Abstract
CRISPR-Cas9-based genetic screens are a powerful new tool in biology. By simply altering the sequence of the single-guide RNA (sgRNA), Cas9 can be reprogrammed to target different sites in the genome with relative ease, but the on-target activity and off-target effects of individual sgRNAs can vary widely. Here, we use recently-devised sgRNA design rules to create human and mouse genome-wide libraries, perform positive and negative selection screens and observe that the use of these rules produced improved results. Additionally, we profile the off-target activity of thousands of sgRNAs and develop a metric to predict off-target sites. We incorporate these findings from large-scale, empirical data to improve our computational design rules and create optimized sgRNA libraries that maximize on-target activity and minimize off-target effects to enable more effective and efficient genetic screens and genome engineering.
INTRODUCTION
The Cas9 protein, an RNA-directed DNA endonuclease, is a powerful tool for manipulating the genome1-4. The ease of programming Cas9 has enabled CRISPR-based genetic screens5, identifying well-established genes and providing novel insight into gene function for multiple phenotypes6-8. Initial libraries were designed with little knowledge of sgRNA activity rules, a critical design parameter, as interpreting screening data requires consistency among multiple sgRNAs targeting the same gene to distinguish true hits from false positives. Inactive and non-specific sgRNAs reduce the effective gene coverage of the library and the accuracy of the hit list.
Many studies indicate that Cas9 off-target activity depends on both sgRNA sequence and experimental conditions10-14. These studies have provided qualitative but incomplete understanding of specificity determinants. Finding generalizable patterns is quite challenging, requiring large datasets to adequately sample the vast number of possible imperfect sgRNA:DNA interactions to reveal sequence features for prediction of off-target activity. Here, we present the design and characterization of human and mouse genome-wide sgRNA libraries based on our previously published rules for predicting on-target efficiency9. Building on screening data generated with the new libraries and large-scale assessment of off-target activity, we develop improved algorithms for on- and off-target activity prediction, allowing further optimization of our genome wide libraries.
RESULTS
Genetic screens with the Avana and Asiago libraries
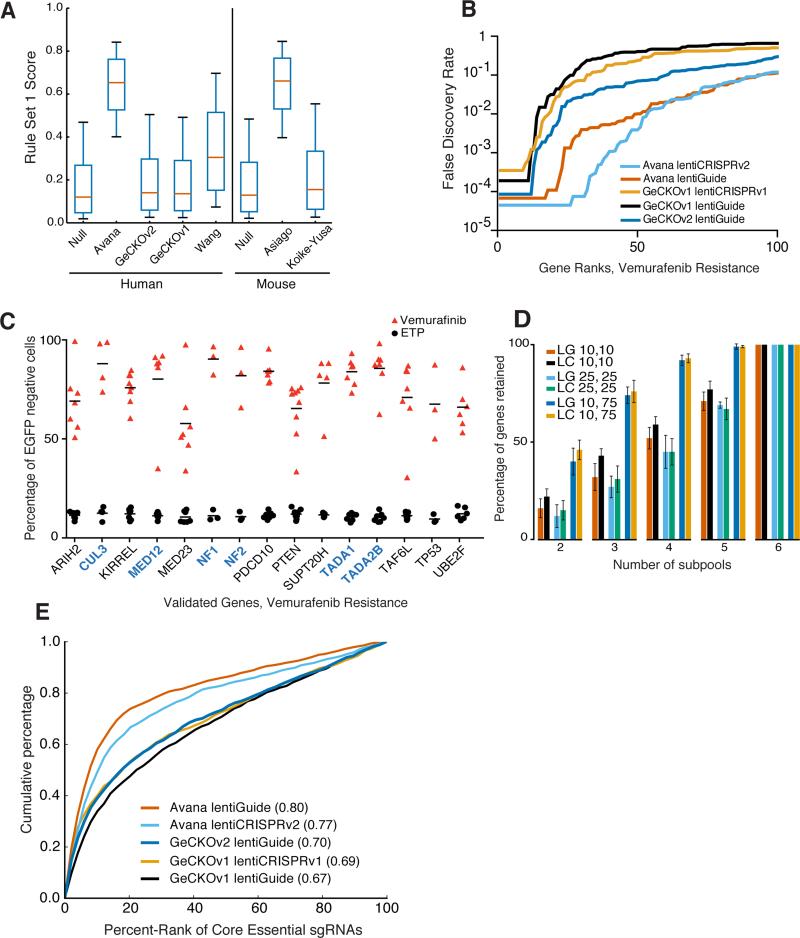
Previously, we examined the activity of 1,841 sgRNAs to determine sequence features leading to increased efficacy and developed rules for improved sgRNA design (Rule Set 1)9. We implemented these rules in human and mouse genome-wide libraries, named Avana and Asiago, respectively, and tested their performance in phenotypic screens. We selected six sgRNAs per gene according to three criteria: Rule Set 1 score, specificity within protein coding regions, and the target site location within the gene (Supplementary Tables 1, 2, 3; Methods). The distribution of Rule Set 1 scores for the previously-published GeCKO6,15 and Koike-Yusa et al.8 libraries resembles the null distribution, as these were not designed with on-target criteria (Fig. 1a). The Wang et al.7 library incorporated early rules for increasing sgRNA activity and has higher Rule Set 1 scores. The Avana and Asiago libraries are, by design, the most enriched for sgRNAs predicted to be active.
Figure 1.
Comparative performance of the Avana library. (a) Distribution of Rule Set 1 scores across libraries. The box represents the 25th, 50th, and 75th percentiles, whiskers show 5th and 95th percentiles. (b) Comparison of the FDR-corrected q-values determined by STARS for the top 100 ranked genes in the vemurafenib resistance assay in A375 cells. (c) Validation of individual sgRNAs for vemurafenib resistance in a competition assay in A375 cells. Horizontal bars represent the average of the individual sgRNAs for each gene. Previously-validated genes are labeled in blue. ETP = early time point. (d) Subsampling analysis of the Avana library. We first identified genes that passed at different FDR thresholds with STARS when all six subpools were analyzed (first number in legend), the average number of retained genes that score at different FDR thresholds following removal of subpools (second number in legend). LG = lentiGuide; LC = lentiCRISPRv2. (e) ROC-AUC analysis of individual sgRNAs targeting core essential genes in dropout screens in A375 cells. AUC values are indicated in parentheses.
We first tested our library using a well-established screening system, resistance to vemurafenib in A375 melanoma cells, which carry the BRAF V600E mutation and are sensitive to MAPK pathway inhibition (Supplementary Fig. 1, 2)6,16,17. We transduced each of the six Avana subpools, each containing 1 sgRNA per gene, in biological replicates at low multiplicity of infection and performed the same screen with the GeCKOv1 and GeCKOv2 libraries; for cells infected with the Avana library, we also applied the MEK-inhibitor selumetinib18,19, another small molecule previously examined in this resistance model (Supplementary Fig. 3, Methods). We ranked sgRNAs by their log2-fold-change relative to their abundance in the plasmid DNA pool and averaged ranks from the two replicates (Supplementary Table 4).
We first used the RIGER algorithm to analyze the GeCKOv1, GeCKOv2, and Avana screens (Supplementary Table 5, Supplementary Fig. 4). The weighted sum option with RIGER, however, incorporates information from only the top two perturbations targeting a gene, discarding additional evidence about specificity of the hit list provided by other sgRNAs. We thus developed STARS, an alternative gene-ranking system to generate false discovery rates (FDR)20, rewarding genes where a high fraction of sgRNAs score, similar to the MAGeCK algorithm21. Using STARS, we observed that six genes previously validated to confer vemurafenib resistance (CUL3, MED12, NF1, NF2, TADA1, TADA2B) all score with FDR < 1% (Supplementary Table 6).
We directly compared the performance of the three libraries in the lentiGuide vector. At FDR < 10%, 27 genes scored with GeCKOv1 whereas 60 genes scored with GeCKOv2 (Fig. 1b), an increase likely due to library size, as GeCKOv1 averages 3 - 4 sgRNAs per gene while GeCKOv2 contains 6 sgRNAs per gene. 92 genes scored at FDR < 10% with Avana, which also contains 6 sgRNAs per gene. Similarly, 94 genes scored with the Avana library screened in lentiCRISPRv2, and 36 genes scored at FDR < 10% across both vectors.
