Abstract
Objective
To evaluate genes involved in androgen receptor (AR) signaling as candidate genes for polycystic ovary syndrome (PCOS).
Design
Two groups of PCOS and control women (discovery and replication cohorts) were genotyped for single nucleotide polymorphisms (SNPs) in eight genes for AR chaperones and co-chaperones: HSPA1A, HSPA8, ST13, STIP1, PTGES3, FKBP4, BAG1, and STUB1. SNPs were tested for association with PCOS status, and with androgenic and metabolic parameters.
Setting
Tertiary referral center.
Patients
Discovery cohort: 354 PCOS and 161 control women. Replication cohort: 397 PCOS and 306 control women.
Interventions
Phenotypic and genotypic assessment.
Main outcome measures
SNP genotypes, association with PCOS status, and androgenic and metabolic parameters.
Results
In the discovery cohort, FKBP4 SNPs rs2968909 and rs4409904 were associated with lower odds of PCOS. This finding was not confirmed in the replication cohort analysis; however, when combining the two cohorts, rs4409904 was associated with lower odds of PCOS. In PCOS subjects in the replication cohort as well as in the combined cohort, rs2968909 was associated with lower BMI.
Conclusions
SNPs in FKBP4, which codes for the AR co-chaperone FKBP52, may be associated with PCOS and BMI in PCOS patients. The remaining genes studied do not appear to be major contributors to the development of PCOS. These findings warrant confirmation in future studies, and genes encoding other androgen pathway components remain to be studied.
Keywords: PCOS, androgen receptor, chaperones, co-chaperones
Introduction
Polycystic ovary syndrome (PCOS) affects 6-8% of reproductive aged women and is characterized by clinical and/or biochemical androgen excess, ovulatory dysfunction, and polycystic ovaries. Genetic factors are thought to play a role in the etiology of PCOS. A central feature of PCOS is androgen excess, manifesting as hirsutism and/or hyperandrogenemia. However, hirsutism and ovulatory dysfunction may occur without significantly elevated androgen levels (1). Among PCOS patients, 20-40% have normal levels of circulating androgens, and even those with hyperandrogenemia often have a mild degree of androgen elevation (1, 2).
In addition, approximately two-thirds of women with PCOS diagnosed by the NIH 1990 (classic) criteria demonstrate insulin resistance and compensatory hyperinsulinemia. Insulin functions as a co-gonadotropin to promote ovarian androgen biosynthesis (3). However, although hyperinsulinism is associated with hyperandrogenism in PCOS, insulin resistance alone is not sufficient for the development of PCOS (4), suggesting that an underlying (genetic) predisposition to hyperandrogenism must also be present.
Given these observations, it appears that women with PCOS are genetically predisposed to heightened androgen sensitivity. This rationale led us to hypothesize that inherited abnormalities in genes involved in the androgen signaling pathway may contribute to the development of PCOS.
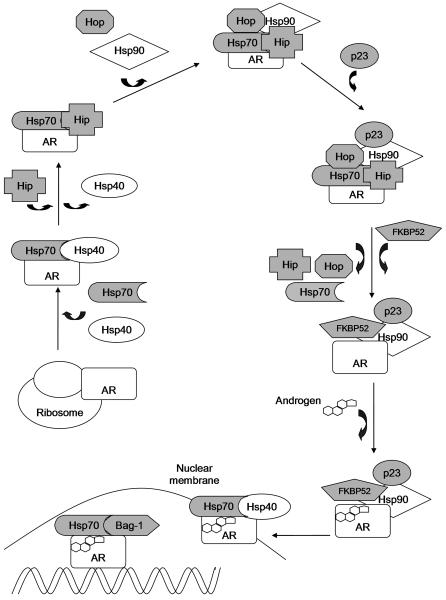
Androgen signaling occurs via the androgen receptor (AR). The AR is a ligand-dependent transcription factor belonging to the nuclear receptor superfamily. When unbound to ligand, the AR is maintained in a state of readiness for ligand-binding through interactions with a large dynamic hetero-complex composed of chaperones and co-chaperones that associate with the AR in an assembly-line fashion (5-11) (Figure 1). Chaperones are molecules that bind directly to the AR to affect its configuration, while co-chaperones bind to and assist in the function of chaperones. Chaperones allow steroid receptors to reach their mature and functional conformation by assisting in receptor folding, and are involved in the intracellular trafficking, transcriptional activity, and degradation of steroid receptors. The chaperone hetero-complex consists of heat shock proteins (Hsp), which act mainly as chaperones, and their co-chaperones. The two principal heat shock proteins are Hsp70 and Hsp90. Upon hormone binding, the AR is transformed into the DNA-binding competent state and transported into the nucleus, where it regulates gene expression. In addition to chaperones and co-chaperones, coregulators (coactivators that enhance gene expression by the receptor and corepressors that inhibit gene expression) also affect the function of nuclear receptors (10, 12, 13). Similar to the chaperone heterocomplex, these coregulators exist in large multiprotein complexes, and the sequential recruitment of coregulators is thought to be required for proper gene expression (12).
