Abstract
Fusarium wilt caused by Fusarium oxysporum f. sp. conglutinans (FOC) is a destructive disease of Brassica crops, which results in severe yield losses. There is little information available about the mechanism of disease resistance. To obtain an overview of the transcriptome profiles in roots of R4P1, a Brassica oleracea variety that is highly resistant to fusarium wilt, we compared the transcriptomes of samples inoculated with FOC and samples inoculated with distilled water. RNA-seq analysis generated more than 136 million 100-bp clean reads, which were assembled into 62,506 unigenes (mean size = 741 bp). Among them, 49,959 (79.92%) genes were identified based on sequence similarity searches, including SwissProt (29,050, 46.47%), Gene Ontology (GO) (33,767, 54.02%), Clusters of Orthologous Groups (KOG) (14,721, 23.55%) and Kyoto Encyclopedia of Genes and Genomes Pathway database (KEGG) (12,974, 20.76%) searches; digital gene expression analysis revealed 885 differentially expressed genes (DEGs) between infected and control samples at 4, 12, 24 and 48 hours after inoculation. The DEGs were assigned to 31 KEGG pathways. Early defense systems, including the MAPK signaling pathway, calcium signaling and salicylic acid-mediated hypersensitive response (SA-mediated HR) were activated after pathogen infection. SA-dependent systemic acquired resistance (SAR), ethylene (ET)- and jasmonic (JA)-mediated pathways and the lignin biosynthesis pathway play important roles in plant resistance. We also analyzed the expression of defense-related genes, such as genes encoding pathogenesis-related (PR) proteins, UDP-glycosyltransferase (UDPG), pleiotropic drug resistance, ATP-binding cassette transporters (PDR-ABC transporters), myrosinase, transcription factors and kinases, which were differentially expressed. The results of this study may contribute to efforts to identify and clone candidate genes associated with disease resistance and to uncover the molecular mechanism underlying FOC resistance in cabbage.
Introduction
Fusarium wilt is a destructive disease that causes great losses to cabbage (Brassica oleracea L. var. capitata) production worldwide. This disease was first identified in the United States by Smith in the 1890s [1], and in the following decades was subsequently found in Japan and several other countries [2]. In recent years, fusarium wilt has been identified in several provinces in China [3–5].
Cabbage fusarium wilt is a soil-borne disease caused by pathogen FOC (Fusarium oxysporum f. sp. Conglutinans), which can remain in soil for years or even decades [6]. This pathogen infects cabbage roots, colonizes and occludes the xylem vessels, and leads to leaf wilt or sometimes wilting of the entire plant, with stunted growth and eventually death. Traditional methods, such as crop rotation and chemical control, have almost no effect on the disease because this pathogen is ubiquitous in soil and is not eradicated by these methods. Consequently, developing resistant cultivars is considered to be the most effective measure to control fusarium wilt in cabbage [7].
Molecular mapping of genes controlling fusarium wilt resistance has been extensively reported over the past five years. In order to determine the inheritance pattern of resistance to Fusarium in cabbage and to clone FOC resistance genes for marker-assisted selection (MAS) in cabbage resistance breeding, Jiang et al. [8] developed the stable SCAR marker S46M48199, which is linked in repulsion to the dominant allele of the fusarium wilt resistance gene FOC-1 at a distance of 2.78 cM in cabbage. Pu et al. [7] mapped fusarium wilt resistance gene Foc-Bo1 to linkage group seven (O7) using both segregation test and quantitative trait locus (QTL) analysis in cabbage, and they eventually cloned FocBo1, which encodes a TIR-NBS-LRR type R gene [9]. Lv et al. [10] developed two InDel markers, M10 and A1, flanking the FOC resistance gene at 1.2 and 0.6 cM, respectively, based on a DH population, ultimately mapping the candidate resistance gene to Bol037156 on chromosome C06; this gene encodes a putative TIR-NBS-LRR type R protein [11]. However, these reports do not provide a comprehensive view of the defense mechanism to FOC in cabbage.
High-throughput RNA sequencing (RNA-seq) technology represents a powerful and efficient method for transcriptome analysis and has led to the discovery of many interesting genes. It has made it possible to monitor disease resistance-related gene expression profiles and to reveal the signal transduction pathways involved in the defense network. Applying genome wide transcriptomics to study host-pathogen interactions has provided insights into the mechanisms underlying disease development, basal defense and gene-for-gene resistance. Several transcriptome profiling studies of plants following inoculation with Fusarium fungus have been reported, including studies in watermelon [12], banana [13], Arabidopsis thaliana [14, 15] and wheat [16]. In these studies, the following have been investigated in plant defense responses: defense-related genes, including pathogenesis-related (PR) genes, phytoalexin, transcription factors, protein kinase and ROS-related genes, resistance (R) genes; and the roles of salicylic acid (SA), jasmonic acid (JA) and ethylene (ET) signaling pathways. Signaling pathways activating disease resistance are commonly classified as gene-for-gene resistance responses, SA-dependent responses, and JA- and ET-dependent responses. All of these studies suggest that JA signaling plays an important role in resistance to Fusarium. Some studies have shown that SA signaling is not involved in resistance to necrotrophic pathogens or necrotrophic phases [13, 15, 16]. However, a study examining the response to F. oxysporum-infection in Arabidopsis [10] showed that genes associated with SA-dependent SAR were induced at 6 DPI (days post infection), when F. oxysporum switches from a biotrophic to a necrotrophic lifestyle [16] and when the SA pathway is not activated. Although study showed that effective defense responses against biotrophic pathogens are associated with the activation of the SA-dependent defense pathway, and JA and ET signaling are activated during defense responses to necrotrophic pathogens [17], there are exceptions to this rule, and the situation is complex. The details of the resistance response to FOC in cabbage remain to be elucidated.
In this study, we performed the first global analysis of transcriptome dynamics during the FOC defense response in cabbage using RNA-seq. Specifically, we analyzed the differential gene expression patterns between inoculated and control roots at various time points. The results of this study will help reveal genes and pathways associated with resistance to FOC in cabbage, which contributes to our understanding of the FOC resistance mechanism.
Materials and Methods
Preparation of material
The fungal strain GLHW1, isolated from diseased cabbage in Shouyang, Shanxi province, China, which belongs to Race 1 [4, 5] was incubated on potato-dextrose agar plates at 26–28°C for 5–7 days, following by growth in potato-lactose broth on a shaker at 125 rpm at 26–28°C for 5–7 days. The suspension concentration was adjusted to 1×106 spores ml-1 with sterile distilled water prior to inoculation.
R4P1 (resistant) and R2P2 (susceptible) seeds were grown in sterilized soil (peat: vermiculite = 1:1) in an artificial climate box at 25°C/18°C day/night temperatures with a 16-h light/8-h dark photoperiod. Seedlings at the three-leaf stage were infected with FOC by root dip inoculation into a suspension of fungal spores for 15 min and then were returned to their original pots, where they were grown at 28°C under the same photoperiod [18]. Control plants were treated in a similar manner but were mock-inoculated with distilled water. Both inoculated and mock-inoculated plant roots were sampled at 4, 12, 24 and 48 hai (hours after inoculation). Five individual seedlings were used per replicate, with a total of three replicates collected per treatment at each sampling time point.
RNA isolation and cDNA library construction
Total RNAs were isolated using the RNAprep Pure Plant Kit (TIANGEN). Twelve RNA samples isolated from R4P1 roots at 4, 12, 24 and 48 hai, including eight inoculated samples (FW4_1, FW4_2, FW12_1, FW12_2, FW24_1, FW24_2, FW48_1 and FW48_2) and four mock-inoculated samples (DW4, DW12, DW24 and DW48), were sent to Novogene Bioinformatics Technology Co. Ltd. (Beijing) for library construction and sequencing.
RNA integrity was confirmed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA) with a minimum RNA integrated number of 8. Sequencing libraries were generated using an Illumina TruSeq™ RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). Briefly, mRNA was purified from 3 μg total RNA using poly-T oligo-attached magnetic beads and fragmented using Illumina proprietary fragmentation buffer at 94°C for 15 min. First-strand cDNA was synthesized using a 6 bp-random primer and SuperScript II. Second-strand cDNA was then synthesized. These cDNA fragments were then subjected to an end repair process, and to adenylation and ligation of adapters P5 and P7. The insert size of the library was restricted to approximately 300 bp using PCR products purified with the AMPure XP system (Beckman Coulter, Beverly, MA, USA).
Illumina sequencing and assembly for transcriptome analysis
Transcriptome sequencing generated 100 bp paired-end (PE) raw reads using the Illumina HiSeq 2000. The raw reads were then filtered by discarding adaptor reads, reads with the ratio of ‘N’ >10% and low quality reads with more than 50% bases of quality value ≤5. The remaining clean reads were then assembled using Trinity software for transcriptome assembly without a reference genome, with min_kmer_cov set to 2 by default and all other parameters set to default values [19].
