ |
| D2 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/+/+ | |
| D3 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/+/+ | |
| D4 | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | ‒/‒/‒ | +/+/+ | |
| D5 | N/‒/‒ | N/‒/‒ | N/‒/‒ | N/‒/‒ | N/‒/‒ | N/‒/‒ | N/‒/‒ | N/+/+ |
Fig 1. Composition of lactic acid bacterium species, expressed in percentage of the total number of isolates, in rye sourdoughs from four Estonian bakeries (Abakery, Bbakery, Cbakery and Dbakery).
Samples are coded according to the description reported in Table 1.
Fig 2. Relative abundance of bacterial species/genera detected in rye sourdoughs from four Estonian bakeries (Abakery, Bbakery, Cbakery and Dbakery) using pyrosequencing of 16S rRNA gene amplicons.
Samples are coded according to the description reported in Table 1.
Table 4. Number of reads, OTUs, expected OTUs at 500 reads and rate of new OTUs at 500 reads obtained from 16S rRNA pyrosequencing of rye sourdoughs from four Estonian bakeries (Abakery, Bbakery, Cbakery and Dbakery).
Samples are coded according to the description reported in Table 1.
| Sample | Raw reads | Reads | OTUs | Expected OTUs at 500 reads | Rate of new OTUs at 500 reads |
|---|---|---|---|---|---|
| A0 | 1929 | 1574 | 15 | 8.745 | 0.008 |
| A1 | NA* | NA | NA | NA | NA |
| A2 | 4332 | 4153 | 10 | 4.941 | 0.003 |
| A3 | 2740 | 2335 | 12 | 6.750 | 0.004 |
| A4 | 2320 | 1952 | 22 | 10.561 | 0.011 |
| Bs | 2417 | 2281 | 7 | 4.643 | 0.001 |
| B0 | 1259 | 303 | 21 | NA | NA |
| B1 | 2462 | 1260 | 12 | 7.414 | 0.007 |
| B2 | 859 | 503 | 18 | 17.940 | 0.020 |
| B3 | 932 | 688 | 14 | 11.589 | 0.014 |
| B4 | 2179 | 1334 | 17 | 9.875 | 0.010 |
| C0 | 2754 | 2749 | 7 | 2.389 | 0.003 |
| C1 | 1410 | 263 | 15 | NA | NA |
| C2 | 1960 | 1922 | 9 | 4.349 | 0.004 |
| C3 | 3569 | 3437 | 12 | 4.149 | 0.004 |
| D1 | 1415 | 1314 | 13 | 7.186 | 0.009 |
| D2 | 2435 | 2356 | 7 | 3.661 | 0.002 |
| D3 | 991 | 593 | 11 | 9.878 | 0.012 |
| D4 | 3122 | 2923 | 10 | 4.197 | 0.002 |
| D5 | 3303 | 2966 | 6 | 2.815 | 0.002 |
* NA–not acquired
After the first sample (Bs) was collected from Bbakery, the sourdough was renewed using fresh sourdough sourced from another bakery (sample B0). The stability of Bbakery sourdough was monitored for six months after renewal. The results of culture dependent analysis revealed that both Lactobacillus amylovorus and Lactobacillus frumenti were dominant species within all sourdough samples collected from Bbakery (Table 3 and Fig 1). The ratio between this species varied during propagation. The same fingerprint type of dominant L. amylovorus was detected throughout the study (data not shown). L. panis entered the sourdough cycle with sample B0 and remained constant within the sourdough at low counts during subsequent propagation. DGGE analysis confirmed the dominance of L. amylovorus and L. frumenti (Table 3 and S1 Fig). On the contrary, L. panis was undetectable by both DGGE and 16S rRNA pyrosequencing (Table 3 and Fig 2). Pyrosequencing analysis revealed that L. frumenti and, especially, L. amylovorus, were the dominant OTUs in the sourdough samples collected at the Bbakery. Additional OTUs (Lactobacillus secaliphilus and Lactobacillus sp.) were found as sub-dominant in all the Bbakery samples. In Cbakery, L. plantarum, Lactobacillus rhamnosus and Lactobacillus casei/paracasei were isolated from the starter sample (C0) (Fig 1). After one year of propagation (C1), L. casei/paracasei persisted and other species (Lactobacillus zymae, L. fermentum, Leuconostoc lactis and Pediococcus acidilactici) were detected. In the sourdough samples taken 21 and 28 months after renewal (samples C2 and C3), all the LAB species previously detected, except for L. zymae, seemed to be replaced by L. pontis. In contrast, DGGE analysis revealed L. pontis as the only species in sample C0 and L. pontis and L. sanfranciscensis in all the remaining sourdough samples (Table 3 and S1 Fig). Overall, pyrosequencing analysis of Cbakery samples was in agreement with DGGE, excluding the lack of L. pontis in C1 and C3 samples and the presence, at high relative abundance, of L. zymae in C1 (Table 3 and Fig 2). Given the discrepancy between culture dependent and independent analyses regarding the presence of L. sanfranciscensis in the samples collected at the Cbakery, the C3 sample was analyzed using two additional media, SDB and mMRS. L. sanfranciscensis could be isolated after an extended incubation time (72 h) of mMRS plates (data not shown).