Figure 1. Molecular chaperones of the androgen receptor.
Shaded chaperone and co-chaperones are those whose genes were included in this study. The first chaperone to interact with the AR as it is translated on the ribosome is Hsp70 and its co-chaperone Hsp40. Together, these proteins prevent irreversible aggregation of the AR in the cytoplasm. Next, Hip, another Hsp70 co-chaperone, prolongs the interaction of the AR and this intermediate chaperone complex. Hop then forms a bridge between Hsp70 and Hsp90 using tetratricopeptide repeat (TPR) domains, which mediate protein-protein interactions. Next, co-chaperone p23 binds to Hsp90 to stabilize the AR in this intermediate conformation, while Hip, Hop, and Hsp70 dissociate. The release of Hop allows other TPR-containing co-chaperones, including FKBP52 and SGTA, to compete for the TPR binding site on Hsp90. FKBP52 binding generates the final complex prior to hormone binding. Upon hormone binding, the AR is transformed into the DNA-binding active state. Hormone binding leads to dissociation of the AR from the chaperone complex. However, Hsp70 and Hsp40 re-associate, and assist in transporting the AR across the nuclear membrane. In the nucleus, Hsp70 and Bag-1 (a Hsp70 co-chaperone) are recruited with the AR to promoter regions of AR target genes. Bag-1 is thought to upregulate AR transcriptional activity, binding directly to the N-terminal domain of the AR which contains its major transactivation function. Degradation of structurally unsound AR occurs via the ubiquitin-proteasome pathway. CHIP (not depicted), a TPR-containing Hsp70 co-chaperone, interacts with the complex at multiple stages to cause dissociation and loss of ligand binding ability. This is followed by ubiquitylation and degradation via the proteasome.
Abnormal function or expression of chaperones and co-chaperones has been shown to affect AR activity, which may contribute to androgen-dependent disease states such as prostate cancer (5). One study found that over-expression of Hsp70 and its co-chaperone Bag-1 in prostate cancer cells enhanced transcriptional activity of the AR (14). Mice deficient in the AR co-chaperone Fkbp52 have phenotypic changes including hypospadias, ambiguous external genitalia, and malformation of the seminal vesicles and prostate (15). Genes encoding components of the chaperone complex are logical candidates to delineate the role of androgen signaling in conditions such as PCOS.
The hypothesis of the present study is that inherited abnormalities in genes encoding AR chaperones and co-chaperones may influence the development or phenotype of PCOS. This was tested by determining whether single nucleotide polymorphisms (SNPs) within such genes are associated with PCOS or androgenic and metabolic parameters in women with PCOS. This first genetic association study of these genes in PCOS also includes a replication effort.
MATERIALS AND METHODS
Subjects and Phenotyping
Discovery cohort
The discovery phase aimed to investigate the role of the selected chaperones and co-chaperones in the androgen signaling pathway as new candidate genes for PCOS. The discovery cohort included 354 unrelated Caucasian women with PCOS and 161 healthy Caucasian controls. These subjects were recruited at two centers, the University of Alabama at Birmingham (242 PCOS and 146 controls) and Cedars-Sinai Medical Center (CSMC, 112 PCOS and 15 controls).
Subjects with PCOS were recruited by offering participation in research studies to patients meeting the following inclusion criteria: premenopausal, non-pregnant, taking no hormonal therapy including oral contraceptives for ≥3 months, and meeting diagnostic criteria for PCOS. PCOS was defined according to the 1990 NIH criteria: oligomenorrhea, hirsutism and/or hyperandrogenemia, and the exclusion of related disorders (16). Parameters for defining hirsutism, hyperandrogenemia, ovulatory dysfunction, and exclusion of related disorders were previously reported (17).
Controls were recruited by word of mouth and posted advertisements. Controls were healthy, eumenorrheic, premenopausal women taking no hormonal therapy for ≥3 months, and having no personal or family history of hirsutism or endocrine disorders. All subjects underwent a physical examination and morning blood sampling in the fasting state per a previously described protocol (17).