We examined the data for new modulators of vemurafenib resistance. With the Avana library, the tumor suppressors PTEN, TP53, and RB1 scored with FDR < 1%, while in the GeCKOv2 library, these genes score with FDR of 78%, 7%, and 2%, respectively (Supplementary Table 6). We thus annotated the hit lists for PanCancer genes, a highly curated and validated set containing many genes whose loss may restore the MAPK pathway or activate alternative survival pathways (Supplementary Table 7)22. We observed that the Avana library identified more PanCancer genes than either version of GeCKO. At FDR < 10%, we identified 4 PanCancer genes with GeCKOv1 (p = 1.1 × 10−5, hypergeometric distribution), 6 genes with GeCKOv2 (p = 2.2 × 10−7), and 10 with Avana (p = 2.9 × 10−11). In the selumetinib screen, Avana identified 20 genes (p = 4.6 × 10−24), including 9 of the 10 genes identified at the same threshold in the comparable vemurafenib dataset (Supplementary Tables 8, 9).
We used a competition assay to validate a selection of newly-identified hits. Cells carrying a single sgRNA were mixed with EGFP-labeled cells; if that sgRNA provided a selective growth advantage, then EGFP-negative cells would enrich over time, as measured by flow cytometry. We tested this assay with 9 positive control sgRNAs targeting validated hits (NF1, NF2, CUL3); 10 negative control sgRNAs with no known target; and 5 sgRNAs to a new hit, TP53. When grown without vemurafenib, the fraction of sgRNA-containing cells decreased slightly, on average 8.4% across all sgRNAs (Supplementary Figure 5). In the presence of vemurafenib, the populations with negative control sgRNAs died and could not be assessed by flow cytometry, whereas positive control and TP53 sgRNAs came to represent an average of 93% and 82% of the population, respectively (Supplementary Figure 5). In a similar competition assay for the 6 previously-validated genes and 9 newly-identified genes, the average representation of sgRNA-containing cells after addition of vemurafenib ranged from 80% to 90% and 58% to 84%, respectively, validating the reproducibility of all 9 selected hits identified by the Avana primary screen (Fig. 1c). Several of these newly-validated hits represent additional members of complexes previously identified23,24.
Although screening more sgRNAs per gene increases the power of a library to detect hits, larger libraries require larger-scale screening, reducing the number of conditions that could be screened and barring models with limited cell numbers. We performed subsampling analysis to determine how decreasing the number of sgRNAs affected the hit list for the vemurafenib screen. The Avana library screened in lentiGuide identified 92 hits at FDR < 10%. For combinations of four 1 sgRNA/gene subpools, on average 52% of genes were recovered at the same FDR (Fig. 1d). At a relaxed FDR threshold of < 75%, 92% of genes were recovered with 4 sgRNAs; similar rates were seen for the Avana library screened in lentiCRISPRv2 (93%) and for the selumetinib screen with both lentiGuide (93%) and lentiCRISPRv2 (93%) (Supplementary Fig. 6). This observation suggests a useful screening strategy, especially for arrayed screens or pooled models where scale-up is costly or prohibitive: perform a primary screen with a small number of sgRNAs at genome-scale, use a relatively relaxed cut-off for hit selection, then perform a secondary screen on hundreds of primary screen hits using additional sgRNAs per gene.
We next examined library performance for negative selection, depletion of sgRNAs targeting proliferation-essential genes. Unlike positive selection, where small fractions of cells can strongly enrich, dropout screens represent a more challenging screening modality, as large fractions of cells receiving perturbations must show the phenotype in order to score. We performed a viability screen using the Avana, GeCKOv1, and GeCKOv2 libraries in A375 cells (Supplementary Fig. 7, Supplementary Tables 10, 11) and analyzed sgRNAs targeting a curated set of 291 core essential genes (Supplementary Table 11)25. To enable comparison across libraries of different sizes, we performed ROC-AUC analysis of individual sgRNAs. The Avana library performed significantly better than either version of the GeCKO library, producing AUC values of 0.77 and 0.80, versus GeCKO AUC values of 0.67 to 0.70 (Fig. 1e). Using STARS, Avana identified 171 of the 291 genes (59%), while GeCKOv2 identified 76 (29%) at FDR < 10% (Supplementary Fig. 8, Supplementary Table 12). Similarly, using Gene Set Enrichment Analysis (GSEA)26, the Avana library identified essential cellular complexes and processes with greater statistical confidence than GeCKOv2 (Supplementary Fig. 9). Combining data from the Avana and GeCKO libraries followed by STARS analysis identified 1,545 and 1,952 essential genes in A375 cells at FDR < 10% and < 25%, respectively, a range consistent with a recent report (Supplementary Table 13)27.
We confirmed the improved negative-selection screen performance of the Avana library in HT29 cells, a colon cancer line (Supplementary Tables 10, 11, Supplementary Fig. 10). Here, we screened the first four subpools of the Avana library and observed a significant improvement in depletion of core essential genes, despite employing fewer sgRNAs: Avana with 4 sgRNAs identified 161 genes, whereas GeCKOv2 with 6 sgRNAs identified 92 genes (Supplementary Table 12).
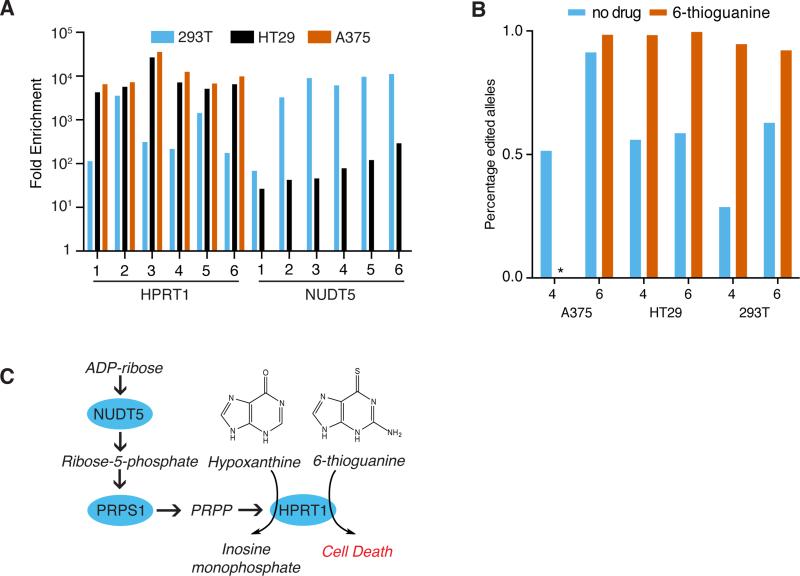
To further test the Avana library in different assays, we screened for resistance to the purine analog 6-thioguanine in three different cell lines. In A375 cells, single sgRNAs dominated each of the six subpools, and all six targeted HPRT1, a gene well-established in this assay (Fig. 2a, Supplementary Table 14)28. The HPRT1 sgRNA in each subpool was enriched at least 700-fold more than the second-ranked sgRNA, suggesting that none of the other ~110,000 sgRNAs in this library led to significant off-target activity at the HPRT1 locus. In HT29 and 293T cells, HPRT1 sgRNAs were highly-enriched, but targeting of NUDT5 also produced 6-thioguanine resistance (Fig. 2a). We validated these observations by individual infections in these 3 cell types with 2 sgRNAs each against HPRT1 and NUDT5 (Supplementary Fig. 11). To understand the differential enrichment of NUDT5 sgRNAs among cell lines, we examined genomic DNA by the TIDE technique29. Both sgRNAs led to modification of NUDT5 in all three cell lines in the absence of selection (Fig. 2b), suggesting that observed phenotypic differences between cell lines were not due to ineffective gene editing.
Figure 2.
