Abstract
Aims/hypothesis
Epidemiological studies in Pima Indians identified elevated levels of HDL-cholesterol (HDL-C) as a protective factor against type 2 diabetes risk in women. We assessed whether HDL-C-associated single-nucleotide polymorphisms (SNPs) also associate with type 2 diabetes in female Pima Indians.
Methods
Twenty-one SNPs in established HDL-C loci were initially analysed in 2,675 full-heritage Pima Indians. SNPs shown to associate with HDL-C (12 SNPs) were assessed for association with type 2 diabetes in 7,710 Pima Indians (55.6% female sex). The CETP locus provided the strongest evidence for association with HDL-C and was further interrogated by analysing tag SNPs.
Results
Twelve of the 21 SNPs analysed had a significant association with HDL-C in Pima Indians; five SNPs representing four loci (CETP, DOCK6, PPP1R3B and ABCA1) reached genome-wide significance. Three SNPs, at CETP, KLF14 and HNF4A, associated with type 2 diabetes only in female participants with the HDL-C-lowering allele increasing diabetes risk (p values: 3.2 × 10 −4 to 7.7 × 10 −5); the association remained significant even after adjustment for HDL-C. Additional analysis across CETP identified rs6499863 as having the strongest association with type 2 diabetes in female participants (p = 5.0 × 10 −6) and this association remained independent of the HDL-C association.
Conclusions/interpretation
SNPs at the CETP, HNF4A and KLF14 locus are associated with HDL-C levels and type 2 diabetes (in female participants). However, since HNF4A and KLF14 are established loci for type 2 diabetes, it is unlikely that HDL-C solely mediates these associations.
Keywords: CETP, HDL-C, HNF4A, KLF14, Lipids, Pima Indians, Type 2 diabetes
Introduction
Elevated levels of HDL-cholesterol (HDL-C) are protective against cardiovascular diseases [1]; however, a less appreciated role of HDL-C is that elevated levels are associated with decreased risk for type 2 diabetes [2–5]. A relationship between HDL-C levels and type 2 diabetes was identified in the Pima Indian population, which suffers from a high prevalence and incidence of type 2 diabetes [3]. In a prospective study of 787 Pima Indians, who were non-diabetic at baseline with a mean follow-up of nearly 10 years, fasting HDL-C levels were significantly protective for development of type 2 diabetes in women (HR 0.35 [95% CI 0.23, 0.54], comparing the 90th with the 10th percentile of the HDL-C distribution, controlled for age, BMI, systolic blood pressure and 2 h glucose) but this relationship was not observed in men [3]. The protective effect of HDL-C remained significant in Pima women even after adjustment for estimates of insulin resistance and alcohol consumption [3]. The Norwegian population-based Finnmark study also identified a female sex-specific protective role for HDL-C in type 2 diabetes, and in the San Antonio Heart Study, non-diabetic individuals who developed diabetes during follow-up had lower baseline HDL-C levels by 0.14 mmol/l, and this effect was stronger in women [5, 6].
Genome-wide association studies (GWAS) have identified a number of genetic loci associated with lipid levels [7, 8]; however, the role of HDL-C loci in the pathophysiology of type 2 diabetes remains unknown. Recent studies have analysed the aggregate effect of dyslipidaemia-associated loci on type 2 diabetes risk and related phenotypes, with conflicting results [9–12]. To follow-up the Pima Indian finding of a female sex-specific protective role for HDL-C in type 2 diabetes, we now identify which established HDL-C loci associate with HDL-C in Pima Indians and assess whether they also associate with type 2 diabetes, and whether the associations are sex specific. We also test for mediation by HDL-C of the association between these loci and type 2 diabetes.
Methods
Study participants
The study population was formed from individuals taking part in a longitudinal study of diabetes from 1965 to 2007 in the Gila River Indian Community in Arizona, whose residents are primarily Pima Indian or Tohono O’odham (a closely related tribe). Community members at age ≥ 5 years were invited for biennial health examinations, which included measurements of BMI, fasting lipid profile and measures of glucose and insulin levels at fasting and in response to a 75 g OGTT. Characteristics of the 7,710 participants included in this study are given in Table 1. Among these, 3,625 were full-heritage and 4,085 were non-full-heritage Pima Indians. All 7,710 individuals were informative for type 2 diabetes, whereas only a subset (n = 5,494) were informative for lipid traits, since lipids were only begun to be measured in 1992 (27 years after the inception of longitudinal study). Given the high prevalence of type 2 diabetes in this population, our lipid analysis included individuals with and without type 2 diabetes, to preserve sample size (i.e. power). However, to minimise any impact of anti-lipidaemic therapy on our results, which may have been prescribed for individuals with diabetes, data from the last examination before the year 2005 were used, when relatively few individuals in this population were using anti-lipidaemic therapy [13]. Data from the last examination when the individual was non-diabetic (n = 5,429) was used for analysis of HOMA-IR and HOMA for beta cell function (HOMA-B). A subset of 561 non-diabetic individuals was also characterised for body fat content and insulin sensitivity in our Clinical Research Center as described elsewhere [14, 15]. All participants gave written informed consent. The study was approved by the institutional review board of the National Institute of Diabetes and Digestive and Kidney Diseases.
Table 1.