Homology annotations were performed using the following public databases: NCBI Nr (non-redundant protein), NCBI Nt (nucleotide sequences), Swiss-Prot, GO (Gene Ontology), KOG (euKaryotic Ortholog Groups) and KEGG (Kyoto Encyclopedia of Genes and Genomes). If the results from different database searches conflicted, they were prioritized in the following order: Nr, Nt, KEGG, Swiss-Prot, GO and KOG. The threshold values for Nr, Nt and Swiss-Prot were 1e-5; that for KOG was 1e-3. And the genome data of Arabidopsis thaliana [20, 21], Brassica rapa [22] and Brassica oleracea [23] were also used for the analysis of unigenes.
Illumina sequencing and mapping for differential gene expression analysis
The SE100 (single-end) sequencing strategy was used for digital gene expression profiling. All clean reads were mapped to the assembled reference sequences with a restriction of no more than 1 bp mismatch. Mapping was conducted using RSEM software [24]. The number of mapped clean reads was calculated and normalized to RPKM values [25]. Read count values are input data used to analyze differential gene expression. For samples with biological replicates, differential expression analysis of two samples was performed using the DESeq R package [26]; genes with an adjusted p-value < 0.05 by DESeq were determined to be differentially expressed. For samples without biological replicates, the DEGseq R package [27] was used; q-value < 0.005 and |log2 (fold change) |>1 were set as the thresholds for differential gene expression. RPKM was used for subsequent analysis, such as correlation analysis of gene expression and DEG cluster analysis.
GO enrichment analysis of the differentially expressed genes (DEGs) was implemented by the GOseq R packages based on Wallenius’ noncentral hypergeometric distribution [28], which can adjust for gene length bias in DEGs. KOBAS [29] software was used to test the statistical enrichment of DEGs in KEGG pathways.
Validation of DEGs by quantitative RT-PCR
DEGs selected were validated using qPCR. The cDNA was synthesized from the same samples used for Illumina sequencing. GAPDH was used as an internal reference. RT-PCR was performed using SYBR Green 1 (TIANGEN) on a LightCycler 480 II Real-Time PCR Detection System (Bio-Rad, USA). The reaction was carried out in a total volume of 20 μL containing 10 μL of 2× SuperReal PreMix Plus, 2 μL of cDNA mix, 0.6 μL of each primer and 6.8 μL of RNase-free ddH2O. The thermal profile for RT-PCR was 95°C for 15 min, followed by 40 cycles of 95°C for 10 s, 60°C for 20 s and 72°C for 20 s. Melting curve analysis was performed at the end of each PCR reaction at 95°C for 5 s, 65°C for 60 s, 97°C (continuous), 40°C for 30 s. Relative expression was calculated using the comparative CT method (2-ΔΔCt method). Genes and primer sequences can be found in S1 Table.
Results
Transcriptome characterization of R4P1 roots after infection with FOC by high-throughput RNA sequencing
All inoculated and non-inoculated samples were equally mixed (designated Trans_BO) and sequenced for transcriptome analysis. After raw read filtering and quality checks, more than 136 million 100-bp clean reads were obtained. Transcriptome assembly was carried out with Trinity software, resulting in the identification of 62,506 unigenes with an average length of 741 bp for annotation (Table 1), total of 49,959 unigenes (79.92% of all unigenes) were identified in at least one database.
Table 1. Statistical results of unigene annotations.
| Database type | Number of unigenes | Percentage (%) |
|---|---|---|
| NT | 34036 | 54.45 |
| NR | 43501 | 69.59 |
| SwissProt | 29050 | 46.47 |
| PFAM | 25716 | 41.14 |
| KO | 12974 | 20.75 |
| GO | 33767 | 54.02 |
| KOG | 14721 | 23.55 |
| Annotated in at least one Database | 49959 | 79.92 |
| Total Unigenes | 62506 | 100 |
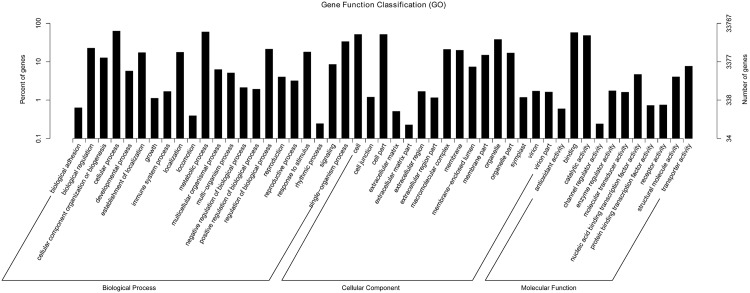
GO enrichment analysis was performed to classify gene functions. A total of 33,767 unigenes were assigned to 49 GO terms in three categories: BP (Biological process), CC (Cellular component) and MF (Molecular Function). A high proportion of DEGs were assigned to cellular process and metabolic process in the BP category, to cell, cell part and organelle in CC and to binding and catalytic activity in MF (Fig 1).
Fig 1. Gene ontology classification of unigenes.
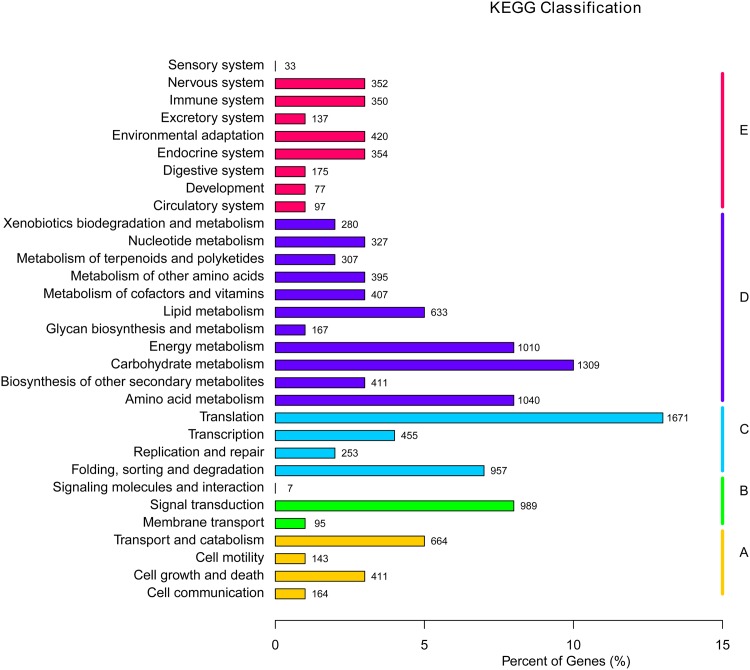
A total of 12,974 unigenes were annotated in GO and were then classified in the KEGG database to analyze the metabolic pathways in which they participate, which were divided into five branches: A (Cellular Processes), B (Environmental Information Processing), C (Genetic Information Processing), D (Metabolism) and E (Organismal Systems). In each of the five branches, the most highly represented pathways were environmental adaption (420, 0.67%), carbohydrate metabolism (1,309, 2.09%), translation (1,671, 2.67%), signal transduction (989, 1.58%) and transport and catabolism (664, 1.06%). Moreover, 350 unigenes (0.56% of all unigenes) participate in the immune system (Fig 2).
Fig 2. KEGG classification of unigenes.
(A) Cellular Processes. (B) Environmental Information Processing. (C) Genetic Information Processing. (D) Metabolism. (E) Organismal Systems.