HPRT1 and NUDT5 confer 6-thioguanine resistance. (a) For each of 6 sgRNAs targeting these genes, fold-enrichment for the indicated sgRNA after two weeks of selection with 6-thioguanine, relative to its starting abundance, assayed in three different cell lines. (b) TIDE analysis of indels for sgRNAs number 4 and 6 from (a) targeting NUDT5 tested in three cell lines. * indicates a sample where no cells survived and thus TIDE analysis could not be performed (c) Schematic of purine metabolism. Proteins are shown in blue circles, small molecules in italics. PRPS1 is also known as PRPP synthetase; PRPP is phosphoribosyl pyrophosphate.
6-thioguanine is a substrate for HPRT1 in the purine salvage pathway, substituting for hypoxanthine and thereby causing toxicity (Fig. 2c). NUDT5 plays an earlier role in purine synthesis, ultimately leading to production of 5-phosphoribosyl diphosphate (PRPP), another substrate for HPRT1-catalyzed production of inosine monophosphate30. Depletion of NUDT5 thus may prevent the toxic effects of 6-thioguanine incorporation (Fig. 2c)31. Differences in phenotypic severity between A375 and HT29 cells might relate to the fact that A375 cells have lost one allele of the NUDT5 gene32 and may have upregulated alternative means of producing PRPP.
Finally, we tested the Asiago mouse library in a model of interferon signaling in which BV2 cells were challenged with interferon gamma and STARS analysis of surviving cells revealed enrichment of well-established mediators of interferon signaling (Table 1, Supplementary Table 15). Nine of the ten most-enriched sgRNAs targeted Jak1, Stat1, Ifngr1, or Ifngr2, and all 4 sgRNAs for each of these genes scored in the top 25 of the 79,641 sgRNAs screened. Interestingly, query of the Molecular Signatures Database (MSigDB)33 with human homologs for the genes identified at FDR < 50% identified a significant enrichment for mitochondrial genes (FDR 8.99×10−12, hypergeometric test) not previously implicated in interferon signaling.
Table 1.
Screening results for mediators of interferon-gamma signaling using the mouse genome-wide Asiago library. Full results are provided in Supplementary Table 15.
| Gene Symbol | sgRNA Ranks | STARS Score | p-value | FDR |
|---|---|---|---|---|
| Jak1 | 4;5;10;13 | 15.15 | < 3.66×10−6 | < 2.61×10−4 |
| Ifngr2 | 7;11;12;15 | 14.90 | < 3.66×10−6 | < 2.61×10−4 |
| Stat1 | 1;2;9;16 | 14.79 | < 3.66×10−6 | < 2.61×10−4 |
| Ifngr1 | 3;6;14;22 | 14.23 | < 3.66×10−6 | < 2.61×10−4 |
| Jak2 | 18;20;55;178 | 10.60 | < 3.66×10−6 | < 2.61×10−4 |
| Ifnar1 | 131;145;273;1176 | 7.32 | 1.10×10−5 | 6.53×10−4 |
| Irf9 | 71;74;196;56950 | 7.23 | 1.10×10−5 | 5.60×10−4 |
| Ifnar2 | 37;137;348;8433 | 6.48 | 7.68×10−5 | 3.43×10−3 |
Rule Set 2 for sgRNA on-target activity
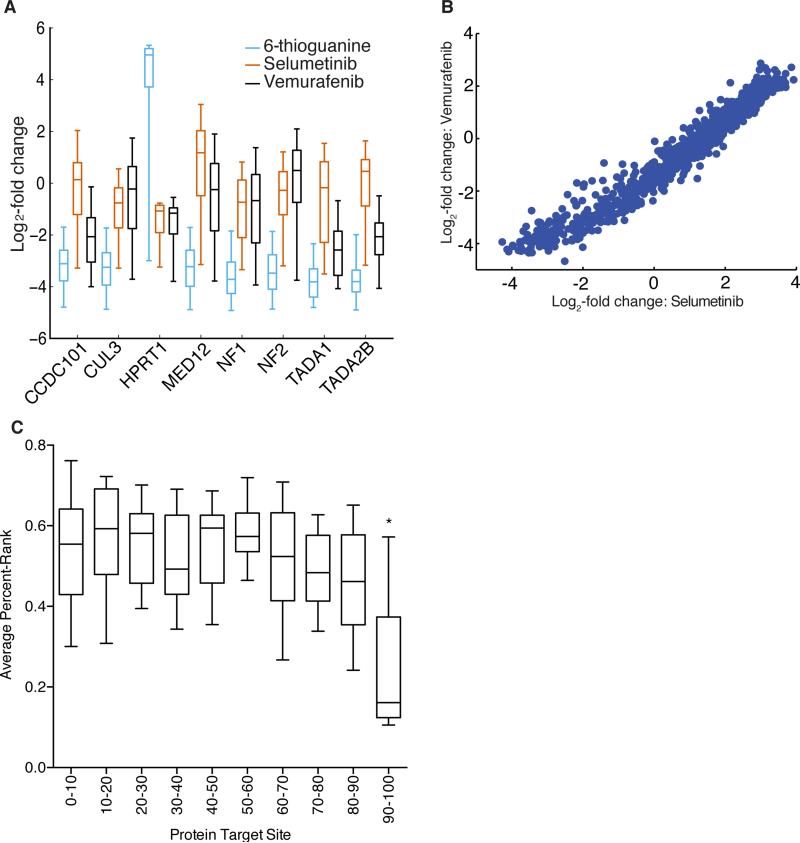
We originally modeled on-target activity with 1,841 sgRNAs targeting 9 human and mouse genes assayed by flow cytometry (FC dataset)9. To improve predictions, we designed a tiling library targeting all possible NGG PAM-containing sites in 15 genes known to confer resistance to vemurafenib, selumetinib, 6-thioguanine, or etoposide. In A375 cells, several genes reported as hits for resistance to etoposide (TOP2A, CDK6)7 and 6-thioguanine (MLH1, MSH2, MSH6, and PMS2)8 in other cell types failed to show significant enrichment under the conditions tested here. For small molecules that yielded resistant cells, we examined sgRNA enrichment (Fig. 3a, Supplementary Table 16). As expected, the activity of individual sgRNAs correlated well between vemurafenib and selumetinib (Fig. 3b). These 2,549 sgRNAs, targeting 8 genes, comprise the RES (resistance) dataset.
Figure 3.
Tiled library screen for resistance genes. (a) Performance of sgRNAs by gene for each of three small molecule challenges. The box represents the 25th, 50th, and 75th percentiles, whiskers show 5th and 95th percentiles, and outliers are shown as individual dots. (b) For sgRNAs targeting MED12, comparison of the log2-fold-change when challenged with vemurafenib and selumetinib. (c) Activity of sgRNAs as a function of target site within the protein, divided by deciles, for 17 proteins. The box represents the 25th, 50th, and 75th percentiles, whiskers show 10th and 90th percentiles. The final decile has a statistically-significant difference in activity (adjusted p-values < 0.02, one-way ANOVA with repeated measures, with Tukey's correction for multiple comparisons).
With combined data for over 4,000 sgRNAs targeting 17 genes, we examined the efficacy of each sgRNA versus its position in the protein coding region (Supplementary Fig. 12). Initial design guidelines recommended targeting close to the N'terminus. Alternatively, Vakoc and colleagues recently reported on improved generation of loss-of-function alleles when targeting specific domains of several well-characterized proteins34. For the 17 genes under consideration here, however, we did not observe discrete regions of the protein-coding sequence that were obviously more-productive target sites for gene inactivation (Supplementary Fig. 12). Overall, we observed that only the C-terminal 10% of the protein-coding region showed a statistically significant reduction in activity (Fig. 3c). An expanded target site window allows more flexibility in sgRNA selection, both to optimize for on-target efficacy and to minimize off-target potential.