Fasting glucose and insulin levels were available for a subset (70%) of subjects in the discovery cohort. This subset did not differ demographically or hormonally from the study subjects overall. Using the fasting glucose and insulin data, the computer-based homeostasis model assessment (HOMA, www.dtu.ox.ac.uk/homa) was used to calculate indices of insulin resistance (HOMA-IR) and insulin secretion/beta-cell function (HOMA-%B). The following parameters were tested for association with SNP genotypes: PCOS status, total testosterone, DHEAS, fasting insulin, fasting glucose, HOMA-IR, HOMA-%B, and body mass index (BMI).
Replication cohort
The second part of the study was the replication phase, where the intent was to reproduce significant results identified in the discovery phase of the study in a larger group of PCOS and control women. The replication cohort consisted of 397 unrelated Caucasian women with PCOS (all meeting 1990 NIH criteria) and 306 Caucasian controls. The subjects were derived from three sources: 380 PCOS subjects and 71 controls previously recruited at Pennsylvania State University (18), 17 PCOS subjects and 2 controls recruited at CSMC, and 233 control women derived from the Cholesterol and Pharmacogenetics (CAP) study (19). The CAP samples consist of general community controls.
All subjects gave written consent to study participation. The study was approved by the Institutional Review Boards of the recruiting centers and CSMC.
Genotyping
Eight genes were selected based on their roles in the AR signaling pathway. These genes encode chaperones and co-chaperones of the AR (Figure 1). Gene selection and Figure 1 were primarily based on the review by Prescott and Coetzee (5). The candidate genes include heat shock 70kDa protein 1A (HSPA1A, on chromosome 6p21.3) and heat shock 70kDa protein 8 (HSPA8, on chromosome 11q24.1), which encode the hsp70 family member chaperones Hsp70 protein 1A and Hsc70, respectively. The following co-chaperone genes were also studied: ST13 (on chromosome 22q13.2, encodes Hsc70 interacting protein (Hip)), STIP1 (on chromosome 11q13, encodes Hsp organizer protein (Hop)), PTGES3, on chromosome 12q13.3, encodes p23), FKBP4 (on chromosome 12p13.33, encodes FK506-binding protein of 52 kDa (FKBP52)), BAG1 (on chromosome 9p12, encodes Bag-1), and STUB1 (on chromosome 16p13.3, encodes carboxy terminus of Hsc70-interacting protein (CHIP)).
SNPs within these genes were selected using the genotype data of the International HapMap database (release 24), which allows selection of a limited number of tagging SNPs to capture the majority of genetic variation within genes (20). The CEU population (Utah residents with ancestry from northern and western Europe) of HapMap was used, as all subjects in our study were Caucasian. Fifty SNPs were selected to capture the common variation in these eight genes plus 50 kb upstream and 5 kb downstream: 11 in FKBP4, 8 in ST13, 7 in HSPA8, 6 in STUB1, 6 in STIP1, 5 in HSPA1A, 4 in PTGES3, and 3 in BAG1. All 50 SNPs were genotyped in the discovery cohort using Golden Gate technology (Illumina, San Diego, CA). Of the 50 SNPs, two failed to genotype properly (rs138349 in ST13 and rs2186571 in STIP1). In the replication cohort, two SNPs in FKBP4 (rs2968909 and rs4409904) were genotyped using Applied Biosystems Taqman Assays-on-Demand (Applied Biosystems, Foster City, CA). All genotyping was conducted at Cedars-Sinai Medical Center.
Statistical analysis
In the primary analysis, SNPs were tested for association with PCOS status. In secondary analyses, SNPs were tested for association with the following quantitative traits: total testosterone, DHEAS, fasting insulin, fasting glucose, HOMA-IR, HOMA-%B, and BMI. The additive genetic model was used in all analyses. The discovery cohort was used to test for association of all 48 SNPs with the presence or absence of PCOS. This was evaluated using logistic regression, adjusting for age, BMI, and recruitment site. SNPs that were found to be significantly associated with PCOS in the discovery cohort were genotyped again in the larger replication cohort. Association of SNPs with quantitative metabolic and androgenic traits were evaluated using analysis of covariance (ANCOVA), adjusting for age, BMI, and site. The analyses where BMI was the dependent variable were adjusted for recruitment site and age only. Quantitative traits were log- or square root-transformed as appropriate to reduce non-normality. Similar analyses were conducted in the replication cohort and in a meta-analysis of both cohorts. Unpaired t-tests were used to compare clinical characteristics between cases and controls.