The sourdough samples from Dbakery contained L. zymae, L. pontis and L. helveticus (Figs 1 and 2 and S1 Fig). The relative proportion of these species in a given sourdough sample depended on the ambient temperature in the bakery. In the wintertime (sample D1), growth of L. zymae was favored, while L. pontis and L. helveticus dominated in the samples (D2, D3, and D4) collected during warmer periods.
Impact of new propagation parameters on the LAB community of Dbakery sourdough
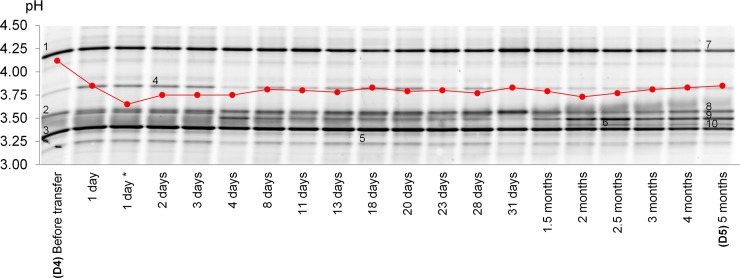
In order to improve the stability of Dbakery sourdough, a new sourdough propagation protocol was applied with a controlled fermentation temperature, prolonged fermentation time and reduced inoculum size. As ascertained through culture independent analyses, L. pontis and L. helveticus species dominated in the sourdough (sample D5) after 5 months of propagation performed under the new protocol (Figs 2 and 3). The pH of the sourdough ranged from 3.73–3.79 during the five months of observation (Fig 3).
Fig 3. pH (red plot) and lactic acid bacterium species detected by DGGE analysis of the 16S rRNA gene amplicons in rye sourdoughs collected at the Dbakery before (D4) and after applying the new propagation protocol.
Time after transfer is indicated below the gel (d–day; m–month). Bands: 1, 7 –Lactobacillus helveticus; 2, 8 –Cereal chloroplast DNA; 3, 4, 5, 10 –Lactobacillus pontis; 6 –Lactobacillus sp.; 9 –Lactobacillus zymae; *—sample collected from sourdough after 36 h storage at 5°C.
Analysis of yeast communities in rye sourdoughs
DGGE analysis of amplified 28S rRNA (S2 Fig) was performed to identify the yeast species in the sourdoughs collected at the four bakeries. Kazachstania telluris was the only yeast species harbored in the freeze-dried starter (A0), as well as in all the sourdough samples collected at the Abakery. No yeast species were detected in Bbakery sourdough. Candida humilis was the only yeast species detected in Cbakery and Dbakery sourdough samples, whereas no yeast species were detected in the starter (C0) used in Cbakery (S2 Fig).