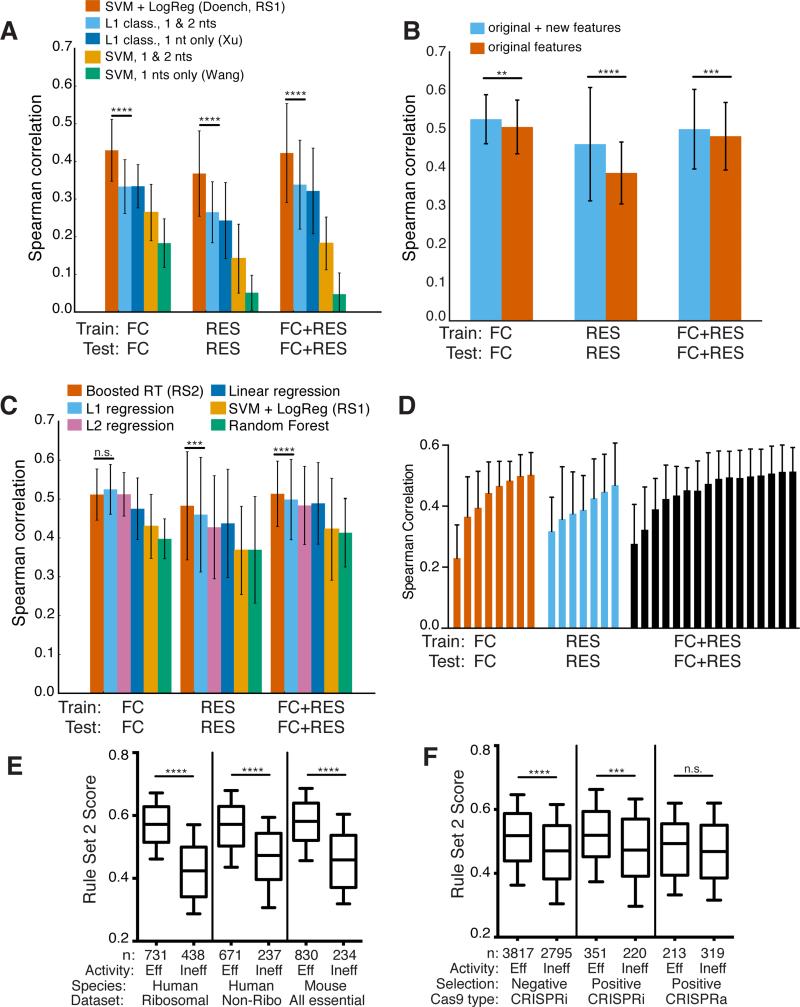
Previously, to generate Rule Set 1, we treated the top 20% of sgRNAs for each gene as highly active and trained on a 20% vs. 80% classification model to identify significant predictive features.9 We compared the performance of this model, which used support vector machine (SVM) with logistic regression, to other classification-based approaches applied to sgRNA activity predictions, SVM7,35 and L1 logistic regression36, evaluating performance with a leave-one-gene-out approach. Assessing the FC, RES, and combined datasets, we observed the best performance from SVM plus logistic regression (Fig. 4a, Supplementary Fig. 13). The SVM-alone or L1 logistic regression models augmented with dinucleotide features, previously found to be informative, performed worse than SVM plus logistic regression on these datasets (Fig. 4a).
Figure 4.
Development of Rule Set 2 for prediction of sgRNA on-target activity. (a) Comparison of classification models. Spearman correlation between measured activity and predicted activity score is plotted. Error bars show the standard deviation across genes with a leave-one-gene-out approach. SVM + LogReg (Rule Set 1), performs better than the next-best model for all three datasets (left to right p-values of 1.8×10−8, 5.2×10−13, and p < 10−16, using the statistical test for differences in Spearman correlation)48. (b) Addition of new features improves performance using L1 linear regression. Significance determined as in (a), with p-values of, left to right, 4.2×10−3, p < 10−16, 2.32×10−4. (c) Comparison of regression models, as well as the best-performing classification model, SVM + LogReg. Significance values are shown for the comparison between gradient-boosted regression trees (Boosted RT) and L1 regression, using the same measure of significance as in (a), p-values of, left to right, 0.054, 4.9×10−4, and 5.3×10−5. (d) Assessment of modeling performance with increasing number of genes used in each training set. Error bars indicate one standard deviation across genes with a leave-one-gene-out approach. (e) Rule Set 2 performance on independently-generated negative selection datasets. From left to right, p-values for the three comparisons are 5.9×10−80, 2.1×10−24, and 3.9×10−35 (two-sample Kolmogorov-Smirnov test). (f) Rule Set 2 performance on independently-generated CRISPRa/i datasets. From left to right, p-values for the three comparisons are 1.8×10−40, 1.1×10−4, and 0.14 (two-sample Kolmogorov-Smirnov test).
Discretizing sgRNA activity into 20% vs. 80% classes, although convenient, likely resulted in considerable information loss, and thus we explored regression models using normalized sgRNA ranks. We initially retained the same evaluation metric used in the derivation of Rule Set 1, AUC, and observed that L1-regularized linear regression (a continuous regression model) outperformed L1-regularized logistic regression (a discrete classification model) when trained on the same FC data with the same features; we observe a similar result with Spearman correlation, a more suitable metric for regression models (Supplementary Fig. 14). These results suggested further exploration of linear regression models.
Another important factor besides the modeling approach for predicting activity is the feature set. Previously, we used single and dinucleotide position-specific nucleotides, and the GC count of the sgRNA9. We hypothesized that additional features, such as position-independent nucleotide counts and the location of the sgRNA target site within the gene, could improve predictions. Biochemical and structural studies indicate that the sgRNA participates in step-wise association with DNA, suggesting that localized thermodynamic properties may also be useful37,38. These additional features improved the L1 regression model for all datasets (Fig. 4b). Microhomology features, suggested to improve sgRNA activity39, were predictive on their own but did not improve performance when added to our final model.
Because regression models and additional features gave improved results, we investigated the performance of five different regression models (see Methods). On the RES data and the combined dataset, the gradient-boosted regression trees model exhibited better Spearman correlation than all others, while for the FC dataset, it performed comparably to L1- and L2-regression models (Fig. 4c). We conclude that the gradient-boosted regression trees model was the best among those compared here. In this model, dinucleotide and single nucleotide identities at each position of the sgRNA continued to contribute most to activity predictions, representing a total of 58% of the Gini importance, a measure of the weight each feature contributes to the overall model (Supplementary Fig. 15, Supplementary Table 17). New features contributed substantially: position-independent counts of single and dinucleotides, location of the sgRNA within the protein, and melting temperatures of different regions gave Gini importance values of 16%, 13%, and 11%, respectively. To gauge if the amount of data had saturated the model, we systematically added genes to the training set, assessed performance, and observed a plateau in performance with the combined dataset (Fig. 4d). Furthermore, when trained and tested on all pairwise combinations of datasets, the model trained with the combined dataset produced the best results across all test sets (Supplementary Fig. 16). We refer to the gradient-bossed regression trees model with the augmented feature set trained on the combined dataset as Rule Set 2.
For further validation of Rule Set 2, we assessed performance on independent negative selection datasets not used in the construction or evaluation of the final model, from two human cell lines7 and mouse embryonic stem cells8, curated by Xu and colleagues7,8,36. Rule Set 1 scores clearly distinguished sgRNAs classified as effective vs. ineffective in all three datasets, with p-values of 1.4×10−32, 1.8×10−16, and 1.1×10−11 (two-sample Kolmogorov-Smirnov test, Supplementary Fig. 17). With Rule Set 2 we observed even more significant distinctions between effective and ineffective sgRNAs, with respective p-values of 5.9×10−80, 2.1×10−24, and 3.9×10−35 (Fig. 4e), illustrating the generalizability of our new modeling approach.
We next examined curated datasets from screens that used CRISPRa/i technology36,40. In the largest CRISPRi dataset, comprising over 5,000 sgRNAs in a negative selection screen, Rule Set 2 exhibited the greatest difference in the distributions of scores for sgRNAs classified as effective vs. ineffective (p-value = 1.8×10−40, two-sample Kolmogorov-Smirnov test, Fig. 4f). A smaller positive selection screen with 571 sgRNAs gave a p-value of 1.1×10−4. In the smallest dataset, 532 sgRNAs from a positive selection CRISPRa screen, Rule Set 2 again favored effective sgRNAs but with a higher p-value (0.14). Together, these observations suggest that commonalities between CRISPR knockout and CRISPRa/i, such as the interactions between the sgRNA and Cas9, and the sgRNA and DNA, represent meaningful components of the predictive power of Rule Set 2.