Significance was taken as P<0.05 for association with PCOS status (the primary analysis) in both the discovery and replication cohorts. For testing association with quantitative parameters in the discovery cohort, significance was taken as P<0.003, given that eight genes were analyzed against two families of traits (androgens and metabolic traits), yielding a Bonferroni correction factor of 16 (0.05/16 = 0.003). For testing association with quantitative parameters in the replication cohort, significance was taken as P<0.025, given that one gene was analyzed against two families of traits, yielding a Bonferroni correction factor of 2. Data are displayed as median (interquartile range). The above analyses were carried out using Statview 5.0 (SAS Institute, Cary, NC).
Using the Genetic Power Calculator program (21), the power for an association study was calculated for case-control design for discrete traits. Assuming a population prevalence of PCOS of 7%, the power was calculated for various allele frequencies and relative risks.
RESULTS
The clinical characteristics of PCOS and control subjects are detailed in Table 1. In both cohorts, women with PCOS were younger and had higher BMI, total testosterone, DHEAS, insulin, HOMA-IR, and HOMA-%B compared to the controls (P<0.0001).
Table 1.
Clinical characteristics of PCOS and control subjects
| Discovery | Replication | |||
|---|---|---|---|---|
| Control (n=161) |
PCOS (n=354) |
Control (n=306) |
PCOS (n=397) |
|
| Age (years) | 33.0 (15.0) | 26.7 (10.2)a | 51.0 (23.8) | 28.0 (9.0)a |
| BMI (kg/m2) | 24.0 (5.5) | 31.2 (14.9)a | 25.9 (8.3) | 35.0 (12.3)a |
| Total testosterone (ng/dl) | 40.0 (26.8) | 70.0 (40.0)a | 29.0 (15.0) | 70.1 (32.0)a |
| DHEAS (μg/dl) | 98.4 (73.3) | 218.8 (183.1)a | 137.1 (62.6) | 210.7 (145.3)a |
| Insulin (μIU/ml) | 7.1 (6.4) | 14.0 (16.2)a | 12.0 (7.5) | 21.0 (16.0)a |
| Glucose (mg/dl) | 85.0 (10.0) | 85.0 (11.0) | 90.3 (12.2) | 87.5 (11.5) |
| HOMA-IR | 0.98 (0.82) | 1.77 (1.91)a | 1.58 (0.96) | 2.70 (1.82)a |
| HOMA-%B | 106.7 (52.2) | 152.5 (100.8)a | 129.5 (58.3) | 196.8 (82.0)a |
P<0.0001 compared to control group. Data are median (interquartile range).
In the discovery cohort, the sample size of 354 cases and 161 controls had excellent power (≥90%) to detect association of risk alleles of frequency ≥0.1 with PCOS at odds ratio ≥3, and fair-to-excellent power (62-93%) to detect association at odds ratio ≥2 (Supplementary Table 1). In the replication cohort, the sample size of 397 cases and 306 controls had good to excellent power (>80%) to detect association of risk alleles of frequency ≥0.1 with PCOS at odds ratio ≥2, and fair-to-excellent power (62-94%) to detect association at odds ratio 1.75. The power calculations revealed lower power to detect association of rare risk alleles (frequency ≤0.1) with PCOS at odds ratios of 1.5.
The minor allele frequencies of the 48 SNPs genotyped in the discovery cohort and their locations within the genes are listed in Supplementary Table 2. All SNPs were in Hardy-Weinberg equilibrium except rs3830140 in STUB1 (P=0.005). Of the SNPs studied in the discovery cohort, two SNPs in the FKBP4 gene had significant association with PCOS status. Rs2968909 (OR=0.62, 95% CI 0.39-0.99, P=0.048) and rs4409904 (OR=0.48, 95% CI 0.29-0.78, P=0.003), were both associated with reduced odds of PCOS. These SNPs are not in linkage disequilibrium (r2=0.18). Rs7290793 in ST13 was also associated with reduced odds of PCOS (OR=0.51, 95% CI 0.27-0.95, P=0.035). There was no association of these three SNPs with any of the quantitative parameters assessed. The remaining SNPs in FKBP4, ST13, STIP1, HSPA1A, HSPA8, STUB1, PTGES3, and BAG1 had no significant association with either PCOS status or the quantitative parameters among PCOS subjects.