Beta-diversity across the sourdough samples
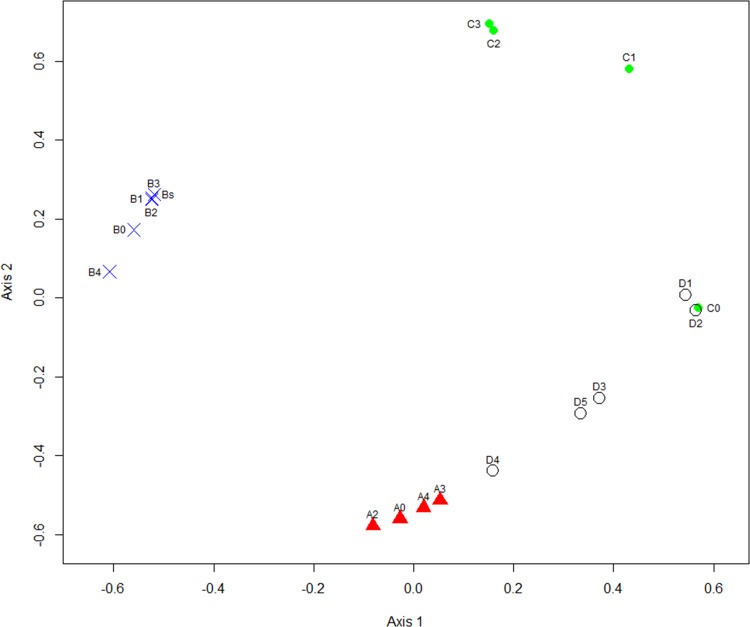
Beta-diversity analysis was performed on the pyrosequencing data to compare diversity between each sourdough sample and to determine the similarity (or difference) in species composition of the samples (Fig 4). Two-dimensional non-metric multidimensional scaling (NMDS) provided a stress value of 0.156 and an R2 value of 0.885. The stress value decreased to 0.064 and the R2 value increased to 0.977 when calculating the NMDS with three dimensions. Sourdough samples from Abakery and Bbakery grouped in two different clusters, both characterized by closeness of the grouped samples, which illustrates the stability of both sourdough propagation processes. On the contrary, samples from both Cbakery and, especially, Dbakery grouped in looser clusters, thus indicating that the bacterial communities within these sourdoughs are less stable (Fig 4). The freeze-dried starter C0 that contained L. pontis groups together with Dbakery samples where L. pontis is prevalent. Abakery and Dbakery sourdough samples can be found in the same quadrant of the NMDS plot because both contain L. helveticus and L. pontis. Samples collected at the Bbakery which employs a higher fermentation temperature, differ significantly from all other sourdoughs.
Fig 4. Two dimensional non-metric multidimensional scaling (NMDS) of sourdough samples from four Estonian bakeries (Abakery, Bbakery, Cbakery and Dbakery).
Discussion
This study evaluates the stability of both the microbial communities and chemical properties of rye sourdoughs from four Estonian bakeries. The bakeries used flour from the same source, but employed different propagation parameters. Our data showed that controlling the propagation conditions stabilized the cell density and distribution of prevalent LAB species in rye sourdoughs during long term propagation. The cell density of culturable LAB fluctuated in sourdoughs fermented at ambient temperature. High LAB cell densities correlate with high titratable acidity and low pH, which are both characteristic of mature rye sourdough and prerequisite for producing rye bread with desirable sensory properties. Bbakery sourdough samples showed the highest acidity because of the highest fermentation temperature. In contrast, samples collected in both Cbakery and Dbakery were characterized by insufficient acidity during the winter, probably due to the combination of low ambient temperature and short fermentation cycle. Adoption of a constant fermentation temperature (30°C) by Dbakery resulted in the stabilization of acid production and LAB cell density, even in this small scale bakery.
Higher stability of LAB communities was found in sourdough fermented at controlled conditions, compared to those fermented at ambient temperature that seemed strongly affected by the season of collection. Generally, the number of cycles of propagation of type II sourdoughs is lower than type I sourdough due to instability of the microbial community. The starter bacteria are often outcompeted by microorganisms contaminating flour and bakery environment. However, the sourdoughs collected in both large-scale bakeries (Abakery and Bbakery) considered in this study showed better stability. This is probably due to the starter preparation chosen, which contained LAB communities that had been previously adapted to the propagation parameters employed in these bakeries. L. helveticus alone or L. amylovorus together with L. frumenti were the dominant LAB species in Abakery and Bbakery, respectively. L. amylovorus and L. frumenti were previously identified as prevalent in other industrial rye sourdoughs propagated at elevated fermentation temperatures [4–5], similar to that (42–44°C) applied in Bbakery, and are characterized by strong thermo- and acid-tolerance [27]. The persistence of L. amylovorus in sourdoughs may be also attributed to its high amylolytic activity and ability to produce bacteriocine amylovorin, a common feature for representatives of this species [28–29]. The same fingerprint type of dominant L. amylovorus was detected throughout the study, including the fresh sourdough (B0) from another bakery belonging to the same corporation. Adaptation to the sourdough environment could be the reason for such remarkable stability.