CFD score to predict sgRNA off-target effects
The off-target effects of sgRNAs have been extensively investigated across many experimental systems and conclusions regarding the extent of off-target activity have varied widely.5 We sought to understand off-target effects under typical pooled-screening conditions in mammalian cells using a library targeting the coding sequence of human CD33 with all possible sgRNAs, regardless of PAM. For all sites with the canonical NGG PAM, in addition to the perfect-match sgRNAs, we introduced three types of sgRNA mutations: first, all 1 nucleotide deletions; second, all 1 nucleotide insertions; third, all 1 nucleotide mismatches to the target DNA, generating a library with 27,897 unique sgRNAs. To these we added 10,618 sgRNAs targeting the mouse Thy1 locus to serve as negative controls. We infected MOLM13 cells and performed flow cytometry to isolate CD33-negative cells (Supplementary Fig. 18, Supplementary Table 18).
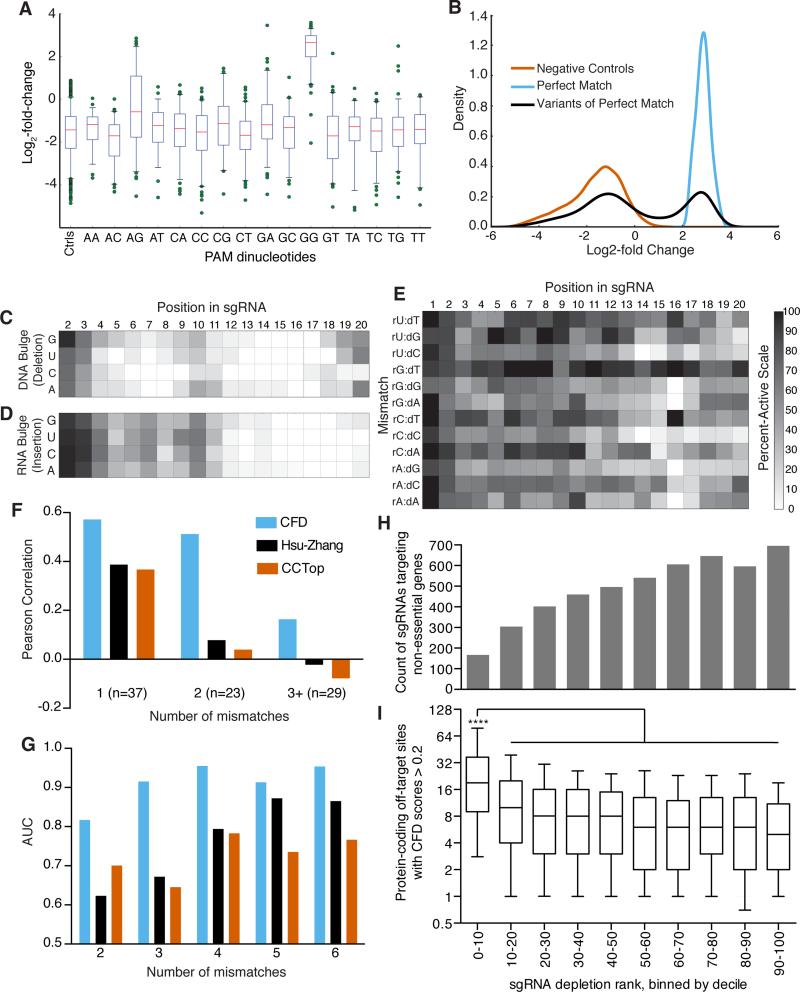
We examined the activity profile of perfect match sgRNAs with an NGG PAM to identify CD33 target sites that led to robust loss-of-function, and subsequent analyses only examined sgRNAs in this region (Supplementary Fig. 19). Using negative control sgRNAs to define inactivity, we compared the activities of sgRNAs with different PAM sequences. As expected, sgRNAs with an NGG PAM had the highest proportion of active sgRNAs, 91% (Fig. 5a). Some alternative PAM sequences led to notable but smaller rates of sgRNA activity: NAG (26%), NCG (11%), and NGA (7%). While alternative PAMs should be avoided to maximize on-target activity, they must be considered as potential off-target sites14.
Figure 5.
CFD score for assessing off-target activity of sgRNAs. (a) Activity of sgRNAs as a function of the final two nucleotides of the PAM. The box represents the 25th, 50th, and 75th percentiles, whiskers show 5th and 95th percentiles, and outliers are shown as individual dots. (b) Distribution of log2-fold change values for three classifications of sgRNAs assessed by flow cytometry for activity against CD33. (c) Heat-map of the percent-active values for all sgRNA:DNA interactions where one nucleotide was removed from the sgRNA, creating a bulged DNA base. (d) Same as in (c) but with an insertion of nucleotide in the sgRNA to create a bulged RNA base. (e) Same as in (c) and (d) but with symmetric mismatches. Grayscale is the same for panels c – e. (f) Comparison of the correlation of 3 off-target scoring metrics to measured off-target activity of 89 sgRNAs with mismatches to the cell surface receptor H2-K (g) AUC values for GUIDE-Seq reads as a function of number of mismatches assessed by three scoring metrics; same color scheme as in (f). (h) Distribution of sgRNAs targeting non-essential genes in a dropout screen in A375 cells. All 109,463 sgRNAs in the Avana library screened in A375 cells were ranked by their depletion, binned by decile, and the count of 4,950 sgRNAs targeting the set of non-essential genes in each bin is plotted. (i) For the sgRNAs targeting non-essential genes plotted in (h), the distribution of the number of off-target sites in protein-coding regions with CFD scores > 0.2. The box represents the 25th, 50th, and 75th percentiles, whiskers show 10th and 90th percentiles. The first bin, with the most-depleted sgRNAs, is statistically significant compared to all other bins, Kruskal-Wallis test, p < 10−4. The x-axis is the same for panels (h) and (i).
To examine sgRNAs with mismatches, deletions, or insertions to their target DNA, we identified 65 perfect-match sgRNAs that produced > 4.8-fold enrichment for CD33 knockout, two standard deviations above the mean of the negative control sgRNAs (Fig. 5b), and their 9,914 associated variant sgRNAs. The activity distribution of these variants was bimodal, providing a clear threshold to classify each as active or inactive. We then calculated the percent-activity for sgRNAs sharing the same nucleotide mutation at the same position, that is, the fraction of individual sgRNA:DNA interactions that led to activity, a calculation enabled by sampling from large numbers of sgRNAs (Supplementary Table 19, Supplementary Fig. 20). We also calculated the delta-log-fold-change (dLFC) to quantify the decrease in activity for each variant sgRNA relative to its perfect match sgRNA (Supplementary Table 19). The two metrics for assessment of mismatch variants, percent-activity and dLFC, were highly correlated (R = −0.97, Pearson correlation coefficient, Supplementary Fig. 21).
We next examined the changes in activity produced by the three different types of variants. Deletions in the RNA, which create a single bulged DNA base, seldom led to activity (Fig. 5c). Of the 76 nucleotide and position combinations (we did not assess nucleotide number 1), only 14 of these types of deletions retained high levels of cleavage using the dLFC metric (p-value > 10−4), 13 of which occurred at positions 2, 3, 19, and 20. Notably, at positions 19 and 20 we observed that deletion of cytidine in the RNA to create a bulged guanine in the DNA was poorly tolerated, with only 4% of sgRNAs showing activity, while deletion of other nucleotides led to an average of 44% of active sgRNAs. Likewise, an extra base in the sgRNA sequence rarely preserved activity; only positions 2 and 3 gave rise to high levels of activity at the same p-value threshold (Fig. 5d). Asymmetric sgRNA:DNA interactions that lead to bulged RNA or DNA bases have been examined for a small number of examples and our results are qualitatively consistent41.
Finally, we examined single-base mismatches between the sgRNA and DNA. Overall, both the position and identities of mismatched nucleotides played major roles in determining activity (Fig. 5e). For example, rG:dT mismatches were generally tolerated even in the PAM-proximal region: across all positions, 256 of 289 (89%) of these interactions were active. Conversely, only 37% of rC:dC interactions preserved sgRNA activity. Other nucleotide mismatches, however, had quite different effects at different positions. For example, 0 of 46 purine:purine mismatches at position 16 were active, compared to 30 of 31 mismatches with a thymidine in the DNA. By contrast, at position 19, 41% of purine:purine mismatches and 48% of mispaired thymidines led to active sgRNAs. These results suggest that each type of mismatch must be evaluated individually and that metrics to score off-target sites must incorporate both the position and identity of imperfectly matched RNA:DNA interactions.