Characteristics of Pima Indians from the Gila River Indian community
| Analysis | Full-heritage Pima Indians | Non-full-heritage Pima Indiansb |
|---|---|---|
| Lipid analysis (n=5,494)a | ||
| n (% IFGc) | 2,717 (10.4) | 2,777 (10.8) |
| Age, yearsd | 41.4±15.2 | 30.7±12.9 |
| Age range, years | 15.0–90.4 | 15.0–87.9 |
| TC, mmol/l | 4.58±1.01 | 4.57±0.98 |
| HDL-C, mmol/l | 1.12±0.36 | 1.20±0.35 |
| LDL-C, mmol/l | 2.63±0.79 | 2.69±0.82 |
| TG, mmol/l | 1.64±1.28 | 1.54±1.24 |
| Type 2 diabetes analysis (N=7,710)e with/without diabetes |
||
| n (% age <15 years) | 1,684 (0.77) / 1,941 (9.8) | 865 (2.5) / 3,220 (19.5) |
| Female sex, % | 63 / 52 | 59 / 53 |
| Age, yearsd | 49.1±14.0 / 32.1±14.6 | 41.2 ± 14.1 / 24.9 ± 11.9 |
| Age range, years | 10.6–90.4 / 5.6–85.3 | 9.4–85.4 / 5.3–87.9 |
| Type 2 diabetes analysis set restricted to individuals with HDL- C data (n=5,494) with/without diabetes |
||
| n | 1,325 / 1,392 | 653 / 2,124 |
| Female sex, % | 64.7 / 53.5 | 60.5 / 56.5 |
| Age, yearsd | 48.7±13.6 / 34.5±13.2 | 41.1±13.2 / 27.6±11.03 |
| Age range, years | 15.5–90.4 / 15.0–81.7 | 15.2–85.3 / 15.0–87.9 |
| HOMA analysis (n=5,429)f | ||
| n (% IFGc) | 2,542 (35) | 2,887 (22.9) |
| Age, yearsd | 39±14.5 | 30.4±12.3 |
| Age range, years | 15.0–85.3 | 6.5–87.9 |
| HOMA-IR, SD unitsg | −0.09±0.8 | −0.30±0.9 |
| HOMA-B, SD unitsg | 0.34±0.7 | 0.25±0.7 |
Data are presented as means ± SD unless stated otherwise
Data used from last examination before the year 2005
Heritage is, on average, 4/8 Pima and an additional 2/8 American Indian of different tribes
IFG is based on the 2005 ADA criteria
Age reported is the age at last examination for the trait reported
These individuals constituted 4,625 sibships of which 68% are singletons
Data from the last visit when the participant was non-diabetic. Data from individuals who were diabetic during the first visit were not included
Three different assays for serum insulin were used over the course of the longitudinal study; to account for differing sensitivities among these assays, the natural logarithms of the HOMA values for all examinations were standardised within assay prior to analysis, and results reported in SD units
IFG, impaired fasting glucose
Single-nucleotide polymorphism selection and genotyping
Fifty-five single-nucleotide polymorphisms (SNPs), representing 47 loci, independently associated with HDL-C levels at genome-wide significance in individuals of European ancestry were initially considered for inclusion in this study [7]. Based on previously analysed whole genome sequence (WGS) data from 335 Pima Indians, 19 SNPs with a minor allele frequency (MAF) ≤ 0.05 in Pima Indians were excluded from our analysis (electronic supplementary material [ESM] Table 1). Since lipid traits are highly correlated, we further omitted SNPs (n = 8) at loci having stronger associations with triacylglycerol (TG), LDL-cholesterol (LDL-C) or total cholesterol (TC) (ESM Table 1) in European ancestry populations [7]. Recent studies identifying genetic determinants of adiposity, a risk factor for type 2 diabetes, show that some of these SNPs are also associated with lipids [16–19]. Therefore, data from the Genetic Investigation of Anthropometric Traits (GIANT) consortium (https://www.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files, accessed 1 September 2015) were used to exclude adiposity-associated variants among the remaining 28 SNPs [20, 21]. Six SNPs had a Bonferroni-corrected significant association (p ≤ 0.0017 based on 0.05/28 SNPs) with BMI or waist–hip ratio (adjusted for BMI) in the GIANT consortium GWAS metadata (2012–2015) and were excluded from our study (ESM Table 2). The remaining 21 SNPs (one SNP, rs2040293, failed quality control [QC] metrics) were analysed for association with lipids in the full-heritage Pima Indian sample and those that had an association (p < 0.05) with HDL-C were genotyped in all remaining samples to increase power for assessing type 2 diabetes association. In the event the index GWAS SNP failed design for genotyping assays, a close proxy was substituted based on Pima Indian WGS data (ESM Table 1). SNPs were genotyped by TaqMan allelic discrimination assays (Applied Biosystems, Carlsbad, CA, USA) or by BeadXpress System (Illumina, San Diego, CA, USA). QC required a genotype call rate > 95%, no deviation from Hardy–Weinberg equilibrium (p > 1.0 × 10 −3) and discrepancy rate < 2.5% for blind duplicates (> 100 for each sample set).
The genomic region (chr16:56985835-57018855) encompassing CETP (chr16:56995835-57017756) was further interrogated with 12 additional tag SNPs (ESM Fig. 1) that captured common variants (MAF > 0.05) at the CETP locus with r2 > 0.8. WGS data from 234 full-heritage Pima Indians were used to identify common variation in this ethnic group and tags were selected by pairwise tagging using the tagger algorithm in Haploview, version 4.2 (Broad Institute, Cambridge, MA, USA).
Statistical analysis
Linear regression models were used to assess association between the continuous traits and genotypes assuming an additive model. Since the SNPs included in this study are from previously established HDL-C loci, p < 0.05 was considered statistically significant for association with HDL-C. The p values and effect estimates for lipid association were adjusted for age, sex, diabetes, admixture estimate, Pima fraction and date of last examination. Previous analyses suggested a high degree of overlap in the genetic determinants of lipid levels between diabetic and non-diabetic individuals in this population [22]. Therefore, we analysed diabetic and non-diabetic individuals together, with adjustment for diabetes, to maximise power. The natural logarithm of HDL-C was analysed, and the regression coefficient was exponentiated to obtain the effect estimate, expressed as a multiplier. The individual estimate of European admixture was derived from 45 markers with large differences in allele frequency between populations [23, 24]. Type 2 diabetes association was analysed in the whole population-based sample of 7,710 Pima Indians by logistic regression; p values and ORs were adjusted for age, sex, birth year, admixture estimate and self-reported Pima fraction. Heterogeneity in HDL-C and type 2 diabetes associations between female and male participants was analysed by including a product term (genotype × sex) in the model. SNPs showing evidence for genotype–sex interaction were analysed separately in male and female participants. Type 2 diabetes association was also analysed in the subset of participants (n = 5,494) used for HDL-C analysis and informative for type 2 diabetes at the time of lipid examination. Models were fit with generalised estimating equations to account for dependence among siblings. An un-weighted multi-allelic genetic risk score (GRS) for HDL-C was created by adding the number of risk alleles (allele lowering HDL-C) across all 21 SNPs analysed in the study, which assumes that each SNP acts independently in an additive manner with equal effect. Linear regression was used to assess the association between the GRS as a continuous variable and traits after adjustment for the aforementioned covariates. A second GRS, restricted to only 12 SNPs having significant (p < 0.05) association with HDL-C in full-heritage Pima Indians, was created to assess the association between these 12 SNPs in combination and HDL-C and type 2 diabetes in the entire sample. Effect estimates and ORs are given per HDL-C-lowering allele.
Results
SNPs associated with lipid levels
Lipid association data for 21 previously established HDL-C SNPs in full-heritage Pima Indian samples are shown in ESM Table 3. Twelve SNPs were significantly (p < 0.05) associated with HDL-C in 2,675 full-heritage Pima Indians and all the associations were directionally consistent with those reported in individuals with European ancestry [7]. These 12 SNPs were further genotyped in the remaining participants (n = 5,035 were informative for type 2 diabetes of which 2,819 also had lipid measures). Re-analysis of the 12 HDL-C-associated SNPs in all participants with lipid measures (n = 5,494, representing 2,717 full and 2,777 non-full-heritage Pima Indians) identified the strongest HDL-C associations for SNPs rs17231506 and rs12720922 at the CETP locus, which reduced HDL-C by approximately 6.5% per risk allele (Table 2, ESM Table 4).
Table 2.