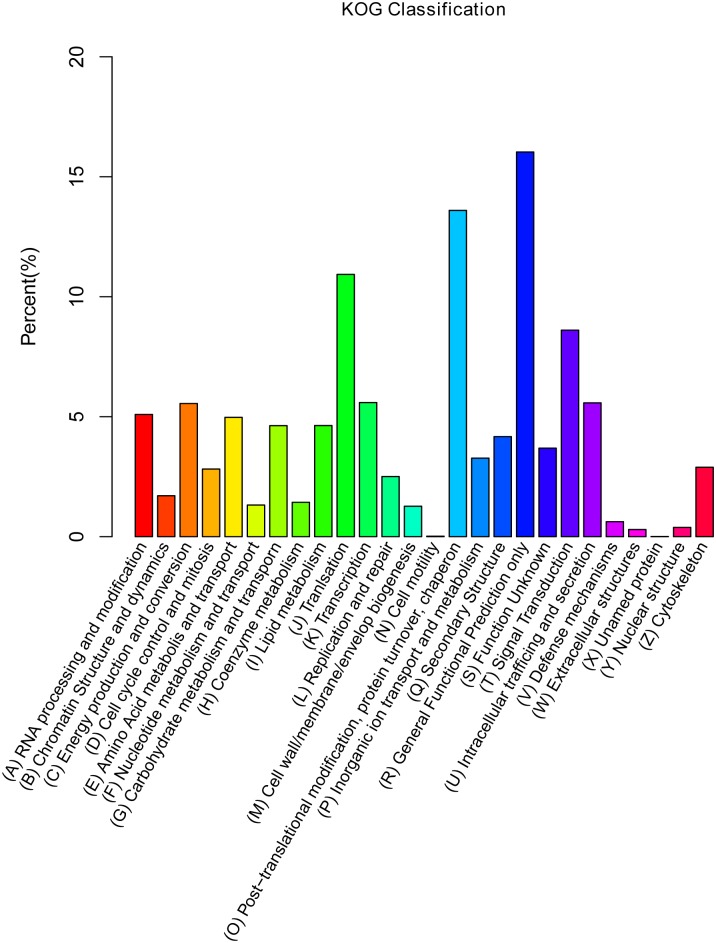
In addition, 14,721 unigenes (23.55%) were assigned to 26 KOG functional categories (Fig 3). Among these genes, ‘general function prediction only’ represents the largest group (2,360, 3.77%), followed by ‘post-translational modification, protein turnover, and chaperones’ (2,002, 3.20%), ‘translation’ (1,609, 2.57%) and ‘signal transduction’ (1,267, 2.03%).
Fig 3. KOG classification.
Digital gene expression library sequencing and annotation
Based on the transcriptome data, 12 libraries were constructed to identify the gene expression profiles of R4P1 roots during FOC infection, including DW4, FW4_1, FW4_2, DW12, FW12_1, FW12_2, DW24, FW24_1, FW24_2, DW48, FW48_1 and FW48_2 (table 2). Each library generated 10.85 to 15.74 million raw reads. After performing a quality check, the number of clean reads ranged from 10.72 to 15.52 million. The minimum and maximum GC percent values of the 12 libraries were 45.72% and 46.70%, respectively. The Q20 percentages were ≥96.50% and the number of reads mapped to 62,506 unigenes using RSEM ranged from 13,832,709 (89.08%) to 13,469,925 (91.13%) [24], confirming that the sequencing data were appropriate for subsequent analysis.
Table 2. Data quality evaluation of sample.
| Sample name | Raw reads | Clean reads | GC (%) | Error (%) | Q20 (%) | Total mapped |
|---|---|---|---|---|---|---|
| DW4 | 14828646 | 14638472 | 46.28 | 0.04 | 96.54 | 13339188(91.12%) |
| FW4_1 | 11124834 | 10985291 | 46.31 | 0.04 | 96.64 | 9970094 (90.76%) |
| FW4_2 | 13629475 | 13435056 | 46.55 | 0.04 | 96.55 | 12162626(90.53%) |
| DW12 | 14950551 | 14780536 | 45.72 | 0.04 | 96.87 | 13469925(91.13%) |
| FW12_1 | 14905611 | 14714091 | 46.11 | 0.04 | 96.84 | 13383604(90.96%) |
| FW12_2 | 10853023 | 10724071 | 46.13 | 0.04 | 96.58 | 9603058 (89.55%) |
| DW24 | 15461231 | 15289907 | 46.51 | 0.04 | 96.68 | 13912721(90.99%) |
| FW24_1 | 15748199 | 15529280 | 46.45 | 0.04 | 96.72 | 13832709(89.08%) |
| FW24_2 | 12921595 | 12747599 | 46.01 | 0.04 | 96.60 | 11423440(89.61%) |
| DW48 | 11949074 | 11785699 | 46.70 | 0.04 | 96.54 | 10655203(90.41%) |
| FW48_1 | 11187930 | 11038671 | 46.63 | 0.04 | 96.58 | 10023240(90.80%) |
| FW48_2 | 15715268 | 15512388 | 46.16 | 0.04 | 96.63 | 14021279(90.39%) |
Reliability analysis of digital gene expression sequencing data
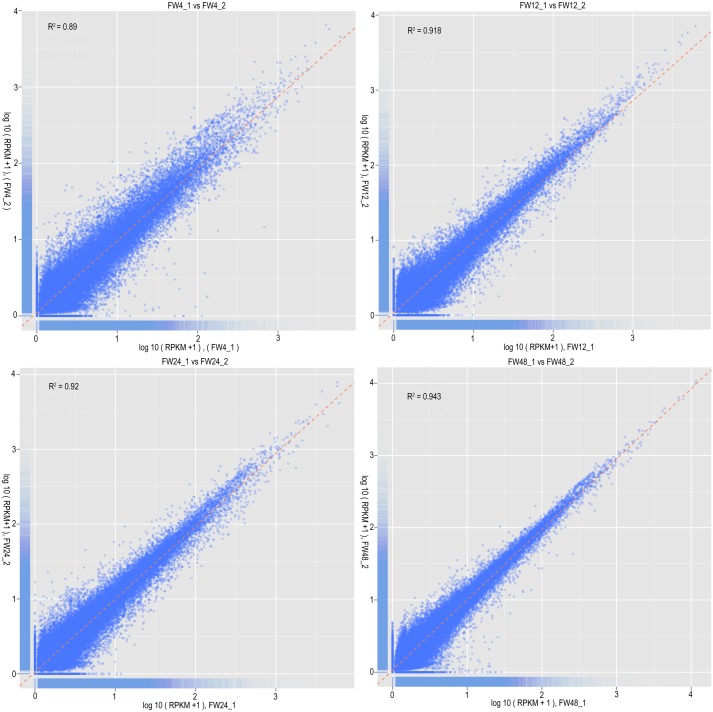
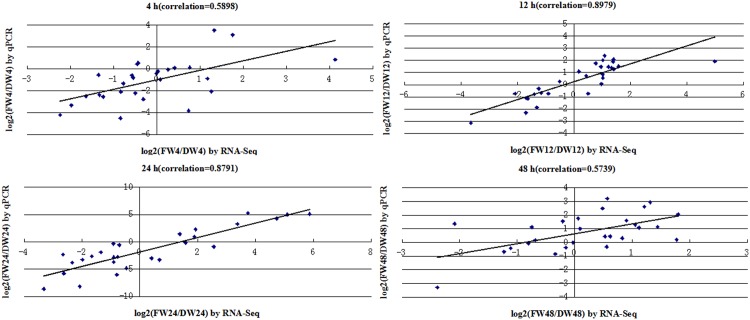
Correlation analysis of gene expression levels between replicate samples is an important technique for verifying experimental reliability and sampling accuracy. We determined the correlation coefficient between two replicate samples based on the RPKM values of genes, which were calculated using the number of read counts and the mapped gene length [25]. The R2 values of the four replicate groups were 0.89, 0.918, 0.92 and 0.943 (Fig 4), respectively, exhibiting remarkable consistency. These results suggest that the replicate samples had high reliability and repeatability, which ensures that digital gene expression analyses reflected actual differences in gene expression between infected and mock-infected materials.
Fig 4. Correlation scatter diagram of gene expression among four treatment groups.
Analyses of DEGs
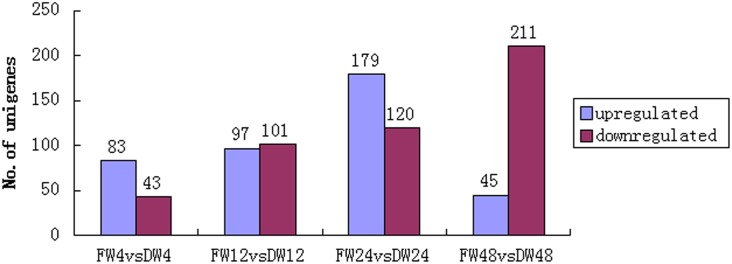
A total of 885 DEGs were identified between infected and mock-infected samples at each inoculation time, with a cut-off value of adjusted p-value < 0.05 and |log2 (fold change) |>1. The numbers of up- or down-regulated genes at different time points after pathogen inoculation are shown in Fig 5. As the time after inoculation increased, the number of down-regulated genes also increased, while there was a slight difference in the number of DEGs and up-regulated genes; the largest numbers of genes in these categories were detected at 24 hai.
Fig 5. Number of differentially expressed genes at different time points after inoculation.