Because two independent SNPs in FKBP4 were associated with PCOS, we decided to focus on this gene in the replication effort. The minor allele frequencies of the two FKBP4 SNPs were similar to the frequencies observed in the discovery cohort (rs2968909, overall 0.17, PCOS 0.15, control 0.19; rs4409904, overall 0.13, PCOS 0.10, control 0.15). Before adjusting for age, BMI, and site, both SNPs were associated with reduced odds of PCOS: rs2968909, OR=0.74, 95% CI 0.56-0.99, P=0.045; rs4409904, OR=0.63, 95% CI 0.45-0.88, P=0.006. However, after adjustment, the statistical significance of the associations was lost: rs2968909 OR=0.84, 95% CI 0.51-1.37, P=0.48; rs4409904 OR=0.74, 95% CI 0.39-1.41, P=0.36. Analysis of the quantitative parameters in the PCOS cases revealed an association of rs2968909 with BMI, with increasing copies of the minor allele G correlating with decreasing BMI: CC 35.70 (12.04), GC 33.28 (12.34), GG 26.73 (16.11) kg/m2; P=0.004. This association with BMI was not observed in the control group. The two FKBP4 SNPs were not significantly associated with the remaining quantitative traits (Supplementary Table 3).
A final analysis of the two FKBP4 SNPs was performed in a meta-analysis of the discovery and replication cohorts. rs2968909 was not significantly associated with PCOS (P=0.054), while rs4409904 was associated with significantly reduced odds of PCOS (OR=0.56, 95% CI 0.39-0.84, P=0.004) after adjusting for age, BMI, and site. Meta-analysis of the quantitative parameters in the PCOS cases revealed a trend for association of rs2968909 with BMI similar to that in the replication cohort, with each additional copy of the minor allele G decreasing BMI by 1.2 kg/m2; P=0.025.
DISCUSSION
This study was based on the general hypothesis that inherited abnormalities in genes involved in androgen signaling may contribute to PCOS. Previous studies examining the association of PCOS with genes involved in androgen biosynthesis and action have lent some support to this hypothesis. Several studies focused on the AR gene. Exon 1 of the AR gene contains a variable length CAG repeat polymorphism, and shorter CAG repeats may increase the transactivation function of the androgen receptor (22). Some studies found associations of this variant with PCOS and androgenic traits including testosterone levels, hirsutism scores, and acne (23-29); however, other studies have not found these associations, and a meta-analysis was negative for association with PCOS (30). Therefore, it is not yet known with certainty whether this polymorphism is a major contributing factor to the development of PCOS.
Variants in other genes that promote androgen signaling have been studied for association with PCOS, including the genes encoding the two 5α-reductase enzymes, which convert testosterone to the more potent dihydrotestosterone in target tissues such as hair follicles. In these studies, variants in both the SRD5A1 and SRD5A2 genes (which encode the type 1 and type 2 isoforms, respectively) were associated with PCOS, and variation in SRD5A1 was associated with the degree of hirsutism in PCOS women (31, 32). Another gene found to be associated with PCOS is small glutamine-rich tetratricopeptide repeat-containing protein alpha (SGTA). SGTA is a co-chaperone that binds to the AR and inhibits its action by holding the AR in the cytoplasm, preventing it from translocating to the nucleus and activating target genes. Variants within SGTA were found to be associated with PCOS and with increased insulin resistance in affected women (33, 34).
Given the above evidence that variation in genes involved in androgen signaling may contribute to PCOS, we aimed to expand the number of genes studied in the androgen signaling pathway. None of these genes had been previously studied as candidate genes for PCOS. The chaperone and co-chaperone proteins encoded by these genes play critical roles in changing the AR’s conformational state, which is crucial to the function of the AR (5). The chaperone complex configures the AR into an inactive intermediate with a high affinity for hormone. Without the chaperone complex, the AR is not accessible to hormone. The cycle of this complex in the proper folding, transport, and transcriptional activity of the AR is depicted in Figure 1.
In our analysis, minor alleles of two SNPs in FKBP4, rs2968909 and rs4409904, were found to be associated with reduced odds of PCOS in the discovery cohort. This association was not confirmed in the replication cohort after adjustment for age, BMI, and site. However, before adjustment, both SNPs had significant protective associations with PCOS. In the meta-analysis, rs4409904 was significantly associated with reduced odds of PCOS. In women with PCOS, the minor allele of SNP rs2968909 was associated with lower BMI, which might explain why adjustment eliminated the association of this SNP with PCOS. These results suggest that the co-chaperone coded by FKBP4, FKBP52, might influence PCOS risk not only by effects on androgen signaling, but also via effects on adiposity, a trait known to exacerbate the PCOS phenotype.