Association of SNPs with HDL-C in Pima Indians from the Gila River Indian community
|
|
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Locusa | Alleles |
HDL-C levels |
|
|
||||
| R/NR | RAF | Full Pima (n=2,717) | Non-full Pima (n=2,777) |
Combined (n=5,494) | |||||
|
|
|
|
|
||||||
| Effect (%)b |
p value | Effect (%)b |
p value | Effect (%)b |
p value | ||||
|
|
|
|
|||||||
| rs17231506 | CETP | C/T | 0.65 | −6.9 | 2.6×1018 | −6.2 | 6.7×10−16 | −6.6 | 2.6×10−32 |
| rs12720922 | CETP | A/G | 0.30 | −6.4 | 1.9×10−17 | −6.8 | 4.2×10−17 | −6.5 | 3.1×10−32 |
| rs56322906 | DOCK6 | A/G | 0.46 | −3.3 | 2.8×10−5 | −3.5 | 4.2×10−6 | −3.5 | 1.4×10−10 |
| rs9987289 | PPP1R3B | A/G | 0.26 | −3.4 | 7.5×10−5 | −3.3 | 4.9×10−5 | −3.4 | 6.2×10−9 |
| rs1883025 | ABCA1 | T/C | 0.37 | −3.0 | 1.7×10−4 | −3.3 | 2.0×10−5 | −3.1 | 1.2×10−8 |
| rs1532085 | LIPC | G/A | 0.62 | −1.8 | 0.02 | −3.2 | 1.2×10−5 | −2.6 | 2.3×10−6 |
| rs838880 | SCARB1 | T/C | 0.59 | −1.8 | 0.02 | −2.7 | 2.6×10−4 | −2.3 | 2.0×10−5 |
| rs4731702 | KLF14 | C/T | 0.43 | −2.5 | 2.8×10−3 | −1.5 | 0.05 | −2.0 | 4.4×10−4 |
| rs7134594 | MMAB | C/T | 0.70 | −2.5 | 4.6×10−3 | −1.3 | 0.10 | −1.9 | 1.4×10−3 |
| rs1077834 | LIPC | A/G | 0.20 | −2.9 | 0.01 | −1.6 | 0.09 | −2.1 | 3.9×10−3 |
| rs10127775 | GALNT2 | A/T | 0.41 | −1.8 | 0.02 | −0.9 | 0.28 | −1.3 | 0.02 |
| rs1800961 | HNF4A | T/C | 0.03 | −6.5 | 7.3×10−3 | −0.7 | 0.71 | −3.1 | 0.05 |
Association with HDL-C was analysed in full-heritage Pima Indians, non-full-heritage Pima Indians and in the combined dataset. Natural logarithm of HDL-C was analysed, and the regression coefficient was exponentiated to obtain the effect estimate, expressed as a multiplier
Locus is based on previously reported data [7], where ever possible a plausible candidate gene is listed
Effect is given as percentage reduction in HDL-C level per copy of the risk allele
NR, non-risk allele; R, risk allele (HDL-C lowering allele); RAF, risk allele frequency in Pima Indians
Analysis of HDL-C-associated SNPs for association with type 2 diabetes
The 12 HDL-C-associated SNPs were further analysed in 7,710 participants informative for type 2 diabetes. SNPs at HNF4A, CETP, KLF14, MMAB and GALNT2 loci had nominal associations with type 2 diabetes (Table 3). Three of these SNPs (in HNF4A, CETP and ABCA1) had significant genotype–sex interaction (p_int_sex < 0.05) for type 2 diabetes association and were re-analysed in male and female participants separately (no genotype–sex interaction observed for the HDL-C associations, p_int_sex > 0.05). Rs4731702 in KLF14 had marginal evidence for genotype–sex interaction (p_int_sex = 0.06) for type 2 diabetes association and was also re-analysed separately in male and female participants. For three of these SNPs (CETP, KLF14 and HNF4A) the HDL-C-lowering allele significantly (Bonferroni correction requires p < 4.2 × 10 −3 based on 0.05/12 SNPs) associated with increased risk of type 2 diabetes in female but not male participants (Table 3). This sex-specific effect was directionally consistent in full and non-full-heritage Pima Indian samples when analysed separately, although the association for the CETP SNP in non-full Pima female participants did not reach statistical significance (p = 0.10, ESM Table 5). In contrast, the HDL-C-lowering allele for the ABCA1 SNP associated with decreased risk for type 2 diabetes in female participants, but did not achieve Bonferroni-corrected significance (Table 3). SNPs in GALNT2 and MMAB did not achieve a Bonferroni-corrected significant association with type 2 diabetes in the combined dataset and showed no evidence for a genotype–sex interaction; thus they were not further analysed separately in male and female participants (Table 3). Similar results were obtained when the 12 HDL-C-associated SNPs were analysed prospectively using a Cox proportional hazard model; the HDL-C-lowering allele of SNPs in CETP, KLF14 and HNF4A were associated with increased risk for type 2 diabetes in female participants and the SNP in ABCA1 was protective for type 2 diabetes in female participants (ESM Table 6).
Table 3.
Association of HDL-C-associated SNPs with type 2 diabetes in Pima Indians from the Gila River Indian Community
|
|
|
|
||||||
|---|---|---|---|---|---|---|---|---|
| SNP ID | Locusa | Combined participants (N=7,710) |
Male participants (n = 3,426) |
Female participants (n = 4,284) |
||||
| OR (95% CI) |
p
value |
p_int_sex | OR (95% CI) | p value | OR (95% CI) | p value | ||
|
|
|
|
||||||
| rs172315 06 |
CETP | 1.03 (0.94, 1.13) |
0.54 | 0.66 | ||||
| rs127209 22 |
CETP | 1.12 (1.02, 1.24) |
0.02 | 0.004 | 0.98 (0.86, 1.14) |
0.81 | 1.25 (1.11, 1.43) |
2.4×10−4 |
| rs563229 06 |
DOCK6 | 1.04 (0.95, 1.14) |
0.38 | 0.27 | ||||
| rs998728 9 |
PPP1R3B | 0.96 (0.87, 1.07) |
0.48 | 0.43 | ||||
| rs188302 5 |
ABCA1 | 0.95 (0.87, 1.04) |
0.28 | 0.01 | 1.08 (0.94, 1.24) |
0.28 | 0.85 (0.76, 0.96) |
9.6×10−3 |
| rs153208 5 |
LIPC | 1.01 (0.92, 1.10) |
0.89 | 0.42 | ||||
| rs838880 | SCARB1 | 0.99 (0.90, 1.08) |
0.78 | 0.77 | ||||
| rs473170 2 |
KLF14 | 1.14 (1.04, 1.25) |
3.5×10 −3 |
0.06 | 1.06 (0.93, 1.21) |
0.40 | 1.25 (1.10, 1.40) |
3.2×10−4 |
| rs713459 4 |
MMAB | 1.11 (1.01, 1.22) |
0.03 | 0.43 | ||||
| rs107783 4 |
LIPC | 0.99 (0.88, 1.11) |
0.81 | 0.19 | ||||
| rs101277 75 |
GALNT2 | 0.90 (0.83, 0.99) |
0.03 | 0.89 | ||||
| rs180096 1 |
HNF4A | 1.38 (1.09, 1.75) |
7.6×10 −3 |
0.006 | 0.91 (0.62, 1.33) |
0.63 | 1.83 (1.35, 2.46) |
7.7×10−5 |
HDL-C associated SNPs were analysed for association with type 2 diabetes in the entire data set of 7,710 Pima Indian participants. SNPs showing evidence of sex interaction (p_int_sex approaching or less than 0.05) for type 2 diabetes association were further analysed separately in male and female participants. ORs are reported per copy of the HDL-C-lowering allele
Locus is based on previously reported data [7] or the closest plausible candidate gene
To evaluate whether the sex-specific associations with type 2 diabetes for SNPs in CETP, KLF14 and HNF4A are mediated by HDL-C levels, we analysed the SNPs in the subset of samples informative for both type 2 diabetes and HDL-C levels (n = 5,494) and additionally adjusted for HDL-C (Table 4). In male participants, the type 2 diabetes associations were not significant with or without adjustment for HDL-C; however, in female participants all three SNPs remained nominally significant even after adjustment, although the effects were attenuated (Table 4). When additionally adjusted for other lipid measures (LDL-C, TG and TC levels) the female sex-specific type 2 diabetes association of SNPs in KLF14 and HNF4A remained nominally significant (n = 2,977; p = 0.03, OR 1.16 and p = 0.02, OR 1.54, respectively) although this adjustment rendered the association of the CETP SNP non-significant (p = 0.14).