Although L. helveticus is not a common dominant species in sourdoughs [4, 13, 16] we found it as dominant bacterial species also in the sourdough of the small scale Dbakery. However, in contrast with Abakery, the sourdough fermentation at ambient temperature in Dbakery prevented the stable prevalence of thermophilic L. helveticus. Indeed, depending on the season, L. pontis and L. zymae prevailed over L. helveticus. L. zymae, a species capable of growing at lower temperatures, has previously been found in both Greek and Belgian wheat sourdoughs, which indicates that it is widely spread [30–31]. New propagation parameters (higher fermentation temperature, decreased inoculum size, prolonged fermentation time, and use of 4°C refrigeration during breaks in production) adopted in Dbakery stabilized the LAB community and triggered an increase in the proportion of L. helveticus and L. pontis. No new species originating from raw materials or bakery environment were detected in the sourdough community of Dbakery even after five months of using the new protocol. This suggests high competitiveness and robustness of the dominant LAB that had adapted to different temperatures and initial sourdough acidity, although the house microbiota of the bakery may have also been the source of these LAB. The importance of house microbiota in the stability of sourdough microbial communities has been shown [12, 32].
Representatives of L. zymae and L. pontis species were also detected among the dominant population of LAB in Cbakery, which utilized sourdough propagation parameters that are very similar to those originally applied in Dbakery. Unfortunately, comparing representatives of L. pontis with those contained in the commercial starter used in this bakery was not possible since no L. pontis was isolated. In contrast with the Dbakery sourdough, L. sanfranciscensis was also identified among prevailing bacteria in sourdough samples from Cbakery. L. sanfranciscensis is frequently found in type I sourdoughs due to its adaptation to sourdough conditions, its small genome, and metabolism [4]. Stable non-competitive association of this maltose-positive LAB species with maltose-negative yeast C. humilis exists in traditional sourdoughs [33]. L. sanfranciscensis species is capable of hydrolyzing maltose by intracellular maltose phosphorylase activity and thereby accumulate glucose in the environment for C. humilis to utilize [34]. C. humilis was the only yeast species identified in the sourdoughs of both small-scale productions Cbakery and Dbakery.
Co-existence of L. helveticus with the yeast species K. telluris was found in the sourdough samples collected at the Abakery. K. telluris (formerly Saccharomyces telluris, Arxiozyma telluris) is mainly known to cause infections in rodents and it may be isolated from soil [35]. Occurrence of this species in sourdough has not been previously reported. However, this thermophilic yeast is able to ferment glucose and grow on glucose, ethanol, and lactic acid [35]. As our identification is based only on culture-independent method (sequencing of 28S rRNA), further research should be carried out to assess the role of this yeast species in the sourdough community. The high fermentation temperature in Bbakery prevented the development of yeasts in the sourdough.
The culture independent methods applied in this study enabled us to identify LAB species (e.g. L. secaliphilus and L. sanfranciscensis) from sourdough and starter samples that were difficult to be cultivated. It has been previously shown that many sourdough LAB are sensitive to oxygen and/or have complex nutrient requirements [27, 36–37]. A wide variety of media should therefore be used to isolate sourdough LAB, since there is no universal medium that is suitable for all LAB. Culture independent methods such as DGGE and pyrosequencing enable one to detect LAB that are difficult to culture on common laboratory media. High throughput sequencing also allows for species identification at the sub-population level and provides quantitative information regarding the relative abundance of species within sourdough [16, 38].
Our data showed that sourdough bacterial communities within large-scale production facilities can be stable for many months using controlled propagation conditions, whereas, fermentation at room temperature leads to seasonal fluctuations in the species composition.