Using these data, we propose the Cutting Frequency Determination (CFD) score to calculate the off-target potential of sgRNA:DNA interactions (see Methods). We tested the ability of the CFD score to predict off-target, imperfect-match activity for an independent dataset of 89 sgRNAs designed to target H2-D, some of which were previously shown to produce effective protein knockout of H2-K, a gene with highly similar sequence9. We compared the CFD score to two previously-described off-target scoring metrics, the CCtop42 and Hsu-Zhang14 scores, both of which consider the position and number, but not identity, of mismatches between the RNA and DNA. The CFD score correlated best with the measured activity of these 89 off-target sgRNAs (Supplementary Table 20). Dividing the dataset by number of mismatches, the CFD score performed best in each subset; with 2 or more mismatches, both the Hsu-Zhang and CCtop scores showed poor correlation to measured activity (Fig. 5f).
To further evaluate the CFD score, we examined results obtained with GUIDE-Seq, an experimental approach for detection of sgRNA target sites43. In those data, 9 sgRNA sequences produced a total of 402 off-target sites. We compared the number of GUIDE-Seq reads, a measure of sgRNA activity, to the calculated off-target scores and observed the best Pearson correlation with the CFD score (R = 0.40), followed by CCtop (0.31) and Hsu-Zhang (0.26). As GUIDE-Seq is an ostensibly unbiased method to detect off-target sites, it is noteworthy that only 11 (2.5%) of the observed sites received CFD scores less than 0.05 and were among the weakest, averaging 100-fold fewer reads than the perfect match sgRNAs.
We next examined the specificity of these off-target scoring metrics. Importantly, we observed that the alignment tool Bowtie244, used by the E-CRISP portal45, does not return all possible mismatched sites, especially as the number of mismatches increases (Supplementary Fig. 22). Thus, the poor performance of the E-CRISP and Zhang portals in identifying off-target sites documented by Joung and colleagues43 may be partly a failure to identify all high-homology candidate sites rather than the scoring of the sites once found. We therefore used Cas-OFFinder46, a comparatively slow but comprehensive tool, to identify all possible sites with 6 or fewer mismatches to the 9 sgRNAs examined by GUIDE-Seq and scored them with the various metrics. Since the number of potential off-target sites increases exponentially with the number of mismatches, we examined each separately (excluding sites with one mismatch, as there were no such sites with zero Guide-Seq reads, precluding a determination of sensitivity). We observed that the CFD score performed best, with AUC values ranging from 0.82 – 0.98, while Hsu-Zhang ranged from 0.61 to 0.87 and CCtop ranged from 0.64 to 0.77 (Fig. 5g). We conclude that the CFD score will enable avoidance of the majority of high-frequency off-target effects.
To determine the impact of off-target activity on screening results, we used a curated set of 927 non-essential genes25. The Avana library contains 4,950 sgRNAs targeting these genes, and we examined their behavior in the viability screen in A375 cells described above. As expected, proportionally far fewer of these sgRNAs showed evidence of strong depletion (Fig. 5h). Using the CFD score, there was a statistically-significant increase in the number of off-target sites predicted for the most-lethal of these sgRNAs (Fig. 5i). This observation suggests that sgRNAs with more predicted off-target sites are more frequently erroneously deplete in a negative selection screen, and thus avoidance of such promiscuous sgRNAs will lead to improved library performance.
Optimized sgRNA libraries: Brunello and Brie
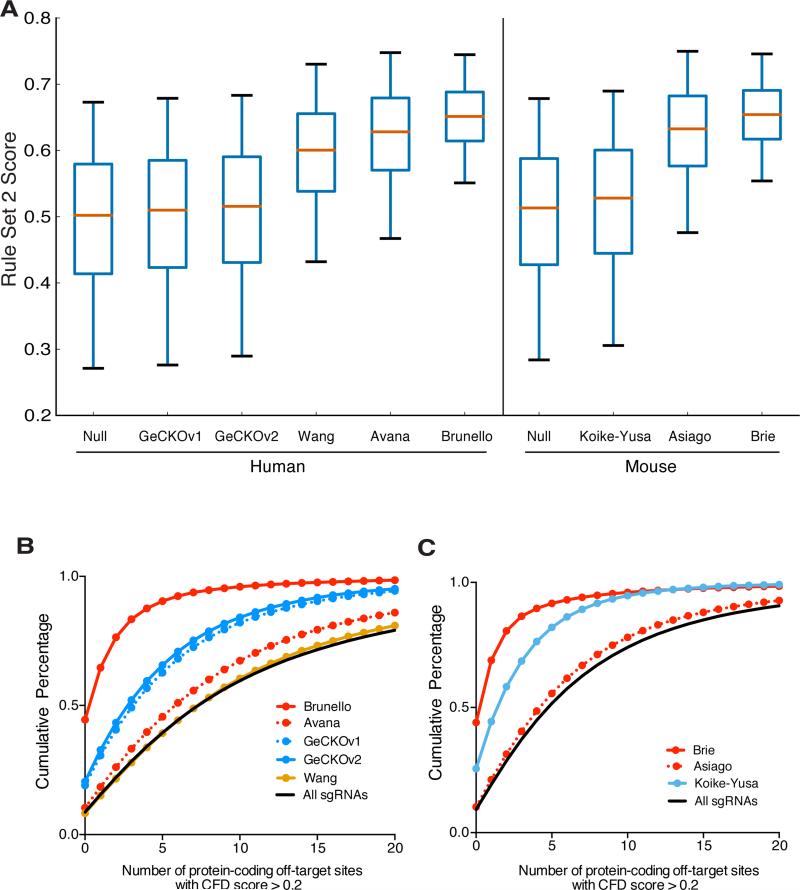
Finally, we incorporated our improved on-target activity predictions, Rule Set 2, with our off-target avoidance metric, the CFD score, to design fully optimized libraries for the human and mouse genomes, named Brunello and Brie, respectively (Supplementary Tables 21, 22; Methods). These new libraries are designed to maximize Rule Set 2 scores and minimize off-target sites with high CFD scores (Fig. 6a, b, c). Brunello and Brie thus likely represent a clear improvement over existing libraries and are available via Addgene. Our web portals incorporate Rule Set 2 and CFD models (http://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design and http://research.microsoft.com/en-us/projects/azimuth/) and provide code for incorporation of these scores into existing tools (Supplementary Information).
Figure 6.
On-target and off-target properties of the Brunello and Brie libraries. (a) Distribution of Rule Set 2 on-target activity scores across libraries. The box represents the 25th, 50th, and 75th percentiles, whiskers show 5th and 95th percentiles. (b) Cumulative distribution of the number of off-target sites with CFD scores > 0.2 in protein-coding regions across human libraries and (c) mouse libraries.
DISCUSSION
Using our previous rules for rational design of sgRNAs, Rule Set 1, we created human and mouse genome-wide libraries and performed genetic screens. In both positive and negative selection screens with the Avana library, expected hits (e.g. PanCancer genes22, core essentials25, and previously-validated targets6) scored as strong hits with multiple sgRNAs, and the screens produced a greater number of statistically-significant genes compared to existing libraries, allowing the identification and confirmation of new genes in these phenotypic assays. Predictions of sgRNA activity with Rule Set 1 were imperfect, however, leaving opportunity for further improvements in library design. By doubling the size of the sgRNA activity dataset and identifying a more effective modeling approach, we developed Rule Set 2, which shows demonstrably improved performance versus Rule Set 1 across multiple datasets from multiple laboratories. Notably, the ability of Rule Set 2 to predict performance extends to CRISPRa and CRISPRi screens. Thus, sgRNA designs based on Rule Set 2 (such as our Brunello and Brie libraries) should deliver substantial additional improvements in library performance.