Table 4.
Association of SNPs in CETP, KLF14 and HNF4A with type 2 diabetes before and after adjustment for HDL-C
|
|
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Locus | Type 2 diabetesa (n=5,494) | Type 2 diabetes_adjHDL-Cb (n=5,494) | ||||||
|
|
|
|
|
||||||
| Male participants (n=2,296) | Female participants (n=3,198) |
Male participants (n=2,296) | Female participants (n=3,198) |
||||||
|
|
|
|
|
||||||
| OR (95% CI) | p value | OR (95% CI) |
p value | OR (95% CI) | p value | OR (95% CI) |
p
value |
||
| rs1272092 2 |
CETP | 0.97 (0.83, 1.14) | 0.72 | 1.31 (1.14, 1.50) |
8.5×10−5 | 0.86 (0.75, 1.05) | 0.15 | 1.20 (1.05, 1.38) |
8.0×10 −3 |
| rs4731702 | KLF14 | 1.01 (0.87, 1.17) | 0.88 | 1.23 (1.07, 1.40) |
2.8×10−3 | 0.97 (0.83, 1.12) | 0.66 | 1.21 (1.06, 1.39) |
5.1×10 −3 |
| rs1800961 |
HNF4
A |
0.83 (0.53, 1.30) | 0.42 | 1.62 (1.16, 2.26) |
4.5×10−3 | 0.81 (0.52, 1.27) | 0.37 | 1.57 (1.12, 2.20) |
8.3×10 −3 |
Type 2 diabetes association was analysed in the subset of samples (n=5,494) informative for both HDL-C levels and type 2 diabetes status. Analysis is shown separately in male and female participants. ORs reported are per copy of the HDL-C-lowering allele
Type 2 diabetes association before adjustment for HDL-C
Type 2 diabetes association after adjustment for HDL-C
Additional analysis at CETP
SNPs in the CETP locus provided the strongest evidence for association with HDL-C. Therefore, an additional 12 tag SNPs in CETP were analysed, along with the previously analysed rs12720922 and rs17231506 variants, for association with HDL-C and type 2 diabetes in full-heritage Pima Indians (Table 5). Rs6499863, which strongly associated with HDL-C (p = 5.6 × 10 −11, effect = −5.4%; Table 5, Fig. 1a), had the strongest evidence for association with type 2 diabetes (p = 4.6 10 × −4, OR 1.26 [1.11, 1.43]; Table 5, Fig. 1b). Rs6499863 had a significant genotype–sex interaction for type 2 diabetes such that this SNP was significantly associated with type 2 diabetes in female (p = 2.8 × 10 −5, OR 1.44 [1.21, 1.71]; Table 5, Fig. 1d) but not male participants (Table 5, Fig. 1c). This SNP was further genotyped and analysed in the non-full-heritage Pima Indian sample where the association with HDL-C (p = 1.3 × 10 −6, effect = −4.9%; Fig. 1a) and type 2 diabetes in female participants (p = 0.04, OR 1.26 [1.01, 1.55]; Fig. 1d) but not in male participants (p = 0.70, OR 1.05 [0.82, 1.34]; Fig. 1c) was replicated. In the full-heritage and non-full-heritage Pima Indian combined sample, rs6499863 robustly associated with HDL-C (p = 3.6 × 10 −15, effect = −4.9%; Fig. 1a) and significantly increased the risk for type 2 diabetes (p = 1.8 × 10 −4, OR 1.22 [1.10, 1.35]; Fig. 1b). The genotype–sex interaction remained significant (p = 0.007) such that this SNP provided evidence for type 2 diabetes association in the combined analysis in female (p = 5.0 × 10 −6, OR 1.36 [1.19, 1.55]; Fig. 1d) but not male participants (Fig. 1c).
Table 5.
Association of SNPs in CETP with HDL-C and type 2 diabetes in full-heritage Pima Indians from the Gila River Indian Community
|
|
|
|
|
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Alleles | HDL-C | Type 2 diabetes | |||||||
|
|
|
|
|
|
||||||
| Full-Pima participants (n=2,717) |
Full-Pima participants (n=3,625) |
Full-Pima male participants (n=1,553) |
Full-Pima female participants (n=2,072) |
|||||||
|
|
|
|
|
|
||||||
| R/NR | RAF | Effect (%) |
p value | OR (95% CI) | p value | OR (95% CI)d | p value | OR (95% CI)d |
p value | |
|
|
|
|
|
|
||||||
| rs17231506 | C/T | 0.67 | −6.9 | 2.6×10 −18 |
1.00 (0.88, 1.13) | 0.97 | 1.04 (0.87, 1.23) |
0.70 | 0.98 (0.83, 1.15) |
0.79 |
| rs12720922 | A/G | 0.33 | −6.4 | 1.9×10 −17 |
1.16 (1.03, 1.30) | 0.01 | 1.01 (0.85, 1.20) |
0.91 | 1.32 (1.12, 1.54) |
5.0×10−4 |
| rs289713 | T/A | 0.58 | −6.3 | 4.7×10 −17 |
1.08 (0.96, 1.22) | 0.20 | 1.05 (0.89, 1.25) |
0.56 | 1.11 (0.95, 1.30) |
0.17 |
| rs708272 | G/A | 0.43 | −5.8 | 2.2×10 −15 |
1.11 (0.99, 1.24) | 0.08 | 1.01 (0.86, 1.19) |
0.94 | 1.22 (1.04, 1.41) |
0.01 |
| rs6499863 | T/C | 0.27 | −5.4 | 5.6×10 −11 |
1.26 (1.11, 1.43) | 4.6×10−
4 |
1.08 (0.90, 1.31) |
0.41 | 1.44 (1.21, 1.71) |
2.8×10−5 |
| rs289742 | C/G | 0.78 | −4.0 | 1.4×10 −5 |
1.02 (0.88, 1.18) | 0.78 | 0.90 (0.74, 1.10) |
0.31 | 1.15 (0.94, 1.39) |
0.18 |
| rs1800774 | T/C | 0.40 | −3.2 | 2.5×10 −5 |
1.05 (0.94, 1.19) | 0.39 | 0.96 (0.80, 1.15) |
0.69 | 1.14 (0.97, 1.32) |
0.12 |
| rs5882 | A/G | 0.47 | −2.9 | 1.4×10 −4 |
1.04 (0.92, 1.17) | 0.53 | 0.93 (0.78, 1.11) |
0.43 | 1.14 (0.98, 1.33) |
0.09 |
| rs5880 | G/C | 0.08 | −5.3 | 3.2×10−4 | 0.80 (0.65, 0.99) | 0.04 | 0.68 (0.48, 0.97) |