Supporting Information
Samples are coded according to the description reported in Table 1. Bands: 1 –Lactobacillus helveticus; 2 –Lactobacillus panis; 3 –Cereal chloroplast DNA; 4 –Lactobacillus pontis; 5 –Lactobacillus amylovorus; 6 –Cereal chloroplast DNA; 7 –Lactobacillus frumenti; 8–10 –Lactobacillus pontis; 11 –Cereal chloroplast DNA; 12 –Lactobacillus pontis; 13 –Lactobacillus sanfranciscensis; 14 –Lactobacillus pontis; 15 –Lactobacillus helveticus; 16 –Lactobacillus pontis; 17 –Cereal chloroplast DNA; 18 –Lactobacillus zymae; 19 –Lactobacillus pontis. Samples are coded according to the description reported in Table 1.
(TIF)
Bands: 1, 2 –Kazachstania telluris; 3 –Cereal DNA; 4, 5, 6, 7 –Candida humilis. Samples are coded according to the description reported in Table 1.
(TIF)
Acknowledgments
The authors would like to thank the four bakeries for providing the sourdough samples.
Data Availability
Pyrosequencing data is available in GenBank database under accession numbers KM972414–KM972548.
Funding Statement
Financial support was provided by the European Regional Development Fund (http://www.struktuurifondid.ee/euroopa-regionaalarengu-fond/) project EU29994 (EV, MB, TT, IS) together with funds from Estonian Research Council (http://www.etag.ee/rahastamine/etf-grandid/) research grant ETF9417 (IS), Ministry of Education and Research (http://www.etag.ee/rahastamine/institutsionaalne-uurimistoetus/) research grant IUT19-27 (TP, IS) and the European Social Fund Doctoral Studies and Internationalization Programme DoRa (http://adm.archimedes.ee/stipendiumid/programm-dora/) administered by SA Archimedes (EV, MB). This work has been partially supported by graduate school „Functional materials and technologies“ receiving funding from the European Social Fund (http://www.struktuurifondid.ee/euroopa-sotsiaalfond-2/) under project 1.2.0401.09-0079 in Estonia (EV). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.De Vuyst L, Neysens P. The sourdough microflora: biodiversity and metabolic interactions. Trends Food Sci Technol. 2005; 16: 43–56. [Google Scholar]
- 2.Hansen AS. Sourdough bread In: Hui YH, Meunier-Goddik L, Hansen AS, Josephsen J, Nip WK, Stanfield PS, Toldra F, editors. Handbook of Food and Beverage Fermentation Technology. New York: Marcel Dekker, Inc.; 2004. pp. 840–870. [Google Scholar]
- 3.Gobbetti M, Corsetti A, Rossi J. The sourdough microflora. Interactions between lactic acid bacteria and yeasts: metabolism of amino acids. World J Microbiol Biotechnol. 1994; 10: 275–279. 10.1007/BF00414862 [DOI] [PubMed] [Google Scholar]
- 4.De Vuyst L, Van Kerrebroeck S, Harth H, Huys G, Daniel H, Weckx S. Microbial ecology of sourdough fermentations: Diverse or uniform? Food Microbiol. 2014; 37: 11–29. 10.1016/j.fm.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 5.Müller M, Wolfrum G, Stolz P, Ehrmann M, Vogel R. Monitoring the growth of Lactobacillus species during rye flour fermentation. Food Microbiol. 2001; 18: 217–227. [Google Scholar]
- 6.Rosenquist H, Hansen A. The microbial stability of two bakery sourdoughs made from conventionally and organically grown rye. Food Microbiol. 2000; 17: 241–250. [Google Scholar]
- 7.Böcker G, Stolz P, Hammes W. Neue Erkenntnisse zum ökosystem saueteig und zur physiologie des saueteig typischen stämme Lactobacillus sanfrancisco und Lactobacillus pontis. Getreide mehl und brot. 1995; 49: 370–374. [Google Scholar]
- 8.Hammes W, Gänzle M. Sourdough breads and related products In: Woods B, editor. Microbiology of Fermented Foods. London: Blackie Academic/Professional; 1998. pp. 199–216. [Google Scholar]