Another major factor driving library performance is specificity. Joung and colleagues compared their experimentally-detected off-target sites to those found by two common off-target prediction algorithms and noted that “neither program identified the vast majority of off-target sites found by GUIDE-seq.”43 Whereas Rule Sets 1 and 2 represent data-driven, quantitative models of on-target sgRNA effectiveness, a quantitative analysis of off-target interactions based on similarly large-scale data has been lacking, and the ability to predict off-target effects therefore quite limited. Our analysis of 9,914 sgRNA variants that represent all single-base mismatches and indels for 65 DNA targets revealed that the pattern of activity effects is complex, and would not be fully apparent by examining smaller numbers of sgRNA:DNA interactions. We used these data to derive the CFD score to predict the likelihood of off-target cutting and showed that it outperformed other off-target scoring metrics. Together, these results provided direction for the creation of even more active and specific libraries for the human and mouse genomes.
The current work provides a resource for the design of improved sgRNA reagents for large-scale screens and small-scale gene editing experiments. As one of the biggest challenges for screens is identifying faithful models for the biology of interest, confirming the generalizability of a finding may require examination of multiple cellular models. Smaller libraries with a high fraction of active and specific sgRNAs, enabled by better on- and off-target activity predictions, will facilitate cost-effective screening across a range of model systems. Directly comparing the effectiveness of CRISPR knockout libraries to other gene-depletion approaches, such as state-of-the-art RNAi47 and CRISPRi40 libraries, will likely reveal different strengths, and thus complete assessment of model systems will benefit from screening with multiple modalities. Additionally, future work will determine if the results obtained using Streptococcus pyogenes Cas9 provide useful lessons regarding the activity of other Cas9 proteins. The experimental and analytical approaches described here illustrate a powerful method to uncover factors contributing to sgRNA activity and specificity and to optimize reagent design for large-scale functional genomics.
Online Methods
Avana and Asiago Libraries
To design these libraries, we targeted protein-coding transcripts annotated by the Consensus Coding Sequence Database (CCDS), totaling 18,675 genes for the human genome and 20,077 genes for the mouse genome. When a gene had more than one CCDS ID, we picked the shortest transcript per gene. We annotated NGG protospacer adjacent motifs (PAMs) on both the plus and minus strands and then selected sgRNAs for inclusion in the library on the basis of three criteria, and divided these criteria into tiers. A most-preferred sgRNA would fulfill the first tier of all three criteria. However, not all sgRNAs can have these properties, and thus to reach a quota of 6 sgRNAs per gene, step-wise relaxation of tiers across criteria was necessary, and the properties of each step-wise round of relaxation are given in Supplementary Table 1. Additionally, we excluded sgRNAs with a BsmBI site in their sequence or with a run of four or more thymidines. We selected up to 6 sgRNAs per gene, and this resulted in a human library (Avana) of 110,257 sgRNAs and a mouse library (Asiago) of 120,453 sgRNAs (Supplementary Tables 2, 3). The final distributions of sgRNAs across these criteria within each tier chosen for inclusion in the Avana and Asiago libraries are provided.
Criterion A: Location of the target site in the protein coding sequence, with the four tiers divided by quartiles of the target: (i) 0 - 25% of the protein coding region, (ii) 25 – 50%, (iii) 50 – 75%, (iv) 75 – 100%.
| Tier | Description | Avana | Asiago |
|---|---|---|---|
| i | 0%-25% | 57% | 57% |
| ii | 25%-50% | 43% | 43% |
| ii | 50%-75% | 0.06% | 0.04% |
| iv | 75%-100% | 0.02% | 0.01% |
Criterion B: To mitigate off-target effects, sequence uniqueness of various lengths, counting from the 3’ end of the sgRNA, e.g. of all possible sgRNAs targeting protein coding genes, the PAM-proximal (i) 13 nts are unique, (ii) 17 nts are unique, (iii) 20 nts are unique or (iv) the sgRNA sequence is not unique.
| Tier | Description | Avana | Asiago |
|---|---|---|---|
| i | Unique 13 nts | 84% | 83% |
| ii | Unique 17 nts | 13% | 13% |
| ii | Unique 20 nts | 0.2% | 0.4% |
| iv | not unique | 3% | 4% |
Criterion C: Rule Set 1 on-target score to maximize gene knockout efficacy, divided into deciles; e.g. (i) score of 0.9 – 1.0, (ii) score of 0.8 – 0.9, etc.
| Tier | Description | Avana | Asiago |
|---|---|---|---|
| i | 0.9-1.0 | 3% | 3% |
| ii | 0.8-0.9 | 14% | 15% |
| iii | 0.7-0.8 | 22% | 23% |
| iv | 0.6-0.7 | 23% | 22% |
| v | 0.5-0.6 | 17% | 16% |
| vi | 0.4-0.5 | 10% | 10% |
| vii | 0.3-0.4 | 6% | 6% |
| viii | 0.2-0.3 | 3% | 3% |
| ix | 0-0.2 | 1% | 1% |
Library Creation
Oligonucleotides were synthesized at the Broad Technology Laboratory (BTL) on a B3 Synthesizer (CustomArray). To each sgRNA sequence, BsmBI recognition sites were appended along with the appropriate overhang sequences (underlined) for cloning into sgRNA expression plasmids. Additional primer sites were appended to allow differential amplification of subsets from the same synthesis pool. The final oligonucleotide sequence was thus: 5’-[Forward Primer]CGTCTCACACCG[sgRNA, 20 nt]GTTTCGAGACG[Reverse Primer].
Unique primer sets were used to amplify individual subpools using 25 μL 2x NEBnext PCR master mix (New England Biolabs), 2 μL of oligonucleotide pool (~40 ng), 5 μL of primer mix at a final concentration of 0.5 μM, and 18 μL water. PCR cycling conditions: 30 seconds at 98°C, 30 seconds at 53°C, 30 seconds at 72°C, for 24 cycles.
| Primer Set | Forward Primer, 5’ – 3’ | Reverse Primer, 5’ – 3’ |
|---|---|---|
| 1 | AGGCACTTGCTCGTACGACG | ATGTGGGCCCGGCACCTTAA |
| 2 | GTGTAACCCGTAGGGCACCT | GTCGAGAGCAGTCCTTCGAC |
| 3 | CAGCGCCAATGGGCTTTCGA | AGCCGCTTAAGAGCCTGTCG |
| 4 | CTACAGGTACCGGTCCTGAG | GTACCTAGCGTGACGATCCG |
| 5 | CATGTTGCCCTGAGGCACAG | CCGTTAGGTCCCGAAAGGCT |
| 6 | GGTCGTCGCATCACAATGCG | TCTCGAGCGCCAATGTGACG |
The resulting amplicons were PCR-purified (Qiagen), digested with Esp3I (Fisher Scientific) and cloned into either lentiGuide (pXPR_003, Addgene 52963) or lentiCRISPRv2 (pXPR_023, Addgene 52961). The ligation product was isopropanol precipitated and electroporated into Stbl4 electrocompetent cells (Life Technologies) and grown at 30°C for 16 hours on agar with 100 μg/mL carbenicillin. Colonies were scraped and plasmid DNA (pDNA) was prepared (HiSpeed Plasmid Maxi, Qiagen). To confirm library representation and distribution, the pDNA was sequenced by Illumina. After mapping of Illumina reads (see below) we calculated the overall fraction of reads that contained intended sgRNAs, which serves as a surrogate for the quality of the oligonucleotide synthesis. By this cloning scheme, only 21 nts of the synthesized oligonucleotide, the prepended G and the 20 nt variable sequence, become incorporated in the final library, in contrast to ligation-independent cloning schemes (e.g. Gibson) in which both the sgRNA and flanking sequences are derived from synthesis. We deem a library to have passed quality control if > 85% of the sequencing reads map to an intended sgRNA, which corresponds to an oligonucleotide synthesis error rate of 0.75% per base or lower (85% = 211-0.0075). A distribution of sgRNA abundance for the subpools, as well as GeCKOv2 for comparison, is given in Supplementary Figure 2.