0.03 | 0.89 (0.69, 1.15) |
0.37 |
| rs708273 | C/T | 0.93 | −2.0 | 0.16 | 1.13 (0.89, 1.44) | 0.31 | 1.16 (0.81, 1.67) |
0.41 | 1.12 (0.83, 1.52) |
0.45 |
| rs820299 | T/C | 0.91 | −1.4 | 0.28 | 1.07 (0.87, 1.33) | 0.52 | 0.96 (0.70, 1.32) |
0.80 | 1.17 (0.90, 1.53) |
0.24 |
| rs8045701 | T/C | 0.69 | −0.3 | 0.74 | 1.03 (0.91, 1.17) |
0.66 | 1.01 (0.83, 1.22) |
0.95 | 1.05 (0.83, 1.23) | 0.59 |
| rs291044 | G/A | 0.93 | −0.4 | 0.76 | 1.10 (0.87, 1.39) | 0.43 | 1.23 (0.79, 1.75) |
0.25 | 0.99 (0.74, 1.35) |
0.97 |
| rs12708970 | G/T | 0.77 | −0.3 | 0.77 | 1.11 (0.97, 1.29) | 0.14 | 1.02 (0.82, 1.27) |
0.85 | 1.20 (1.01, 1.44) |
0.04 |
Type 2 diabetes association results are presented both separately in male and female participants and in the combined sample of full-heritage Pima Indians. HDL-C values were log transformed for calculating p value and effect estimates. Effect is given as percentage reduction per copy of the risk allele. ORs are given per copy of the HDL-C-lowering allele
NR, non-risk allele; R, risk allele (HDL-C-lowering allele); RAF, risk allele frequency in Pima Indians
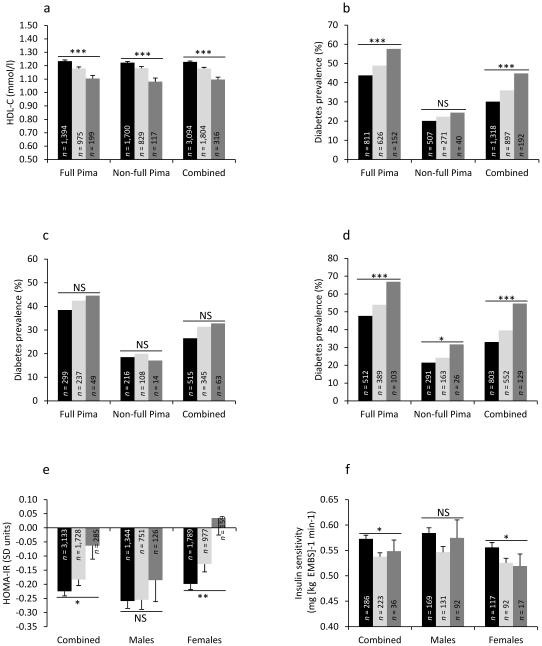
Fig. 1. Association of rs6499863 (CETP) with HDL-C, type 2 diabetes prevalence and diabetes related traits in Pima Indians.
(a) HDL-C levels based on rs6499863 genotypes in full-Pima and non-full Pima Indians and combined dataset. (b–d) Diabetes prevalence based on rs6499863 genotypes before (b) and after stratification by sex (c, male participants; d, female participants) in full-Pima and non-full Pima Indians and combined. (e–f) Measures of HOMA-IR (e) and insulin sensitivity (f) based on rs6499863 genotypes in combined dataset before and after stratification by sex. For HOMA-IR, p value and effect estimates were adjusted for age, sex, BMI, admixture estimate and Pima fraction. For insulin sensitivity, p values and effect estimate were adjusted for age, sex, body fatness and admixture estimate. Black bars, CC genotype; light-grey bars, CT genotype; dark-grey bars, TT genotype. Error bar, SEM, *p<0.05, **p<0.01 and ***p<0.001
Among female participants informative for both HDL-C levels and type 2 diabetes, the association of rs6499863 with type 2 diabetes was significant before and after adjustment for HDL-C levels (n = 3,044; pbefore adjustment = 9.9 × 10 −8, OR 1.49 [1.29, 1.72] and pafter adjustment = 9.9 × 10 −6, OR 1.40 [1.20, 1.62]), and remained significant when adjusted for other lipid measures (TC, LDL-C and TG) (n = 2,936, pafter adjustment = 6.7 × 10 −4, OR 1.31 [1.12, 1.53]). Rs6499863 also associated with estimates of insulin resistance (HOMA-IR) (p = 0.02, β[SD units] = 0.04; Fig. 1e), where the association was restricted to female participants (p = 0.005, β[SD units] = 0.06; Fig. 1e). Rs6499863 further associated with insulin sensitivity as assessed by the euglycaemic–hyperinsulinaemic clamp technique in 561 non-diabetic individuals (p = 0.02, effect = −4% per allele; Fig. 1f), where the association was only found in female participants (p = 0.03, effect = −4% per allele; Fig. 1f).
GRS analysis of HDL-C SNPs in Pima Indians
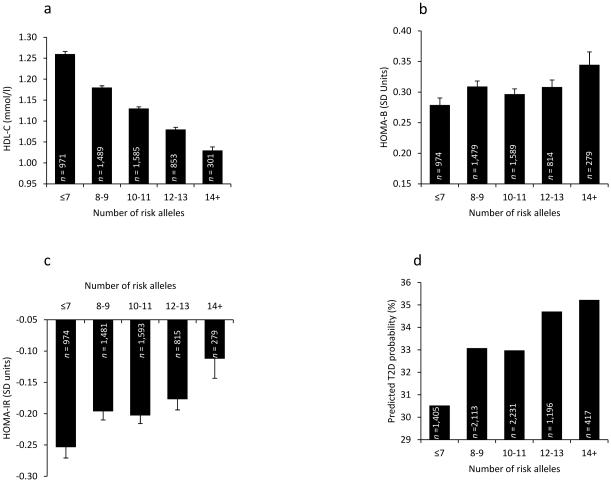
A multi-allelic GRS, which reflected the aggregate effect of all 21 established SNPs analysed (GRS21), associated with HDL-C such that there was an average decrease of HDL-C by 1.2% per risk allele (p = 1.9 × 10 −14). However GRS21 was not associated with HOMA-IR, HOMA-B or type 2 diabetes. This analysis was restricted to only full-heritage Pima Indians since we did not genotype all 21 variants in the entire sample. A refined GRS (GRS12) created from the 12 SNPs significantly associated with HDL-C in full-heritage Pima Indians, and therefore genotyped in the entire sample, strongly associated with HDL-C in the combined analysis (p = 1.4 × 10 −47, effect = −2.2%; Fig. 2a). GRS12 also positively associated with type 2 diabetes and HOMA-IR when analysed in the non-full-heritage Pima samples (n = 4,004, p = 0.002, OR 1.06 [1.02, 1.10] and n = 2,728, p = 0.03, β[SD units] = 0.012 per risk allele, respectively) and in the entire dataset (p = 0.04, OR 1.03 [1.00, 1.05] and p = 0.003, β[SD units] = 0.01, respectively, Fig. 2 c, d), but not with HOMA-B (Fig. 2b). However, the GRS did not have a significant interaction with sex for the traits analysed. When rs4731702 (KLF14) and rs1800961 (HNF4A) (established loci for type 2 diabetes) were removed from the GRS12 analysis, the association with HOMA-IR remained significant (p = 0.02) but the association with type 2 diabetes was rendered non-significant (p = 0.60).