- 9.Meroth C, Walter J, Hertel C, Brandt M, Hammes W. Monitoring the bacterial population dynamics in sourdough fermentation processes by using PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2003; 69: 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siragusa S, Di Cagno R, Ercolini D, Minervini F, Gobbetti M, De Angelis M. Taxonomic structure and monitoring of the dominant population of lactic acid bacteria during wheat flour sourdough type I propagation using Lactobacillus sanfranciscensis starters. Appl Environ Microbiol. 2009; 75: 1099–1109. 10.1128/AEM.01524-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minervini F, Lattanzi A, De Angelis M, Di Cagno R, Gobbetti M. Influence of artisan bakery- or laboratory-propagated sourdoughs on the diversity of lactic acid bacterium and yeast microbiotas. Appl Environ Microbiol. 2012; 78: 5328–5340. 10.1128/AEM.00572-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minervini F, De Angelis M, Di Cagno R, Gobbetti M. Ecological parameters influencing microbial diversity and stability of traditional sourdough. Int J Food Microbiol. 2014; 171: 136–146. 10.1016/j.ijfoodmicro.2013.11.021 [DOI] [PubMed] [Google Scholar]
- 13.Scheirlinck I, Van der Meulen R, Van Schoor A, Vancanneyt M, De Vuyst L, Vandamme P et al. Influence of geographical origin and flour type on diversity of lactic acid bacteria in traditional Belgian sourdoughs. Appl Environ Microbiol. 2007; 73: 6262–6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valjakka TT, Kerojoki H, Katina K. Sourdough Bread in Finland and Eastern Europe In: Kulp K, Lorenz K, editors. Handbook of Dough Fermentations. New York: Marcel Dekker, Inc., 2003. pp. 269–297. [Google Scholar]
- 15.Kline L, Sugihara T, Miller MW. Microorganisms of the San Francisco sour dough bread process. II. Isolation and characterization of undescribed bacterial species responsible for the souring activity. Appl Microbiol. 1971; 21: 459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viiard E, Mihhalevski A, Rühka T, Paalme T, Sarand I. Evaluation of the microbial community in industrial rye sourdough upon continuous back–slopping propagation revealed Lactobacillus helveticus as the dominant species. J Appl Microbiol. 2013; 114: 404–412. 10.1111/jam.12045 [DOI] [PubMed] [Google Scholar]
- 17.Frank J, Reich C, Sharma S, Weisbaum J, Wilson B, Olsen G. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008; 74: 2461–2470. 10.1128/AEM.02272-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weisburg W, Barns S, Pelletier D, Lane D. Ribosomal DNA amplification for phylogenetic study. J Bacteriol 1991; 173: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gafan G, Spratt D. Denaturing gradient gel electrophoresis gel expansion (DGGEGE)–an attempt to resolve the limitations of co-migration in the DGGE of complex polymicrobial communities. FEMS Microbiol Lett. 2005; 253: 303–307 [DOI] [PubMed] [Google Scholar]
- 20.Meroth C, Hammes W, Hertel C. Identification and population dynamics of yeasts in sourdough fermentation processes by PCR-denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2003; 69: 7453–7461. ÕIGE [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKenna P, Hoffmann C, Minkah N, Aye P, Lackner A, Liu Z et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008; 4(2). 10.1371/journal.ppat.0040020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armougom F, Raoult D. Exploring microbial diversity using 16S rRNA high-throughput methods. J Comput Sci Syst Biol. 2009; 2: 74–92. [Google Scholar]
- 23.Schloss P, Westcott S, Ryabin T, Hall J, Hartmann M, Hollister EB et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009; 75: 7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schloss P, Gevers D, Westcott S. Reducing the Effects of PCR Amplification and Sequencing Artifacts on 16S rRNA-Based Studies. PLoS ONE. 2011; 6 10.1371/journal.pone.0027310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prusse E, Quast C, Knittel K, Fuchs B, Ludwig W, Peplies J et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007; 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yue J, Clayton M. A similarity measure based on species proportions. Commun Stat Theor Methods. 2005; 34: 2123–2131. [Google Scholar]