If these FKBP4 variants, both of which are located upstream of the gene, were to have true association with PCOS, they may be causal or they may be in linkage disequilibrium with functional variants elsewhere in the gene or in its promoter. Potential residence of SNPs in regions of predicted regulatory function was evaluated using data from the ENCODE (Encyclopedia of DNA Elements) Project implemented in the UCSC Genome Browser (Supplementary Figure 1) (35, 36). DNase hypersensitivity sites in fibroblasts and induced pluripotent stem cells overlap the SNP rs4409904, indicating this locus is transcriptionally active. This SNP is also on the shore of an estrogen receptor alpha binding motif that is occupied in the breast cancer cell line T-47D, indicating this transcription factor binding site may play a role in hormone dependent signaling. Absence of detected regulatory or transcriptional activity around rs2968909 suggests it may not be a functional variant. However, this variant is in strong linkage disequilibrium (r2>0.8) with a number of other markers, particularly rs10047621, which overlaps active promoter marks, enhancer marks, transcription factor binding sites and alters the motifs for 11 transcription factors, and therefore likely influences transcription of FKBP4.
Our study has several strengths. The genes studied have not been previously assessed for association with PCOS, and therefore our analysis contributes to the fund of knowledge about the genetic basis of PCOS. A major strength is the inclusion of two cohorts of women in the study. Replicating positive findings is a critical aspect of performing genetic association studies, as replication decreases the chance of false positive findings. In addition, our analysis included a well-characterized, homogenous group of PCOS cases in both cohorts, with all patients meeting the 1990 NIH criteria. This minimizes the heterogeneity that arises when different diagnostic criteria for PCOS are used. Consequently, however, our results may not be generalizable to women who meet 2003 Rotterdam criteria for PCOS (37) but not 1990 NIH criteria (e.g. those with oligo-ovulation and polycystic ovarian morphology without hyperandrogenism) or to non-White race groups.
While our cohort sizes were not small, an even larger number of cases and controls would be required to detect more modest associations between SNPs and odds of PCOS. The candidate gene approach of this study presents a potential limitation. This approach relies on the a priori knowledge of the functional role of a relatively small number of possible gene candidates. This is in contrast to the genome-wide association study (GWAS) approach, of which two have been conducted in Chinese PCOS cohorts (38, 39) and one in European cohorts (40). While FKBP4 was not a signal in these GWAS, we should note that a benefit of a hypothesis-driven study of candidate genes is the lower P value penalty resulting from fewer tests performed than in GWAS.
In summary, our initial data suggested an association of two SNPs in FKBP4 with reduced odds of PCOS; however, while this finding was not robustly reproduced in the replication cohort, the meta-analyses did show a protective association of rs4409904 with PCOS and of rs2968909 with lower BMI in affected women. Additional studies of FKBP4, particularly in larger cohorts, are needed. While variations in the other genes examined do not appear to be major contributors to the development of PCOS, minor effects cannot be excluded. Future studies investigating the role of other AR pathway genes including coactivators and corepressors of the AR, as well as other genes relevant to androgen sensitivity, are required to fully elucidate the role of inherited abnormalities in androgen sensitivity in the pathogenesis of PCOS.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by National Institutes of Health Grants R01-HD29364 and K24-HD01346 (to R.A.), Grant R01-DK79888 (to M.O.G.), Grant U54-HD034449 (to R.S.L.), Grant U19-HL069757 (to R.M.K.); National Center for Research Resources Grant M01-RR00425 (to the Cedars-Sinai General Clinical Research Center), Grant U54 RR026071 (to Penn State Clinical and Translational Science Institute); the Winnick Clinical Scholars Award (to M.O.G.); and an endowment from the Helping Hand of Los Angeles, Inc. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant P30-DK063491 to the Southern California Diabetes Research Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Conflicts of Interest: A.K, M.R.J., A.C., R.M.K., Y.I.C., R.S.L., R.A., and M.O.G. have nothing to declare.
Capsule:
Single nucleotide polymorphisms in the FKBP4 gene, which codes for a co-chaperone of the androgen receptor, may be associated with PCOS and body mass index.