Fig. 2. Relationship between GRS12 and HDL-C levels, HOMA-B, HOMA-IR and prevalence of type 2 diabetes in Pima Indians.
GRS calculated as number of risk alleles and plotted to include reasonable number of participants in each category. (a) Association with HDL-C (p=1.4 × 10 −47, effect= −2.2%). (b) Association with HOMA-B (p=0.21). (c) Association with HOMA-IR (p=0.003, β [SD units]=0.01). (d) Association with predicted diabetes probability (p=0.04, OR 1.03 [1.00, 1.05]), diabetes association NS when rs4731702 and rs1800961 are removed from GRS12. Error bar, SEM. T2D, type 2 diabetes
Discussion
We assessed whether SNPs associated with increased HDL-C also associate with decreased risk for type 2 diabetes in female Pima Indians. Twenty-one primarily HDL-C-associated SNPs identified in other ethnic groups were initially included in our analysis; a recent meta-analysis has identified 24 additional loci [8], which were not included in the current study. Twelve SNPs significantly associated with HDL-C were analysed in the entire data set for type 2 diabetes association. However, a nominal association of the other nine variants with HDL-C and/or type 2 diabetes cannot be ruled out. In our type 2 diabetes analysis (both cross-sectional and prospective), we found that three of the 12 SNPs (in HNF4A, CETP and KLF14) associated with both HDL-C and type 2 diabetes, where the HDL-C-increasing allele conveyed protection against type 2 diabetes only in female participants. Sex hormones affect several enzymes involved in HDL-C metabolism, and women have higher levels of HDL-C than men [3, 25]. Oestrogen increases the production of ApoA1 and raises HDL2 levels [26, 27] and among the different sub-fractions of HDL-C, the HDL2a subfraction has the strongest protective role against the development of type 2 diabetes in Pima Indian women [28]. Therefore, the influence of sex hormones on HDL-C metabolism, in particular HDL2a, may potentially explain the observation of a protective role for HDL-C in female but not male participants in this study.
The established HDL-C variant rs1800961 in HNF4A leads to a threonine139isoluceine (p.Thr139Ile) amino acid change in the DNA-binding domain of HNF4A. Although different mutations in HNF4A have been shown to cause MODY 1 [29, 30], the p.Thr139Ile variant has been reported to associate with type 2 diabetes [31]. In our study, the isoleucine allele was associated with lower HDL-C and increased diabetes risk in female participants, the association remaining significant when additionally adjusted for lipid levels. HNF4A is expressed in both pancreatic beta cells, where it controls the expression of insulin and proteins involved in insulin secretion, and in liver, where it regulates the expression of apolipoproteins and affects lipid physiology [32–34]. Given the known effect of mutations in HNF4A on beta cell function [35] and association of the p.Thr139Ile variant with HDL-C, it is unclear whether the association of this variant with type 2 diabetes is due to a primary effect on beta cells or on lipid metabolism in liver or both. In vitro functional studies provide evidence that the p.Thr139Ile variant leads to a loss of function mutation in hepatocytes but not in beta cell lines, suggesting a primary effect on lipid physiology in liver [36]. However, the current observation and the known effect of mutations in HNF4A on beta cell function suggest an association of this mutation with type 2 diabetes independent of its association with lipids.
Another established HDL-C variant, rs4731702, associated with HDL-C and type 2 diabetes in female Pima Indians. However, when adjusted for lipid levels, the type 2 diabetes association remained nominally significant. This observation is not surprising as this SNP maps to the imprinted maternally expressed KLF14 locus and previous studies have shown an association of this SNP with type 2 diabetes after accounting for parent of origin effect [37–38]. Therefore, the association of this SNP with type 2 diabetes is unlikely to be mediated by HDL-C levels.
Among the SNPs in the CETP locus, rs17231506 and rs12720922 had the strongest associations with HDL-C, and conditional analysis identified genome-wide significant independent association for both SNPs, conditional on the effect of the other (n = 5,025, p = 2.8 × 10 11 and p = 2.0 × 10 −8, respectively). However, only rs12720922 associated with type 2 diabetes in female participants. This association remained significant when adjustment was made for HDL-C levels but not for other lipid measures as well. To capture additional information across this locus, additional tag SNPs were analysed. Of these, rs6499863 had the strongest association with type 2 diabetes in female participants and had a genome-wide significant association with HDL-C. SNPs rs6499863 and rs12720922 are in moderate linkage disequilibrium (LD) in Pima Indians (r2 = 0.65, n = 234), but not in individuals of European ancestry (r2 < 0.2, http://www.broadinstitute.org/mammals/haploreg/haploreg_v3.php, accessed 1 September 2015). Conditioning rs6499863 for rs12720922 greatly reduces its association with HDL-C (n = 5,032, p = 0.03), whereas rs12720922 still remains genome-wide significant (p = 4.2 × 10 −15). However, a conditional analysis that included both variants for association with type 2 diabetes identified an association only for rs6499863 (n = 3,937, p = 0.017, OR 1.33 [1.05, 1.67]) and not rs12720922 (p = 0.77) with type 2 diabetes in female participants in the combined dataset. It should also be noted that the association of rs6499863 with type 2 diabetes in female participants remained significant even after adjustment for lipid levels.