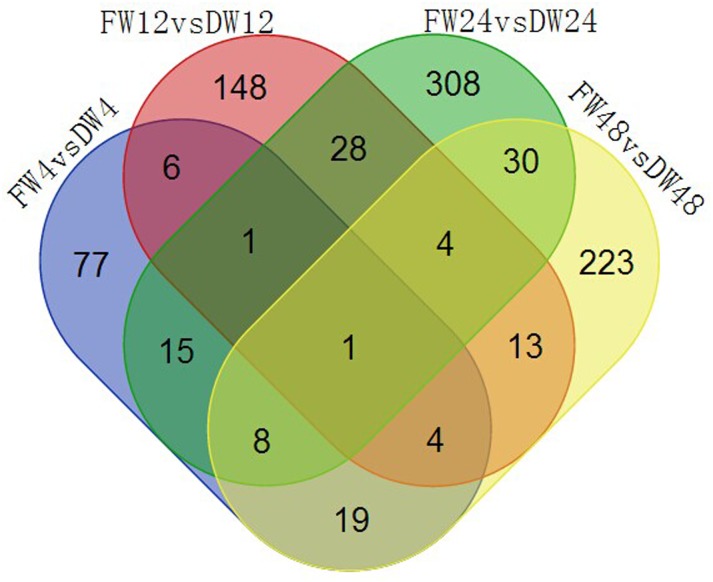
The gene comp37314_c0 was commonly detected in the four groups (Fig 6). This gene encodes a chitinase A, indicating that chitinase plays an important role in the FOC defense system. The number of unique genes in the FW24 vs DW24 group was the highest as that of DEGs and up-regulated genes, suggesting that 24 hai may be a critical time in the disease resistance response in cabbage.
Fig 6. Venn diagram of differentially expressed genes.
Validation of RNA-seq-based DEGs by qRT-PCR
We selected 27 defense-related genes for validation using qPCR. The input data were relative fold changes of log2 (FW_RPKM/DW_RPKM) based on RNA-seq and log2 (2-ΔΔCt) based on qPCR for each DEG between infected and mock-infected roots at four stages of treatment (S1 Table), which were compared using a method described for cotton [30]. Correlation coefficients were calculated to assess the correlation between the two platforms, yielding values of 0.5898, 0.8979, 0.8791 and 0.5739 at 4 h, 12 h, 24 h and 48 h, respectively (Fig 7, correlation is significant at the 0.01 level). The tested genes displayed the same expression of up-regulation or down-regulation by the RNA-seq and qPCR, however, the fold changes varies because of the two different methods for gene quantification. So the lower correlations at 4 h and 48 h may be allowed. In summary, these results indicated that the relative expression of fold changes between the two methods showed a moderate correlation.
Fig 7. Correlation between RNA-seq and qRT-PCR data at different time points.
Global gene regulation in response to FOC
MAPK signaling pathway
The MAPK signal transduction pathway is an early defense response observed upon pathogen attack. We detected seven DEGs that participate in the MAPK signaling pathway according to KEGG enrichment analysis (S2 Table), including four genes encoding heat shock 70 kDa protein 5 (HSC70b), which were significantly induced at 4 hai, and slightly regulated at 12, 24 hai and repressed at 48 hai. MAPKKK13 gene (comp57145_c0) and PDR-type ABC transporter gene (comp20414_c0) were up-regulated after pathogen infection and were induced approximately 58-fold and 5.9-fold at 24 hai, respectively. Gene (comp25940_c0) encoding lipid transfer protein (LTP) was down-regulated after pathogen infection at the four time points. GO analysis showed that comp25940_c0 participated in negative regulation of defense response and MAPK cascade. The expression patterns of most of these genes confirm the early role played by the MAPK signaling pathway in response to FOC in cabbage.
Calcium signaling
Calcium signaling plays important roles in triggering the biosynthesis of SA, JA and ET [31]. The PAMP (pathogen-associated molecular patterns) signaling system generates specific Ca2+ signals in the cytosol, and the calmodulin-binding protein CBP60g participates in activating SA biosynthesis. NO mediated by Ca2+ influx plays an important role in JA biosynthesis. Ca2+ signaling activates ACC synthase, which involved in the biosynthesis of ET. Calcium sensors and Ca2+-dependent protein kinases are involved in ET signaling. We detected 12 DEGs associated with calcium signaling (S2 Table). Ca2+ ATPase is a transport protein in the plasma membrane that regulates the amount of Ca2+ within a cell to transport the surrounding signal [32] Ca2+ ATPase gene (comp29882_c0) identified here was up-regulated at 4, 12 and 24 hai and down-regulated at 48 hai. Two calmodulin protein genes and three calmodulin-binding protein (CBP) genes were highly induced at 4 hai. Gene aquaporin TIP (tonoplast intrinsic protein, comp39047_c0) was highly induced in FW12. MYB2 gene (comp35012_c0) was highly induced in FW4 and FW24. In addition, CBP CML37 (comp37685_c0) participates in the ET biosynthetic process, which helps confirm that Ca2+ signaling plays a role in ET biosynthesis. These results indicated that Ca2+ (as a secondary messenger) and subsequent Ca2+ signaling is activated during the early infection stage
JA, ET and SA signaling pathways
Three signaling molecules regulate signaling pathways associated with plant defense responses including JA, ET and SA. Eight genes associated with the JA pathway were identified (S2 Table), including three genes responsible for alpha-linolenic acid metabolism (a necessary pathway in JA biosynthesis) and five genes involved in JA mediated signaling pathways, including a lipid transfer protein (LTP) gene (comp25940_c0), an PDR-type ABC transporter G family gene (comp20414_c0), a CYP94A1 gene (comp44973_c0) and a myrosinase-binding protein-like (MBP) gene (comp41986_c0) and myrosinase gene (comp35843_c0). One LOX (lipoxygenase) gene (comp37329_c0), encoding a critical enzyme in the JA biosynthesis pathway, was highly induced in FW12 and FW48, whereas LTP gene (comp25940_c0) which is necessary in transduction of lipid molecules such as JA was repressed after infection, particularly in FW12 and FW24. Myrosinase gene (comp35843_c0) and JA-induced MBP gene (omp41986_c0) and were significantly down-regulated in FW12 and FW24.
Seven genes involved in SA-dependent SAR were differentially expressed (S2 Table), most of which were highly induced in FW4 and repressed in FW48 when compared to the mock-treated roots. These genes include three transcription factor genes (comp40515_c0, comp41745_c0 and comp28250_c1), two genes encoding unknown proteins (comp51677_c0 and comp33538_c0) and two genes (comp20414_c0 and comp25940_c0) participated in both SA biosynthesis and SA-dependent SAR. In addition, two UDPG genes (comp34450_c0 and comp45408_c0) that participate in the SA-mediated HR were induced at the infection roots and were highly induced in FW24 with fold change of 4.9 and 34.2, respectively (S2 Table).
Six DEGs that participate in ET-mediated signaling and four genes that function in ET biosynthesis were detected, including two genes (comp37878_c0, comp38040_c0) encoding E3 ubiquitin-protein ligase that function in the both pathways (S2 Table). All these genes were highly induced in FW4, slightly induced in FW24 and repressed in FW12 and FW48, except for UDPG (comp35166_c0), which was down-regulated in FW4 and up-regulated in the other infected roots especially in FW12 and FW24. Moreover, twenty ET-responsive transcription factor (ERF) genes were differentially expressed, most of which were highly induced at 4 hai (S3 Table). Ubiquitin-ligase genes respond to chitin, a plant-defense elicitor, play a role in the plant defense response [33]. These results suggest that the ET-mediated signaling pathway is activated upon pathogen infection, which is consistent with the early role of the ET signaling pathway in response to F. oxysporum infection in Arabidopsis [14].
Lignin biosynthesis
In many cases, during plant-pathogen incompatible interactions, transcriptional profiling studies have revealed genes involved in the biosynthesis and modification of cell wall components [30]. Lignin deposited during plant-pathogen interactions forms a physical barrier against infection. In this study, we identified 11 DEGs that participate in phenylpropanoid-lignin pathway (S2 Table), including key enzymes in lignin biosynthesis: peroxidase (POD, comp34233_c0, comp37399_c0, comp28296_c0, comp16630_c0), ferulic acid 5-hydroxylase (F5H, comp35066_c0), caffeic acid 3-O-methyltransferase (COMT, comp30914_c0) and pyridoxal-phosphate dependent enzyme (comp35737_c0), glycosyl hydrolase (comp35141_c0, comp30472_c0,comp35843_c0), as well as a gene that participates in xylem and phloem pattern formation (comp44045_c0). Three POD genes and genes encoding glycosyl hydrolase and pyridoxal-phosphate-dependent enzyme involved in the biosynthesis of L-phenylalanine (intermediate of lignin biosynthesis) were significantly repressed in FW4, FW12 and FW24 but induced in FW48. Another POD gene (comp28296_c0) was highly induced in FW12 and FW48 but repressed in FW4 and FW24. F5H gene (comp35066_c0) was induced in FW12, FW24 and FW48. The COMT gene (comp30914_c0) was significantly repressed after pathogen inoculation. Overall, these regulated genes indicate that the lignin biosynthesis pathway is activated during the defense response to FOC. In addition, myrosinase gene (comp35843_c0) identified here participates in both the phenylpropanoid pathway and the JA-mediated pathway, suggesting that the two pathways interact in the disease resistance.