References
- 1.Azziz R, Carmina E, Dewailly D, Diamanti-Kandarakis E, Escobar-Morreale HF, Futterweit W, et al. Positions statement: criteria for defining polycystic ovary syndrome as a predominantly hyperandrogenic syndrome: an Androgen Excess Society guideline. J Clin Endocrinol Metab. 2006;91:4237–45. doi: 10.1210/jc.2006-0178. [DOI] [PubMed] [Google Scholar]
- 2.Chang WY, Knochenhauer ES, Bartolucci AA, Azziz R. Phenotypic spectrum of polycystic ovary syndrome: clinical and biochemical characterization of the three major clinical subgroups. Fertil Steril. 2005;83:1717–23. doi: 10.1016/j.fertnstert.2005.01.096. [DOI] [PubMed] [Google Scholar]
- 3.Nestler JE, Jakubowicz DJ, de Vargas AF, Brik C, Quintero N, Medina F. Insulin stimulates testosterone biosynthesis by human thecal cells from women with polycystic ovary syndrome by activating its own receptor and using inositolglycan mediators as the signal transduction system. J Clin Endocrinol Metab. 1998;83:2001–5. doi: 10.1210/jcem.83.6.4886. [DOI] [PubMed] [Google Scholar]
- 4.Korhonen S, Hippelainen M, Niskanen L, Vanhala M, Saarikoski S. Relationship of the metabolic syndrome and obesity to polycystic ovary syndrome: a controlled, population-based study. Am J Obstet Gynecol. 2001;184:289–96. doi: 10.1067/mob.2001.109596. [DOI] [PubMed] [Google Scholar]
- 5.Prescott J, Coetzee GA. Molecular chaperones throughout the life cycle of the androgen receptor. Cancer Lett. 2006;231:12–9. doi: 10.1016/j.canlet.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 6.Heinlein CA, Chang C. Role of chaperones in nuclear translocation and transactivation of steroid receptors. Endocrine. 2001;14:143–9. doi: 10.1385/ENDO:14:2:143. [DOI] [PubMed] [Google Scholar]
- 7.Pratt WB, Toft DO. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 2003;228:111–33. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 8.Smith DF. Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones. 2004;9:109–21. doi: 10.1379/CSC-31.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–70. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev. 2007;28:778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- 11.Hessenkemper W, Baniahmad A. Chaperones for proper androgen action - a plethora of assistance to androgen receptor function. Horm Mol Biol Clin Investig. 2012;11:321–8. doi: 10.1515/hmbci-2012-0031. [DOI] [PubMed] [Google Scholar]
- 12.Lonard DM, Kumar R, O'Malley BW. Minireview: the SRC family of coactivators: an entree to understanding a subset of polygenic diseases? Mol Endocrinol. 2010;24:279–85. doi: 10.1210/me.2009-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dasgupta S, Lonard DM, O'Malley BW. Nuclear receptor coactivators: master regulators of human health and disease. Annu Rev Med. 2014;65:279–92. doi: 10.1146/annurev-med-051812-145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shatkina L, Mink S, Rogatsch H, Klocker H, Langer G, Nestl A, et al. The cochaperone Bag-1L enhances androgen receptor action via interaction with the NH2-terminal region of the receptor. Mol Cell Biol. 2003;23:7189–97. doi: 10.1128/MCB.23.20.7189-7197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yong W, Yang Z, Periyasamy S, Chen H, Yucel S, Li W, et al. Essential role for Co-chaperone Fkbp52 but not Fkbp51 in androgen receptor-mediated signaling and physiology. J Biol Chem. 2007;282:5026–36. doi: 10.1074/jbc.M609360200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zawadzki JK, Dunaif A. Diagnostic criteria for polycystic ovary syndrome: towards a rational approach. In: Dunaif A, Givens JR, Haseltine F, Merriam GR, editors. Polycystic Ovary Syndrome. Blackwell Scientific Publications; Cambridge: 1992. pp. 377–84. [Google Scholar]
- 17.Azziz R, Woods KS, Reyna R, Key TJ, Knochenhauer ES, Yildiz BO. The prevalence and features of the polycystic ovary syndrome in an unselected population. J Clin Endocrinol Metab. 2004;89:2745–9. doi: 10.1210/jc.2003-032046. [DOI] [PubMed] [Google Scholar]
- 18.Legro RS, Driscoll D, Strauss JF, 3rd, Fox J, Dunaif A. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci U S A. 1998;95:14956–60. doi: 10.1073/pnas.95.25.14956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon JA, Lin F, Hulley SB, Blanche PJ, Waters D, Shiboski S, et al. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the Cholesterol and Pharmacogenetics (CAP) Study. Am J Cardiol. 2006;97:843–50. doi: 10.1016/j.amjcard.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 20.International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 21.Purcell S, Cherny SS, Sham PC. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–50. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 22.Tut TG, Ghadessy FJ, Trifiro MA, Pinsky L, Yong EL. Long polyglutamine tracts in the androgen receptor are associated with reduced trans-activation, impaired sperm production, and male infertility. J Clin Endocrinol Metab. 1997;82:3777–82. doi: 10.1210/jcem.82.11.4385. [DOI] [PubMed] [Google Scholar]