This observation raises the possibility that the CETP locus harbours independent signals for HDL-C and type 2 diabetes. Since the female sex-specific association of rs6499863 with type 2 diabetes does not achieve genome-wide significance, and CETP has not been identified as a type 2 diabetes locus in large GWAS studies [39, 40], this hypothesis requires validation. However, the current observation is consistent with a previously identified association of the Taq1B (rs708272) polymorphism in CETP and higher plasma cholesteryl ester transfer protein (CETP) activity, HDL-C levels, HOMA-IR, metabolic syndrome and type 2 diabetes [41]. This variant associated with metabolic syndrome independent of its association with insulin resistance and HDL-C [42]. More importantly, supportive evidence for distinct effects of CETP on lipid and glucose metabolism comes from a post hoc analysis of clinical trial data for the CETP inhibitor drug torcetrapib, the results of which identified a very significant effect on plasma glucose levels and other glycaemic traits that did not correlate completely with the drug’s effect on HDL-C levels [43]. Similarly, in a safety trial for the CETP inhibitor anacetrapib, which also increases HDL-C levels, individuals taking the drug had a trend towards decreased HbA1c levels [44]. Although epidemiological studies have shown that lower HDL-C is associated with reduced insulin sensitivity and increased risk for type 2 diabetes, the causal nature of these relationships is uncertain. If lower HDL-C caused increased risk for diabetes then one would expect genetic variants influencing HDL-C concentrations to be associated with risk for type 2 diabetes, and this is the basis for Mendelian randomisation analyses, which attempt to estimate the causal effects. These analyses require the assumption that the genetic variants capture the causal relationships and that the associations are not confounded by population structure, LD between variants influencing each trait or the influence of other mechanisms such as pleiotropy. As discussed by Fall et al [12], these assumptions, particularly the lack of pleiotropy, are problematic for the relationship between lipids and diabetes risk. Furthermore, in the present analyses the CETP locus contained separate variants for HDL-C and diabetes risk, which were in moderate LD. We have thus not attempted a formal Mendelian randomisation analysis, and the estimates may not reflect causal effect of HDL-C on diabetes risk. Our GRS analysis using all 21 established HDL-C variants genotyped in full-heritage Pima Indians identified significant association with HDL-C but not with HOMA-IR and type 2 diabetes. When using a refined GRS (GRS12) we observed a significant association between genetic variants associated with reduced HDL-C and increased risk of type 2 diabetes and increased HOMA-IR. It is difficult to determine whether the differences between these analyses reflect differences in power across the different samples, heterogeneity in effects across variants or overfitting bias due to selection of those with strongest HDL-C association in Pimas. It should also be noted that when the KLF14 and HNF4A SNPs were removed from the GRS analysis the type 2 diabetes association did not remain significant, suggesting that this association was largely influenced by these two variants. Another limitation of these analyses is that the causal variants influencing HDL-C are generally unknown and these may not be as well captured in Pima Indians due to differing LD patterns compared with European populations.
In conclusion, the results presented here and the additional literature evidence with respect to the role of CETP in type 2 diabetes risk are worthy of follow-up. Although the type 2 diabetes association did not reach genome-wide significance, previously reported evidence supports our observation. We also observed female sex-specific type 2 diabetes associations with established HDL-C SNPs in HNF4A and KLF14, but the current evidence is insufficient to conclude that these associations were mediated by differences in HDL-C.
Supplementary Material
Acknowledgements
The authors thank the volunteers who participated in the study and also thank the staff of Phoenix Epidemiology and Clinical Research Branch who helped with the study. This study used the computational resources of the Biowulf system at the National Institutes of Health, Bethesda, MD, USA (for a description please see https://helix.nih.gov/).
Funding
The study was supported by the intramural research programme of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Abbreviations
- CETP
Cholesteryl ester transfer protein, plasma
- GIANT
Genetic investigation of anthropometric traits
- GRS
Genetic risk score
- GRS21
GRS created using genetic information from 21 SNPs.
- GRS12
GRS created using genetic information from 12 SNPs.
- GWAS
Genome-wide association studies
- HDL-C
HDL-cholesterol
- HOMA-B
HOMA for beta cell function
- LD
Linkage disequilibrium
- LDL-C
LDL-cholesterol
- MAF
Minor allele frequency
- SNP
Single-nucleotide polymorphism
- QC
Quality control
- TC
Total cholesterol
- TG
Triacylglycerol
- WGS
Whole genome sequence
Footnotes
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
LJB was responsible for the integrity of the work as a whole. AKN and LJB contributed to the study design. AKN, NAM, MK, WCK, CB, RLH and LJB contributed to data acquisition, and SK, PP and RLH contributed to data analysis. All authors contributed to data interpretation and manuscript drafting and approved the final version of the paper.
References
- 1.Cooney MT, Dudina A, De Bacquer D, et al. HDL cholesterol protects against cardiovascular disease in both genders, at all ages and at all levels of risk. Atherosclerosis. 2009;206:611–616. doi: 10.1016/j.atherosclerosis.2009.02.041. [DOI] [PubMed] [Google Scholar]
- 2.Drew BG, Rye KA, Duffy SJ, Barter P, Kingwell BA. The emerging role of HDL in glucose metabolism. Nat Rev Endocrinol. 2012;8:237–245. doi: 10.1038/nrendo.2011.235. [DOI] [PubMed] [Google Scholar]
- 3.Fagot-Campagna A, Narayan KM, Hanson RL, et al. Plasma lipoproteins and incidence of non-insulin-dependent diabetes mellitus in Pima Indians: protective effect of HDL cholesterol in women. Atherosclerosis. 1997;128:113–119. doi: 10.1016/s0021-9150(96)05978-3. [DOI] [PubMed] [Google Scholar]
- 4.Hirano M, Nakanishi S, Kubota M, et al. Low high-density lipoprotein cholesterol level is a significant risk factor for development of type 2 diabetes: data from the Hawaii-Los Angeles-Hiroshima study. J Diabetes Investig. 2014;5:501–506. doi: 10.1111/jdi.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Njølstad I, Arnesen E, Lund-Larsen PG, et al. Sex differences in risk factors for clinical diabetes mellitus in a general population: a 12-year follow-up of the Finnmark Study. Am J Epidemiol. 1998;147:49–58. doi: 10.1093/oxfordjournals.aje.a009366. [DOI] [PubMed] [Google Scholar]
- 6.Haffner SM, Stern MP, Hazuda HP, Mitchell BD, Patterson JK. Cardiovascular risk factors in confirmed prediabetic individuals. Does the clock for coronary heart disease start ticking before the onset of clinical diabetes? JAMA. 1990;263:2893–2898. doi: 10.1001/jama.263.21.2893. [DOI] [PubMed] [Google Scholar]