Transcription factors (TFs)
Defense-related genes are normally regulated by TFs, which play direct or indirect roles in different signaling pathways [14]. Among the 885 DEGs, a total of 47 differentially regulated TF genes were identified (S3 Table), including genes encoding WRKY (1), MYC (1), CxHy zinc finger (4), bHLH (4), NAC (4), MYB (5), HSF (8) and ERF (20). Most TF genes were highly up-regulated at 4 hai but down-regulated at 12, 24 and 48 hai.
Protein kinases
Protein phosphorylation and dephosphorylation are important steps in many signal transduction pathways. Protein kinases involved these process help complete the signal transmission. We identified eight differentially expressed protein kinase genes associated with the defense system (S4 Table), including genes encoding MEKK (1), serine/threonine protein kinase (3), leucine-rich repeat (LRR) transmembrane protein kinase (2) and lectin protein kinase (2). The MEKK gene (comp57145_c0) was induced in all infected roots and up-regulated about 58.2 fold at 24 hai. One LRR-type kinase gene (comp19030_c0) was up-regulated at 12, 24 and 48 hai and significantly up-regulated at 24 hai.
Detoxifying-related proteins
UDPG [34] and the PDR-type ABC transporters [35] detoxify deoxynivalenol (DON), a virulence factor, during infection by Fusarium species. In this study, we detected DEGs encoding ten UDPG and three PDR-type ABC transporters (S5 Table). All the genes were highly induced in FW24 except for two UDPG genes (comp29479_c1, comp5003_c0).
Defense genes respond to FOC infection
PR genes are important in disease resistance. The antifungal thaumatin-like protein (PR5) cause osmotic breakage of transmembrane pores on the fungal plasma membrane; the Bet v I family protein (PR10) are involved in the synthesis of compounds such as antibiotics [36]. The chitinase family PR3 hydrolyze the β-1,4 glycosidic linked to the N-acetylglucosamine residues of chitin to participate in the disease resistance [37]. Six PRs were differentially expressed (S6 Table), including one PR5 gene (comp35815_c0), four PR10 genes (comp17904_c0, comp17904_c1, comp20529_c0, comp20089_c0) and one PR3 gene (chitinase, comp21138_c0). PR5 gene was highly induced in FW12 and FW48 and repressed in FW4 and FW24. PR10 gene (comp20089_c0) was induced in FW4 and FW12 and repressed in FW24 and FW48. The other genes were all down-regulated after inoculation.
Chitinases, which function as hydrolytic enzymes in plants, break down chitin (the major component of the cell walls of fungi) into mono- and oligomers to destroy the fungi and prevent fungal infection. Studies supported that chitinases were antifungal to inhibit growth of fungal hyphae [38, 39]. Four genes encoding chitinase were identified by DGE analysis (S6 Table). Among them, comp37238_c0 and comp37314_c0 were highly induced in all inoculated samples, Comp32071_c0 and comp37179_c0 were only induced in FW12.
Cytochrome P450 genes were improved to have a role in response to F. oxysporum infection in Arabidopsis [14]. Five cytochrome P450 genes involved in JA, lignin and indole glucosinolate pathways, which are required for the innate immune response, were differentially expressed (S6 Table). comp43877_c0 (CYP82F1), which is involved in the indole glucosinolate metabolic process, was repressed in infected roots except for FW48. comp120471_c0 (CYP71A12), which participates in induced systemic resistance, was highly induced in FW24 and highly repressed in FW48. For genes involved in JA-metabolic process, comp31152_c0 (CYP94B1) was induced in all inoculated samples except for FW24 and comp44973_c0 (CYP94C1) was down-regulated in all inoculated samples. comp35066_c0 (F5H, CYP84A1) involved in lignin biosynthesis was discussed above.
Glucosinolate metabolism is a required component of the plant defense response against microbial pathogens [40]. The hydrolysis of glucosinolates is catalyzed by myrosinases, forming nitrile, isothiocyanate, amine, epithionitrile, thiocyanate, oxazolidine-2-thione and so on [41]. In Brassicales plants, the glucosinolate-myrosinase defense system produces toxic volatile compounds during pathogen attack [42], and several products have demonstrated in vitro toxicity to mycelial growth in cereal root pathogens [43]. Certain types of MyAPs (myrosinase-associated proteins), the ESPs (epithiospecifiers), function as myrosinase cofactors, which are required to modulate the specificity of myrosinases towards the production of particular enzyme products [44]. MBPs (myrosinase-binding proteins) may bind to carbohydrates present in fungal pathogens through their lectin domain to enhance their defense reactions [45]. One myrosinase gene (comp35843_c0), two MyAP genes (comp38098_c0, comp19088_c0) and three MBP genes (comp32249_c0, comp32249_c1, comp41986_c0) were down-regulated in all the infected roots (S6 Table). Results implied a role of these genes in response to FOC infection.
Expression profiles of DEGs selected in R4P1 and R2P2 by qRT-PCR
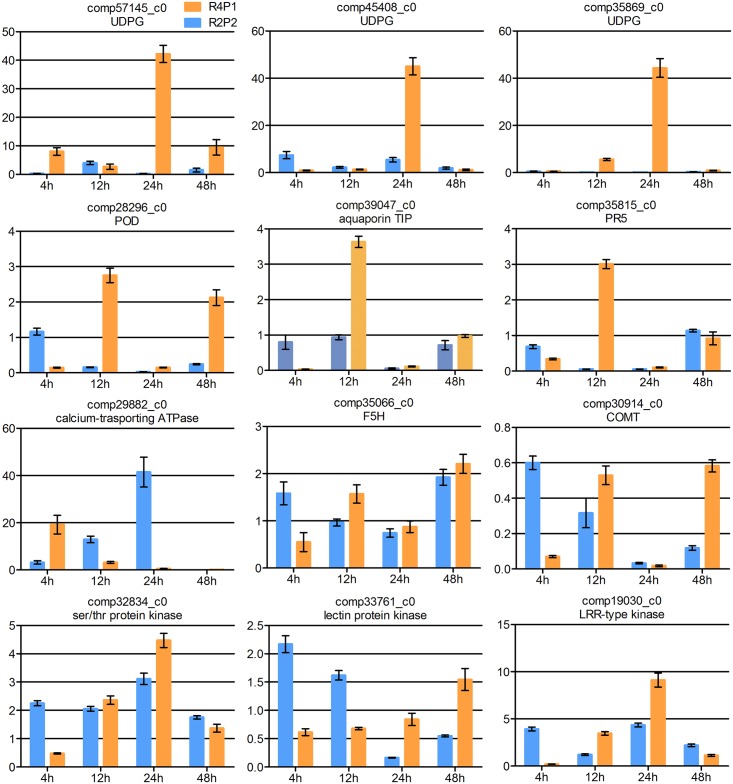
Some of the DEGs were selected to characterize the gene expression profiles between R4P1 (resistant) and R2P2 (susceptible) after FOC inoculation by qRT-PCR (Fig 8). Data were shown in S8 Table. MAPKKK13 (comp57145_c0) and UDPG (comp45408_c0, comp35869_c0) were highly induced more than 40-fold change in R4P1 at 24 hai, while showed low expression level at other times in R4P1 and in R2P2. POD (comp28296_c0), aquaporin TIP (comp39047_c0) and PR5 (comp35815_c0) displayed high expression level at 12 hai or 48 hai but were down-regulated at other times in R4P1 and in R2P2. Calcium-binding ATPase (como29882_c0) showed high expression level at 4 hai in R4P1 compared to R2P2. The expression patterns of aquaporin TIP and Calcium-binding ATPase validated the early role of calcium signaling played in resistance to FOC in cabbage. The expression of the other genes showed an increase trend in R4P1 and a decrease trend in R2P2, and most genes showed a higher expression level in R4P1 except for the 4 hai. These results indicated that these DEGs play roles in disease resistance to FOC in cabbage.
Fig 8. Expression profiles of DEGs in R4P1 and R2P2 by qRT-PCR.
The expression levels on the y-axis were relative to non-inoculated sample from each genotype after normalization with GAPDH gene. Error bars represent the SD for two independent experiment, and three technical replicates.
Discussion
Examining the transcriptomes of R4P1 roots during FOC infection provides comprehensive knowledge for FOC resistance-related gene discovery
In this study, using Illumina sequencing, we identified 62,506 unigenes from R4P1 roots, 79.92% of which were identified in at least one database. The presence of unigenes that could not be annotated might be due to the relatively short lengths of their assembled sequences. It is particularly notable that the number of assembled unigenes (62,506) does not match the 45,758 genes in the recently released cabbage genome sequence. Reasons are the following: (1) most of unigenes obtained were parts of corresponding genes; (2) some were different regions from the same gene; (3) some were discarded from further analysis because of their short size or unsatisfactory alignment [16]. Nonetheless, the transcriptome data offered an overview of the gene expression profiles of FOC-inoculated roots of R4P1 and a valuable set of genes with which to investigate FOC resistance-related genes.