- 27.Ehrmann MA, Brandt M, Stolz P, Vogel RF, Korakli M. Lactobacillus secaliphilus sp. nov., isolated from type II sourdough fermentation. Int J Syst Evol Microbiol. 2007; 57: 745–750. [DOI] [PubMed] [Google Scholar]
- 28.De Vuyst L, Avonts L, Hoste B, Vancanneyt P, Swings J, Callewaert R. The lactobin A and amylovorin L471 encoding genes are identical, and their distribution seems to be restricted to the species Lactobacillus amylovorus that is of interest for cereal fermentations. Int J Food Microbiol. 2004; 90: 93–106. [DOI] [PubMed] [Google Scholar]
- 29.Leroy F, De Winter T, Adriany T, Neysens P, De Vuyst L. Sugars relevant for sourdough fermentation stimulate growth of and bacteriocin production by Lactobacillus amylovorus DCE 471. Int J Food Microbiol. 2007; 112: 102–111. [DOI] [PubMed] [Google Scholar]
- 30.De Vuyst L, Schrijvers V, Paramithiotis S, Hoste B, Vancanneyt M, Swings J et al. The biodiversity of lactic acid bacteria in Greek traditional wheat sourdoughs is reflected in both composition and metabolite formation. Appl Environ Microbiol. 2002; 68: 6059–6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vancanneyt M, Neysens P, De Wachter M, Engelbeen K, Snauwaert C, Cleenwerck I et al. Lactobacillus acidifarinae sp. nov. and Lactobacillus zymae sp. nov., from wheat sourdoughs. Int J Syst Evol Microbiol. 2005; 55: 615–620. [DOI] [PubMed] [Google Scholar]
- 32.Scheirlinck I, Van der Meulen R, De Vuyst L, Vandamme P, Huys G. Molecular source tracking of predominant lactic acid bacteria in traditional Belgian sourdoughs and their production environments. J Appl Microbiol. 2009; 106: 1081–1092. 10.1111/j.1365-2672.2008.04094.x [DOI] [PubMed] [Google Scholar]
- 33.Gänzle M, Ehmann M, Hammes W. Modeling of growth of Lactobacillus sanfranciscensis and Candida milleri in response to process parameters of sourdough fermentation. Appl Environ Microbiol. 1998; 64: 2616–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stolz P, Böcker G. Technology, properties and applications of sourdough products. Adv Food Sci. 1996; 18: 234–236. [Google Scholar]
- 35.Kurtzman C, Robnett C, Ward J, Brayton C, Gorelick P, Walsh T. Multigene phylogenetic analysis of pathogenic candida species in the Kazachstania (Arxiozyma) telluris complex and description of their ascosporic states as Kazachstania bovina sp. nov., K. heterogenica sp. nov., K. pintolopesii sp. nov., and K. slooffiae. J Clin Microbiol. 2005; 43: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mihhalevski A, Sarand I, Viiard E, Salumets A, Paalme T. Growth characterization of individual rye sourdough bacteria by isothermal microcalorimetry. J Appl Microbiol. 2011; 110: 529–540. 10.1111/j.1365-2672.2010.04904.x [DOI] [PubMed] [Google Scholar]
- 37.Vera A, Rigobello V, Demarigny Y. Comparative study of culture media used for sourdough lactobacilli. Food microbiol. 2009; 26: 728–733. 10.1016/j.fm.2009.07.010 [DOI] [PubMed] [Google Scholar]
- 38.Ercolini D. High-Throughput Sequencing and Metagenomics: Moving Forward in the Culture-Independent Analysis of Food Microbial Ecology. Appl Environ Microbiol. 2013; 79: 3148–3155. 10.1128/AEM.00256-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Samples are coded according to the description reported in Table 1. Bands: 1 –Lactobacillus helveticus; 2 –Lactobacillus panis; 3 –Cereal chloroplast DNA; 4 –Lactobacillus pontis; 5 –Lactobacillus amylovorus; 6 –Cereal chloroplast DNA; 7 –Lactobacillus frumenti; 8–10 –Lactobacillus pontis; 11 –Cereal chloroplast DNA; 12 –Lactobacillus pontis; 13 –Lactobacillus sanfranciscensis; 14 –Lactobacillus pontis; 15 –Lactobacillus helveticus; 16 –Lactobacillus pontis; 17 –Cereal chloroplast DNA; 18 –Lactobacillus zymae; 19 –Lactobacillus pontis. Samples are coded according to the description reported in Table 1.
(TIF)
Bands: 1, 2 –Kazachstania telluris; 3 –Cereal DNA; 4, 5, 6, 7 –Candida humilis. Samples are coded according to the description reported in Table 1.
(TIF)
Data Availability Statement
Pyrosequencing data is available in GenBank database under accession numbers KM972414–KM972548.