- 7.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Global Lipids Genetics Consortium. Willer CJ, Schmidt EM, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi Q, Liang L, Doria A, Hu FB, Qi L. Genetic predisposition to dyslipidemia and type 2 diabetes risk in two prospective cohorts. Diabetes. 2012;61:745–752. doi: 10.2337/db11-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klimentidis YC, Wineinger NE, Vazquez AI, de Los Campos G. Multiple metabolic genetic risk scores and type 2 diabetes risk in three racial/ethnic groups. J Clin Endocrinol Metab. 2014;99:E1814–E1818. doi: 10.1210/jc.2014-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li N, van der Sijde MR, LifeLines Cohort Study Group et al. Pleiotropic effects of lipid genes on plasma glucose, HbA1c, and HOMA-IR levels. Diabetes. 2014;63:3149–3158. doi: 10.2337/db13-1800. [DOI] [PubMed] [Google Scholar]
- 12.Fall T, Xie W, Poon W, et al. Using genetic variants to assess the relationship between circulating lipids and type 2 diabetes. Diabetes. 2015;64:2676–2684. doi: 10.2337/db14-1710. [DOI] [PubMed] [Google Scholar]
- 13.Looker HC, Krakoff J, Andre V, et al. Secular trends in treatment and control of type 2 diabetes in an American Indian population: a 30-year longitudinal study. Diabetes Care. 2010;33:2383–2389. doi: 10.2337/dc10-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tataranni PA, Ravussin E. Use of dual-energy X-ray absorptiometry in obese individuals. Am J Clin Nutr. 1995;62:730–734. doi: 10.1093/ajcn/62.4.730. [DOI] [PubMed] [Google Scholar]
- 15.Lillioja S, Mott DM, Spraul M, et al. Insulin resistance and insulin secretory dysfunction as precursors of non-insulin-dependent diabetes mellitus. Prospective studies of Pima Indians. N Engl J Med. 1993;329:1988–1992. doi: 10.1056/NEJM199312303292703. [DOI] [PubMed] [Google Scholar]
- 16.Rana JS, Li TY, Manson JE, Hu FB. Adiposity compared with physical inactivity and risk of type 2 diabetes in women. Diabetes Care. 2007;30:53–58. doi: 10.2337/dc06-1456. [DOI] [PubMed] [Google Scholar]
- 17.Manning AK, Hivert MF, Scott RA, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu CT, Monda KL, Taylor KC, Lange L, et al. Genome-wide association of body fat distribution in African ancestry populations suggests new loci. PLoS Genet. 2013;9:e1003681. doi: 10.1371/journal.pgen.1003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamina C, Forer L, Schönherr S, et al. Evaluation of gene-obesity interaction effects on cholesterol levels: a genetic predisposition score on HDL-cholesterol is modified by obesity. Atherosclerosis. 2012;225:363–369. doi: 10.1016/j.atherosclerosis.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 20.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shungin D, Winkler TW, Croteau-Chonka DC, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imperatore G, Knowler WC, Pettitt DJ, et al. A locus influencing total serum cholesterol on chromosome 19p: results from an autosomal genomic scan of serum lipid concentrations in Pima Indians. Arterioscler Thromb Vasc Biol. 2000;20:2651–2656. doi: 10.1161/01.atv.20.12.2651. [DOI] [PubMed] [Google Scholar]
- 23.Tian C, Hinds DA, Shigeta R, et al. A genomewide single-nucleotide-polymorphism panel for Mexican American admixture mapping. Am J Hum Genet. 2007;80:1014–1023. doi: 10.1086/513522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanis CL, Chakraborty R, Ferrell RE, Schull WJ. Individual admixture estimates: disease associations and individual risk of diabetes and gallbladder disease among Mexican-Americans in Starr County, Texas. Am J Phys Anthropol. 1986;70:433–441. doi: 10.1002/ajpa.1330700404. [DOI] [PubMed] [Google Scholar]
- 25.Haffner SM, Valdez RA. Endogenous sex hormones: impact on lipids, lipoproteins, and insulin. Am J Med. 1995;98:40S–47S. doi: 10.1016/s0002-9343(99)80058-8. [DOI] [PubMed] [Google Scholar]
- 26.Lamon-Fava S, Ordovas JM, Schaefer EJ. Estrogen increases apolipoprotein (apo) A-I secretion in hep G2 cells by modulating transcription of the apo A-I gene promoter. Arterioscler Thromb Vasc Biol. 1999;19:2960–2965. doi: 10.1161/01.atv.19.12.2960. 1999. [DOI] [PubMed] [Google Scholar]
- 27.Fåhraeus L, Wallentin L. High density lipoprotein subfractions during oral and cutaneous administration of 17 beta-estradiol to menopausal women. J Clin Endocrinol Metab. 1983;56:797–801. doi: 10.1210/jcem-56-4-797. [DOI] [PubMed] [Google Scholar]
- 28.Fagot-Campagna A, Knowler WC, Narayan KM, Hanson RL, Saaddine J, Howard BV. HDL cholesterol subfractions and risk of developing type 2 diabetes among Pima Indians. Diabetes Care. 1999;22:271–274. doi: 10.2337/diacare.22.2.271. [DOI] [PubMed] [Google Scholar]
- 29.Yamagata K. Roles of HNF1α and HNF4α in pancreatic β-cells: lessons from a monogenic form of diabetes (MODY) Vitam Horm. 2014;95:407–423. doi: 10.1016/B978-0-12-800174-5.00016-8. [DOI] [PubMed] [Google Scholar]
- 30.Bonnefond A, Froguel P. Rare and common genetic events in type 2 diabetes: what should biologists know? Cell Metab. 2015;2:357–368. doi: 10.1016/j.cmet.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 31.Jafar-Mohammadi B, Groves CJ, Gjesing AP, et al. A role for coding functional variants in HNF4A in type 2 diabetes susceptibility. Diabetologia. 2011;54:111–119. doi: 10.1007/s00125-010-1916-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sladek FM, Zhong WM, Lai E, Darnell JE., Jr Liver enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Maechler P, Antinozzi PA, Hagenfeldt KA, Wollheim CB. Hepatocyte nuclear factor 4alpha regulates the expression of pancreatic beta -cell genes implicated in glucose metabolism and nutrient-induced insulin secretion. J Biol Chem. 2000;275:35953–35959. doi: 10.1074/jbc.M006612200. [DOI] [PubMed] [Google Scholar]
- 34.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor4(nuclearreceptor2A1)is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393–1414. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamagata K, Furuta H, Oda N, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Q, Yamagata K, Miura A, et al. T130I mutation in HNF-4 gene is a loss-of-function mutation in hepatocytes and is associated with late-onset type 2 diabetes mellitus in Japanese subjects. Diabetologia. 2003;46:567–573. doi: 10.1007/s00125-003-1067-y. [DOI] [PubMed] [Google Scholar]
- 37.Kong A, Steinthorsdottir V, Masson G, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanson RL, Guo T, Muller YL, et al. Strong parent-of-origin effects in the association of KCNQ1 variants with type 2 diabetes in American Indians. Diabetes. 2013;62:2984–2991. doi: 10.2337/db12-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahajan A, Go MS, Zhang W, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.López-Ríos L, Nóvoa FJ, Chirino R, Varillas F, Boronat-Cortés M, Wägner AM. Interaction between cholesteryl ester transfer protein and hepatic lipase encoding genes and the risk of type 2 diabetes: results from the Telde study. PloS One. 2011;6:e27208. doi: 10.1371/journal.pone.0027208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandhofer A, Tatarczyk T, Laimer M, et al. The Taq1B-variant in the cholesteryl ester-transfer protein gene and the risk of metabolic syndrome. Obesity (Silver Spring) 2008;16:919–922. doi: 10.1038/oby.2007.130. [DOI] [PubMed] [Google Scholar]
- 43.Barter PJ, Rye KA, Tardif JC, et al. Effect of torcetrapib on glucose, insulin, and hemoglobin A1c in subjects in the Investigation of Lipid Level Management to Understand its Impact in Atherosclerotic Events (ILLUMINATE) trial. Circulation. 2011;124:555–562. doi: 10.1161/CIRCULATIONAHA.111.018259. [DOI] [PubMed] [Google Scholar]
- 44.Cannon CP, Shah S, Dansky HM, et al. Safety of anacetrapib in patients with or at high risk for coronary heart disease. N Engl J Med. 2010;363:2406–2415. doi: 10.1056/NEJMoa1009744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.