- 23.Hickey T, Chandy A, Norman RJ. The androgen receptor CAG repeat polymorphism and X-chromosome inactivation in Australian Caucasian women with infertility related to polycystic ovary syndrome. J Clin Endocrinol Metab. 2002;87:161–5. doi: 10.1210/jcem.87.1.8137. [DOI] [PubMed] [Google Scholar]
- 24.Kim JJ, Choung SH, Choi YM, Yoon SH, Kim SH, Moon SY. Androgen receptor gene CAG repeat polymorphism in women with polycystic ovary syndrome. Fertil Steril. 2008;90:2318–23. doi: 10.1016/j.fertnstert.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 25.Mifsud A, Ramirez S, Yong EL. Androgen receptor gene CAG trinucleotide repeats in anovulatory infertility and polycystic ovaries. J Clin Endocrinol Metab. 2000;85:3484–8. doi: 10.1210/jcem.85.9.6832. [DOI] [PubMed] [Google Scholar]
- 26.Ramos Cirilo PD, Rosa FE, Moreira Ferraz MF, Rainho CA, Pontes A, Rogatto SR. Genetic polymorphisms associated with steroids metabolism and insulin action in polycystic ovary syndrome. Gynecol Endocrinol. 2012;28:190–4. doi: 10.3109/09513590.2011.593661. [DOI] [PubMed] [Google Scholar]
- 27.Shah NA, Antoine HJ, Pall M, Taylor KD, Azziz R, Goodarzi MO. Association of androgen receptor CAG repeat polymorphism and polycystic ovary syndrome. J Clin Endocrinol Metab. 2008;93:1939–45. doi: 10.1210/jc.2008-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Nieuwerburgh F, Stoop D, Cabri P, Dhont M, Deforce D, De Sutter P. Shorter CAG repeats in the androgen receptor gene may enhance hyperandrogenicity in polycystic ovary syndrome. Gynecol Endocrinol. 2008;24:669–73. doi: 10.1080/09513590802342841. [DOI] [PubMed] [Google Scholar]
- 29.Xita N, Georgiou I, Lazaros L, Psofaki V, Kolios G, Tsatsoulis A. The role of sex hormone-binding globulin and androgen receptor gene variants in the development of polycystic ovary syndrome. Hum Reprod. 2008;23:693–8. doi: 10.1093/humrep/dem382. [DOI] [PubMed] [Google Scholar]
- 30.Wang R, Goodarzi MO, Xiong T, Wang D, Azziz R, Zhang H. Negative association between androgen receptor gene CAG repeat polymorphism and polycystic ovary syndrome? A systematic review and meta-analysis. Mol Hum Reprod. 2012;18:498–509. doi: 10.1093/molehr/gas024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodarzi MO, Shah NA, Antoine HJ, Pall M, Guo X, Azziz R. Variants in the 5alpha-reductase type 1 and type 2 genes are associated with polycystic ovary syndrome and the severity of hirsutism in affected women. J Clin Endocrinol Metab. 2006;91:4085–91. doi: 10.1210/jc.2006-0227. [DOI] [PubMed] [Google Scholar]
- 32.Graupp M, Wehr E, Schweighofer N, Pieber TR, Obermayer-Pietsch B. Association of genetic variants in the two isoforms of 5alpha-reductase, SRD5A1 and SRD5A2, in lean patients with polycystic ovary syndrome. Eur J Obstet Gynecol Reprod Biol. 2011;157:175–9. doi: 10.1016/j.ejogrb.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 33.Ewens KG, Stewart DR, Ankener W, Urbanek M, McAllister JM, Chen C, et al. Family-based analysis of candidate genes for polycystic ovary syndrome. J Clin Endocrinol Metab. 2010;95:2306–15. doi: 10.1210/jc.2009-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodarzi MO, Xu N, Cui J, Guo X, Chen YI, Azziz R. Small glutamine-rich tetratricopeptide repeat-containing protein alpha (SGTA), a candidate gene for polycystic ovary syndrome. Hum Reprod. 2008;23:1214–9. doi: 10.1093/humrep/den065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril. 2004;81:19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Li Z, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55–9. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- 39.Shi Y, Zhao H, Cao Y, Yang D, Li Z, Zhang B, et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. 2012;44:1020–5. doi: 10.1038/ng.2384. [DOI] [PubMed] [Google Scholar]
- 40.Hayes MG, Urbanek M, Ehrmann DA, Armstrong LL, Lee JY, Sisk R, et al. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nat Commun. 2015;6:7502. doi: 10.1038/ncomms8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.