MAPK signaling pathway functions in the response to FOC
Recognition of bacterial elicitor flg22 by receptor kinase FLS2 in Arabidopsis activates the cascade reaction MEKK1-MKK4/MKK5-MPK3/MPK6-WRKY22/WRKY29 [14, 46]. The MKK4/MKK5-MPK3/MPK6 cascade in Botrytis cinerea-infected Arabidopsis plants regulates the biosynthesis of camalexin, which is the major phytoalexin in Arabidopsis [47]. In the current study, however, no such MAPK cascade associated with the defense response to FOC was detected, which is consistent with the results of a study in Arabidopsis, cotton and tomato [48]. However, we identified seven genes that participate in MAPK signaling pathway in this study. Among them, four Hsp70b genes were annotated in BP description as viral process or response to virus to trigger the MAPK signaling pathway, which indicated that HSC70b proteins were upstream in MAPK signaling pathway. Indeed, the expression of two HSC70 isoforms were upregulated by pathogen infection, while loss-of-function mutants of individual cytosolic HSC70 genes do not display defense phenotypes [49] LTP genes (comp25940_c0) and PDR-type ABC transporter gene (comp20414_c0) participating in MAPK cascade to negatively regulate the defense response function downstream of this pathway. Although only one MAKKK13 (MEKK13) gene was identified, the analysis above implied that there may be another MAPK cascade in FOC resistance in cabbage.
SA-dependent pathways function in FOC resistance in Brassica oleracea
During the disease resistance response, SA triggers the expression of a number of genes, which can be divided into two groups: group 1, genes that function immediately/early in the HR that were independent of NPR1 (nonexpresser PR genes 1), such as those encoding glycosyltransferases and glutathione S-transferases; group 2, genes that function late in the SAR, such as PRs, which require NPR1 induction [50]. The HR occurs at the site of attack, where NPR1 is degraded to remove its inhibitory effect on effector-triggered cell death and defense, but it accumulates in neighboring cells to promote cell survival and SA-mediated resistance [51], indicating that NPR1 is not involved in the HR.
We identified two differentially expressed SA-induced UDPG genes (comp45408_c0 and comp34450_c0), whose induction by SA is independent of NPR1. In addition, comp34450_c0 is homologous with pathogen-inducible AtSGT1, whose expression is an early disease response in Arabidopsis that may be involved in the accumulation of glucosyl SA during the disease process [52]. SGT1 is an early, essential component of R gene-triggered disease resistance and can interact with the LRR domains of certain NBS-LRR proteins [53–55]. Furthermore, the interaction between SGT1 and HSC70 regulates the immune responses in Arabidopsis [49], and in this study, we identified four HSC70 genes that function in the MAPK signaling pathway, which suggests that crosstalk might occur between the MAPK signaling pathway and the SA-UDPG-dependent HR pathway.
In this study, our results indicate that SA-dependent SAR is involved in cabbage defense response against FOC. However, there is no evidence that these genes are involved in the EDS/PAD4-SA-NPR1-PR signaling pathway. In addition, F. oxysporum is considered to be a hemibiotrophic pathogen, as it begins its infection cycle as a biotroph and subsequently becomes a necrotroph [15]. The SA-dependent defense pathway is activated in the defense response against biotrophic pathogens or in the biotrophic phase [17]. Most genes that participate in SA-dependent SAR were highly induced in FW4, FW12 and FW24 and repressed in FW48, indicating that 48 hai is likely to be the turning point between the biotrophic phase and the necrotrophic phase of the FOC in R4P1 in this study.
Notably, the candidate FOC resistance gene (comp30068_c1) identified in this study (S6 Table), encoding a TIR-NB-LRR-type resistance gene, annotated as SNC1 (Suppressor of NPR1-1, Constitutive1), is involved in SA-dependent SAR. Mutant plants constitutively express PR genes, and PRs in the SA-mediated SAR require NPR1, which function downstream of SA[51]. EDS1 (enhanced disease susceptibility 1), PAD4 (phytoalexin deficient 4) and MOS3 (modifier of SNC1,3) are required in SNC1 resistance signaling. EDS1, an essential component of resistance specified by TIR-NB-LRR, functions upstream of SA-dependent defense responses and acts as a pathogen effector target in Arabidopsis [56]. EDS1 interacts with its positive co-regulator PAD4, resulting in mobilization of the SA defense pathway in rice and Arabidopsis [57]. Studies in rice and Arabidopsis suggest that EDS1 and PAD4 form a dimeric protein complex, which might be important for triggering the SA signaling pathway in plants [58, 59]. The analysis also suggested that the TIR-NB-LRR-EDS/PAD4-SA-NPR1-PR pathway may function in the defense response of cabbage to FOC. Moreover, study verified that R gene-mediated resistance associated with activation of an SA-dependent signaling pathway induces the expression of certain PR proteins, which contribute to resistance in Arabidopsis [17]. However, the classic marker genes of this pathway, including the candidate resistance R gene, EDS1, PAD4 and NPR1, were not differentially expressed after inoculation, perhaps because these genes were constitutively expressed in cabbage roots or because, this pathway may specifically function in foliar tissues, as observed in Arabidopsis [60]. Therefore, further research is required to determine the exact mode of action of SA-mediated fusarium wilt resistance in cabbage. Nonetheless, the results indicate that two SA-mediated signaling pathways are involved in the disease resistance system, including SA-mediated HR and SA-mediated SAR.
The JA-mediated signaling pathway plays an important role in FOC resistance in cabbage
JA signaling pathway is important for disease resistance, which is distinct from the classic SA-dependent SAR [61]. JA biosynthesis and JA-mediated signaling pathways are important components of the fungal resistance system in plants [12–14, 30]. Our data demonstrate that the JA-mediated signaling pathway also contributes to FOC resistance in cabbage. Myrosinase gene (comp35843_c0) is homologous to Arabidopsis gene TGG1 (Thioglucoside glucohydrolase1), whose expression and activity are controlled by COI1 [62]. JA-induced MBP (comp41986_c0) gene depends on COI1 for expression [63]. COI1 is a key player in JA perception and JA-mediated transduction pathway, although COI1 was not differentially expressed in this study. It implied that Myrosinase gene (comp35843_c0) and JA-induced MBP (comp41986_c0) gene identified here were downstream of the JA-mediated signaling. Myrosinases, Brassicaceae-specific β-glucosidases that interact with MBPs, are responsible for the hydrolysis of glucosinolate defense compounds in Brassica crops [46, 64]. When plants are infected with fungi, the non-toxic glucosinolates are hydrolyzed by myrosinases into toxic compounds for disease resistance, including isothiocyanates, thiocyanates, nitriles and epithionitriles [65]. Due to the important role played by the Brassicaceae-specific glucosinolates-myrosinase system in fungal defense, the identification of myrosinase and MBP genes helps confirm that the JA signaling pathway plays significant role in FOC resistance in cabbage. Moreover, MBP (comp41986_c0) is associated with Arabidopsis RIN4 (RPM1-interacting protein 4), which is a negative regulator of plant immunity, and proteolytic cleavage and phosphorylation of which by bacterial effectors activates two RIN4-associated R proteins (RPS2 and RPM1) resulting in the ETI (effector-triggered immunity) defensive reaction and further restriction of colonization by pathogens [14, 66]. This result highlights the function of MBP (comp41986_c0) in disease resistance in cabbage.
In addition, the PDR-ABC transporter gene comp20414_c0 and the lipid-transfer protein gene comp25940_c0 are involved in the MAPK cascade, as well as JA- and SA-mediated signaling pathways, indicating that these genes participate in disease resistance to FOC in cabbage by sharing different signaling pathways. Indeed, in cucumber, the expression of CsPDR12 significantly increases upon the addition of JA, SA or ABA, suggesting that CsPDR12 is involved in a phytohormone-mediated response of plants to different stimuli by sharing different signaling pathways [67].
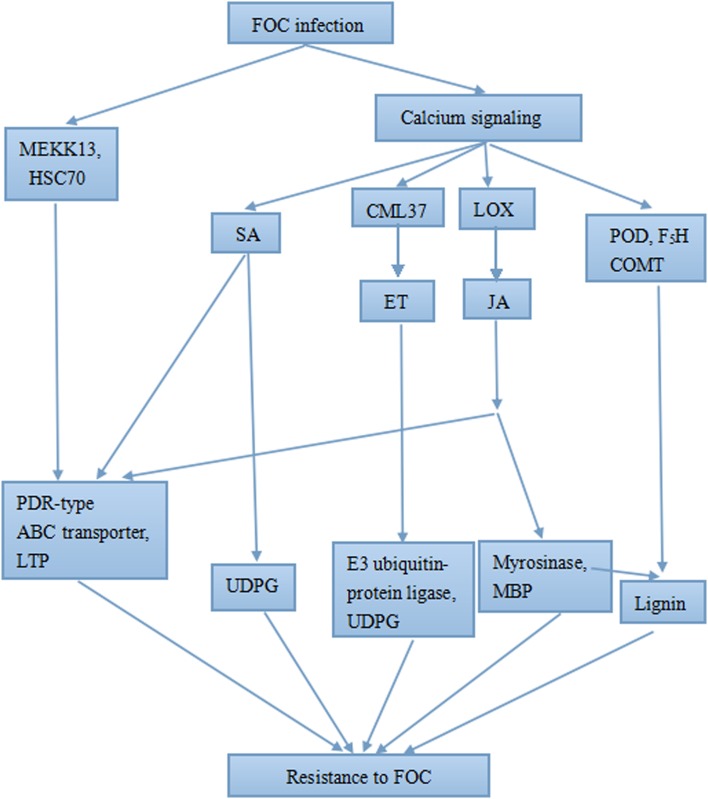
Much is known about the ET-, SA- and JA-mediated signaling systems [68], which allowed us to construct a schematic diagram describing the FOC resistance response in R4P1(Fig 9).
Fig 9. Schematic representation of the response of R4P1 to FOC infection.
MEKK13: MAPKKK13; HSC70: heat shock 70 kDa protein; LTP: lipid-transfer protein; MBP: myrosinase-binding protein.
Resistance genes in response to FOC in cabbage
In this study, many NBS-LRR genes were identified, including 213 TIR-NBS-LRR genes and 65 CC-NBS-LRR genes (S7 Table). These genes were not differentially expressed in inoculated roots compared to mock-inoculated roots under the filter conditions, and most of them were suppressed after infection, which is consistent with the results of a study in bananas, which showed that the expression of most NBS-LRR resistance proteins were quite low in resistant plants [13]. The slight downregulation of the candidate resistance gene comp30068_c1 in all the infected roots may be the result of the interaction of R gene-pathogen, which activates defense responses. Studies showed that there were insertion/deletion variations of the FOC-resistance gene between resistant cabbage material and susceptible materials [9, 11]. The results indicated that the R genes displayed constitutive expression in resistant material.
In conclusion, our transcriptome data provide a comprehensive overview of the gene expression profiles at the four stages of FOC infection, which will facilitate further analysis of the molecular mechanism underlying FOC resistance in cabbage. We proposed a putative network underlying this response in R4P1 (Fig 9). Early defense systems, including calcium signaling, MAPK signaling and SA-UDPG HR, were activated, and SA-dependent SAR, JA- and ET-mediated pathways and the lignin biosynthesis pathway were activated as well, suggesting that they play significant roles in FOC resistance in cabbage. Finally, our results suggest that these signaling pathway are not independent, instead interacting, which demonstrates that R4P1 utilizes different, effective defense pathways comprising a complex resistance network in response to FOC infection.
Supporting Information
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Acknowledgments
This work was financially supported by grants from the National Natural Science Foundation of China (31171958), the Key Projects in the National Science & Technology Pillar Program during the Twelfth Five-Year Plan Period (2012BAD02B01, 2014BAD01B08), and the Beijing Science and Technology Commission Project (Z141105002314020). We thank Elixigen Corporation (Huntington Beach, California, USA) for helping in proofreading and editing the English of final manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by grants from the National Natural Science Foundation of China (31171958), the Key Projects in the National Science & Technology Pillar Program during the Twelfth Five-Year Plan Period (2012BAD02B01, 2014BAD01B08), and the Beijing Science and Technology Commission Project (Z141105002314020).
References
- 1.Smith EF. The fungus infection of agricultural soils in the United States. Sci Am Suppl. 1899; 48:19981–19982. [Google Scholar]
- 2.Nomura Y, Kato K, Takeuchi S. Studies on the method of early selection of the resistance of cabbage to the yellows disease. Jpn Center Agric Exp Rep. 1976; 24:141–182 (in Japanese). [Google Scholar]
- 3.Li M, Zhang T, Li X, Yan H. Fusarium wilt disease on crucifer vegetable and its pathogenic identification. Plant Protect. 2003; 29(6):44–45 (in Chinese). [Google Scholar]
- 4.Kang J, Tian R, Geng L, Chen Y, Jian Y, Ding Y. Screening of cabbage germplasm resources with resistance to Fusarium wilt and analysis on distribution of resistant gene. 2010; (2):15–20 (in Chinese). [Google Scholar]
- 5.Lv H, Fang Z, Yang L, Xie B, Liu Y, Zhuang M, et al. Research on screening of resistant resources to Fusarium wilt and inheritance of the resistant gene in cabbage. Acta Horticult Sinica. 2011; 38(5):875–885 (in Chinese). [Google Scholar]
- 6.Snyder WC, Hansen HN. The species concept in Fusarium. Am J Bot. 1940; 27:64–67. [Google Scholar]
- 7.Pu ZJ, Shimizu M, Zhang YJ, Nagaoka T, Hayashi T, Hori H, et al. Genetic mapping of a fusarium wilt resistance gene in B. oleracea. Mol Breeding. 2012; 30:809–818. [Google Scholar]
- 8.Jiang M, Zhao Y, Xie JM, Tian RP, Chen YY, Kang JG. Development of A SCAR Marker for Fusarium Wilt Resistance in Cabbage. Scientia Agricultura Sinica. 2011; 44(14):3053–3059. [Google Scholar]
- 9.Shimizu M, Pu ZJ, Kawanabe T, Kitashiba H, Matsumoto S, Ebe Y, et al. Map-based cloning of a candidate gene conferring Fusarium yellows resistance in Brassica oleracea. Theor Appl Genet. 2015;128:119–130. 10.1007/s00122-014-2416-6 [DOI] [PubMed] [Google Scholar]
- 10.Lv HH, Yang LM, Kang JG, Wang QB, Wang XW, Fang ZY, et al. Development of InDel markers linked to Fusarium wilt resistance in cabbage. Mol Breeding. 2013; 32:961–967. [Google Scholar]
- 11.Lv HH, Fang ZY, Yang LM, Zhang YY, Wang QB, Liu YM, et al. Mapping and analysis of a novel oleracea candidate Fusarium wilt resistance gene FOC1 in Brassica. BMC Genomics. 2014; 15:1094 10.1186/1471-2164-15-1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lü GY, Guo SG, Zhang HY, Geng LH, Song FM, Fei ZJ, et al. Transcriptional profiling of watermelon during its incompatible interaction with Fusarium oxysporum f. sp. niveum. Plant Pathol. 2011; 131:585–601. [Google Scholar]
- 13.Li CY, Deng GM, Yang J, Altus Viljoen, Jin Y, Kuang RB, et al. Transcriptome profiling of resistant and susceptible Cavendish banana roots following inoculation with Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genomics. 2012; 13:374 10.1186/1471-2164-13-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu QH, Stephen S, Kazan K, Jin GL, Fan LJ, Taylor J, et al. Characterization of the defense transcriptome responsive to Fusarium oxysporum-infection in Arabidopsis using RNA-seq. Gene. 2013; 512(2): 259–266. 10.1016/j.gene.2012.10.036 [DOI] [PubMed] [Google Scholar]
- 15.Lyons R, Stiller J, Powell J, Rusu A, Manners JM, Kazan K. Fusarium oxysporum Triggers Tissue-Specific Transcriptional Reprogramming in Arabidopsis thaliana. PLOS ONE. 2015; 10(4):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao J, Jin XH, Jia XP, Wang HY, Cao AZ, Zhao WP, et al. Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genomics. 2013; 14:197 10.1186/1471-2164-14-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology. 2001; 43, 205–222. [DOI] [PubMed] [Google Scholar]
- 18.Tian RP, Kang JG, Geng LH, Xie JM, Jian YC, Ding YH. Study on the method of Fusarium wilts resistance in cabbage. Chinese Agricultural Science Bulletin. 2009; 25(4): 39–42 (in Chinese). [Google Scholar]
- 19.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. Full length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011; 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin XY, Kaul S, Rounsley S, Shea TP, Benito M- I, Town CD, et al. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999; 402: 761–777. [DOI] [PubMed] [Google Scholar]
- 21.Ecker JR, Theologis A, Federspiel NA, Palm CJ, Osborne BI, Shinn P, et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000; 408: 796–815. [DOI] [PubMed] [Google Scholar]
- 22.Wang XW, Wang HZ, Wang J, Sun RF, Wu J, Liu SY, et al. The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics. 2011; 43: 1035–1039. 10.1038/ng.919 [DOI] [PubMed] [Google Scholar]
- 23.Liu SY, Liu YM, Yang XH, Tong CB, Edwards D, Parkin IAP, et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nature Communications. 2014; 5(5): 3930–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortazavi A, Williams BA, McCue K, Schaeffer L. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008; 5(7), 621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 26.Anders S and Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010; 11, R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010; 26, 136–8. 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- 28.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biology. 2010; 11(2): R14 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mao XZ, Cai T, Olyarchuk JG, Wei LP. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005; 21, 3787–3793. [DOI] [PubMed] [Google Scholar]
- 30.Xu L, Zhu LF, Tu LL, Liu LL, Yuan DJ, Jin L, et al. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. Journal of Experimental Botany. 2011; 62 (15):5607–5621. 10.1093/jxb/err245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vidhyasekaran P. Plant Hormone Signaling Systems in Plant Innate Immunity. Springer; Netherlands: 2015; 27–244. [Google Scholar]
- 32.Trewavas A. Le calcium, C’est la vie: calcium makes waves. The Plant Cell. 1999; 120(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Libault Marc, Wan JR, Czechowski Tomasz, Udvardi, et al. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Molecular Plant-Microbe Interactions. 2007; 20(8):900–11. [DOI] [PubMed] [Google Scholar]
- 34.Poppenberger B, Berthiller F, Lucyshyn D, Sieberer T, Schuhmacher R, Krska R, et al. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J Biol Chem. 2003; 278: 47905–47914. [DOI] [PubMed] [Google Scholar]
- 35.Shang Y, Xiao J, Ma L, Wang H, Qi Z, Chen P, et al. Characterization of a PDR type ABC transporter gene from wheat (Triticum aestivum L.). Chinese Sci Bull. 2009;54(18):3249–3257. [Google Scholar]
- 36.Agarwal P, Agarwal PK. Pathogenesis related-10 proteins are small, structurally similar but with diverse role in stress signaling. Mol Biol Rep. 2014; 41:599–611. 10.1007/s11033-013-2897-4 [DOI] [PubMed] [Google Scholar]
- 37.Van Loon LC and Van Strein EA. The families of pathogenesis-related protein, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol. 1999;55:85–97. [Google Scholar]
- 38.Collinge DB, Kragh KM, Mikkelsen JD, Nielsen KK, Rasmussen U, Vad K Plant chitinases. Plant J. 1993;3:31–40. [DOI] [PubMed] [Google Scholar]
- 39.Derckel JP, Audran JC, Haye B, Lambert B, Legendre L. Characterization, induction by wounding and salicylic acid, and activity against Botrytis cinerea of chitinases and b-1,3-glucanases of ripening grape berries. Physiol Plant. 1998;104:56–64. [Google Scholar]
- 40.Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009; 323, 95–101. 10.1126/science.1164627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGregor DI. Glucosinolate content of developing rapeseed (Brassica napus L. Midas) seedlings. Can J Plant Sci. 1988; 68:367–380. [Google Scholar]
- 42.Rahmanpou S, Backhouse D, Nonhebel HM. Reaction of glucosinolate-myrosinase defence system in Brassica plants to pathogenicity factor of Sclerotinia sclerotiorum. Eur J Plant Pathol. 2010; 128:429–433. [Google Scholar]
- 43.Sarwar M, Kirkegaard JA, Wong PTW, Desmarchelier JM. Biofumigation potential of brassicas. III. In vitro toxicity of isothiocyanates to soil-borne fungal pathogens. Plant Soil. 1998; 201: 103–112. [Google Scholar]
- 44.Bernardi R, Negri A, Ronchi S, Palmieri S. Isolation of the epithiospecifier protein from oil-rape(Brassica napus ssp. oleifera) seed and its characterization. FEBS Lett. 2000; 467:296–298. [DOI] [PubMed] [Google Scholar]
- 45.Rask L, Andreasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J. Myrosinase:gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol. 2000; 42:93–113. [PubMed] [Google Scholar]
- 46.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, et al. MAP kinase signaling cascade in Arabidopsis innate immunity. Nature. 2002; 415, 977–983. [DOI] [PubMed] [Google Scholar]
- 47.Ren DT, Liu YD, Yang KY, Han L, Mao GH, Giazebrook J, et al. A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2008; 105, 5638–5643. 10.1073/pnas.0711301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berrocal-Lobo M, Molina A. Ethylene response factor 1 mediates Arabidopsis resistance to the soil borne fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 2004; 17, 763–770. [DOI] [PubMed] [Google Scholar]
- 49.Noel LD, Cagna G, Stuttmann J, Wirthmuller L, Betsuyaku S, Claus-Peter W, et al. Interaction between SGT1 and Cytosolic/Nuclear HSC70 Chaperones Regulates Arabidopsis Immune Responses. The Plant Cell. 2007; 19: 4061–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanco F, Garreton V, Frey N, Dominguez C, Perez-Acle T, Van der Straeten D, et al. Identification of NPR1-dependent and independent genes early induced by salicylic acid treatment in Arabidopsis.Plant Molecular Biology. 2005; 59:927–944. [DOI] [PubMed] [Google Scholar]
- 51.Yan SP and Dong XN. Perception of the plant immune signal salicylic acid. Plant Biology. 2014; 20:64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song JT. Induction of a salicylic acid glucosyltransferase, AtSGT1, is an early disease response in Arabidopsis thaliana. Molecules and Cells. 2006; 22(2):233–238. [PubMed] [Google Scholar]
- 53.Bieri S, Mauch S, Shen QH, Peart J, Devoto A, Casais C, et al. RAR1 positively controls steady state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell. 2004;16: 3480–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ. Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell. 2005;17: 1268–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002; 295(5562): 2073–2076. [DOI] [PubMed] [Google Scholar]
- 56.Falk A, Feys BJ, Frost LN, Jones JDG, Daniels MJ, Parker JE. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Plant Biology. 1999; 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rietz S, Stamm A, Malonek S, Wanger S, Becker D. Different roles of enhanced disease susceptibility 1 (EDS1) bound to and dissociated from phytoalexin deficient 4 (PAD4) in Arabidopsis immunity. New Phytol. 2011; 191, 107–119. 10.1111/j.1469-8137.2011.03675.x [DOI] [PubMed] [Google Scholar]
- 58.Weirmer M, Feys BJ, Parker JE. Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 2005; 8: 383–389. [DOI] [PubMed] [Google Scholar]
- 59.Singh I and Shah K. In silico study of interaction between rice proteins enhanced disease susceptibility 1 and phytoalexin deficient 4, the regulators of salicylic acid signalling pathway. J. Biosci. 2012; 37: 563–571. [DOI] [PubMed] [Google Scholar]
- 60.Edgar CI, McGrath KC, Dombrecht B, Manners JM, Maclean DC, Schenk PM, et al. Salicylic acid mediates resistance to the vascular wilt pathogen Fusarium oxysporum in the model host Arabidopsis thaliana. Aust Plant Pathol. 2006; 35: 581–591. [Google Scholar]
- 61.Dong XN. SA, JA, ethylene, and disease resistance in plants. Plant Biology. 1996; 1:316–323. [DOI] [PubMed] [Google Scholar]
- 62.Capella AN, Menossi M, Arruda P, Benedetti CE. COI1 affects myrosinase activity and controls the expression of two flower-specific myrosinase-binding protein homologues in Arabidopsis. Plana. 2001; 213:691–699. [DOI] [PubMed] [Google Scholar]
- 63.Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wouding and insect feeding in Arabidopsis. Plant Cell. 2000; 12:707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagano AJ, Fukao Y, Fujiwara M, Nishimura M, Hara-Nishimura I. Antagonistic Jacalin-Related Lectins Regulate the Size of ER Body-Type β-Glucosidase Complexes in Arabidopsis thaliana. Plant Cell Physiol. 2008; 49(6): 969–980. 10.1093/pcp/pcn075 [DOI] [PubMed] [Google Scholar]
- 65.Mithem RF, Magrath R. Glucosinolates and resistance to Leptosphaeria maculans in wild and cultivated Brassica species. Plant Breed. 1992; 108: 60–68. [Google Scholar]
- 66.Liu J, Elmore JM, Coaker G. Investigating the functions of the RIN4 protein complex during plant innate immune responses. Plant Signal. Behav. 2009; 4, 1107–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Migocka M, Papierniak A, Warzybok A, Kłobus G. CsPDR8 and CsPDR12, two of the 16 pleiotropic drug resistance genes in cucumber, are transcriptionally regulated by phytohormones and auxin herbicide in roots. Plant Growth Regul. 2012; 67:171–184. [Google Scholar]
- 68.M’etraux J-P, Nawrath C, Genoud T. Systemic acquired resistance. Euphytica. 2002; 124: 237–243. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.