Cell Culture
Stock cell lines were maintained without added antibiotics and supplemented with 1% penicillin/streptomycin during screens. A375, HT29, and MOLM13 cells were obtained from the Cancer Cell Line Encyclopedia32. 293T cells were obtained from ATCC (CRL-3216). BV2 cells are a mouse microglial cell line that has been described49,50. All cells tested negative for mycoplasma contamination. Cas9 derivatives were made by introducing the lentivector pLX_311-Cas9, which expresses blasticidin resistance from the SV40 promoter and Cas9 from the EF1a promoter, as described9.
| Cell Line | Media | Puromycin | Blasticidin | Polybrene |
|---|---|---|---|---|
| A375 | RPMI + 10% FBS | 1 μg/mL | 5 μg/mL | 1 μg/mL |
| 293T | DMEM + 10% FBS | 1 μg/mL | 5 μg/mL | 1 μg/mL |
| HT29 | DMEM + 10% FBS | 1 μg/mL | 5 μg/mL | 1 μg/mL |
| MOLM13 | RPMI + 10% FBS | 2 μg/mL | 5 μg/mL | 4 μg/mL |
| BV2 | DMEM + 10% FBS + 1% HEPES | 2.5 μg/mL | 4 μg/mL | --- |
Virus Production
293T cells were plated at a density of 1.5e6 cells per well (2 mL volume) 24 hours pre-transfection in a 6-well dish. Transfection was performed using TransIT-LT1 Transfection Reagent (Mirus) according to the manufacturer's protocol. Briefly, two solutions were prepared for each well. One solution contained 8.25 μL of LT1 diluted in 66.75 μL of Opti-Mem (Corning) and incubated at room temperature for 5 minutes. The second solution contained 250 ng pCMV-VSVG (Addgene 8454), 1250 ng psPAX2 (Addgene 12260), and 1250 ng transfer vector (e.g. lentiGuide) in a final volume of 75 μL with Opti-MEM. The two solutions were combined and incubated at room temperature for 20 - 30 minutes. During this incubation period, the media on the 293T cells was changed. The transfection mixture was added dropwise to the cells, and then plates were centrifuged at room temperature at 1000 × g for 30 minutes and returned to 37°C. 6 – 8 hours post-centrifugation, transfection media was removed, leaving ~0.5 mL in each well, and replaced with 5.5 mL viral harvest media (DMEM + 10% FBS + 1% BSA).
Virus was harvested 24 hours post-transfection, the media was replenished, and a second harvest occurred at 48 hours post-transfection.
Determination of Infection Conditions for Pooled Screens
Optimal infection conditions were determined for each batch of virus prep in each cell line in order to achieve 30 - 50% infection efficiency, corresponding to a multiplicity of infection (MOI) of ~ 0.5 – 1. Infections were performed in 12-well plate format with 3.0e6 cells per well for adherent lines and 2.5e6 cells per well for suspension lines. Optimal conditions were determined by infecting cells with different virus volumes (0, 50, 100, 300, and 500 μL for lentiGuide virus; 0, 100, 200, 400, and 600 μL for lentiCRISPRv2 virus) followed by trypsinization and replating equal numbers of cells per each virus volume into 2 wells of a 6-well plate, each with complete medium, one supplemented with the appropriate concentration of puromycin. Cells were counted 3 - 5 days post selection to determine the infection efficiency, comparing survival with and without puromycin selection. Volumes of virus that yielded ~30 – 50% infection efficiency were used for screening.
Screening
Screening-scale infections were performed with the pre-determined volume of virus in the same 12-well format as the viral titration described above, and pooled 4 - 6 hours post-centrifugation. Based on library size, infections were performed with enough cells to achieve a representation of ~400 cells per sgRNA in the library following puromycin selection, unless otherwise noted. Cells were selected with puromycin for 5-7 days following infection to remove uninfected cells. A flowchart demonstrates the experimental timeline (Supplementary Fig. 3).
Vemurafenib and selumetinib resistance screens
Cas9-containing cells were infected in two biological replicates with the Avana and GeCKO libraries in lentiGuide; unmodified, parental cells were infected in two biological replicates with the Avana library in lentiCRISPRv2. Small molecules were added to puromycin-selected cells 7 days post-infection. Cells either received a media change or were passaged every two or three days over the course of the screen in complete media supplemented with 1% penicillin/streptomycin. Vemurafenib (PLX-4032, Selleckchem, S1267) was screened at a final concentration of 2 μM. Selumetinib (AZD-6244, Selleckchem, S1008) was screened at a final concentration of 200 nM. Surviving cells were harvested after 14 days of small molecule treatment. For analysis, the log2-fold-change of each sgRNA was determined relative to the starting plasmid DNA (pDNA) pool for each biological replicate, which we have previously validated as representative of an early time point in pooled screens6.
6-thiogunaine resistance screens, Avana library
Cas9-containing cells were infected in two biological replicates with the Avana library in lentiGuide. 6-thioguanine (2 μg/mL final concentration, Sigma A4660) was added to puromycin-selected cells 7 days post-infection. Cells either received a media change or were passaged every two or three days over the course of the screen in complete media supplemented with 1% penicillin/streptomycin. Surviving cells were harvested after 14 days of small molecule treatment. For analysis, the log2-fold-change of each sgRNA was determined relative to the starting plasmid DNA (pDNA) pool for each biological replicate.
Interferon-gamma resistance screen, Asiago library
BV2-Cas9 cells were infected in a single biological replicate with four different subpools of the Asiago library in lentiGuide. Cells were challenged with interferon gamma (R&D systems, 485-MI-100) at a final concentration of 10 units/mL eight days post-infection. Two days after interferon gamma treatment, surviving cells were harvested. For analysis, the log2-fold-change of each sgRNA was determined relative to mock-challenged cells.
Resistance screens, tiling library for further on-target activity modeling
A375-Cas9 cells were infected in four biological replicates. Small molecules were added to puromycin-selected cells 7 days post-infection. Cells either received a media change or were passaged every two or three days over the course of the screen in complete media supplemented with 1% penicillin/streptomycin. Vemurafenib (PLX-4032, Selleckchem, S1267) was screened at a final concentration of 2 μM. Selumetinib (AZD-6244, Selleckchem, S1008) was screened at a final concentration of 200 nM. 6-thioguanine (Sigma A4660) was screened at a final concentration of 2 μg/mL. Etoposide (Sigma E1383) was screened at a final concentration of 1 μg/mL. Surviving cells were harvested after 14 days of small molecule treatment. For analysis, the log2-fold-change of each sgRNA was determined relative to control cells treated with DMSO.
Off-target library screen
MOLM13-Cas9 cells were infected in one biological replicate at 1000 cells per sgRNA, and were selected with puromycin in complete media supplemented with 1% penicillin/streptomycin for the length of the screen. 21 days post-infection, cells were co-stained with PE conjugated anti-CD33 (Miltenyi Biotec #130-091-732) and DAPI, and then sorted on a BD-FACS Aria II. Gates were set on the BD FACSDiVa software using a CD-33 DAPI positive control and a DAPI-only negative control. The cell population was first gated to exclude debris; two additional gates were applied to select singlets and a fourth to select viable cells via DAPI. Finally, the population was gated in order to sort for PE negative cells (Supplementary Figure 18). For analysis, the log2-fold-change of each sgRNA was determined relative to the pDNA.
Dropout screens, Avana and GeCKOv2 libraries
Cas9-containing cells were infected in two or three biological replicates per cell line per library in lentiGuide, and selected with puromycin. Cells were passaged every two or three days for three weeks in complete media supplemented with 1% penicillin/streptomycin. For analysis, the log2-fold-change of each sgRNA was determined relative to the starting plasmid DNA (pDNA) pool for each biological replicate.
Screen analysis
For analysis, the log2-fold-change of each sgRNA was determined relative to the starting plasmid DNA (pDNA) pool for each biological replicate unless otherwise stated. The pDNA has been shown to serve as a good surrogate for an early time point, and is more cost effective when performing many screens6. We then determined the percent-rank of each sgRNA: each sgRNA was ranked by log2-fold change, and this number was then divided by the total number of sgRNAs in the pool to determine a percent-rank. These percent-rank values were then averaged across biological replicates. For performing RIGER and STARS analysis, the percent-rank values across subpools were merged. In the small number of cases where an sgRNA sequence was present in more than one subpool, the average of the percent-rank values was used. An exception to the above analysis method is subpool 5 in the selumetinib screen, where one replicate each of lentiGuide and lentiCRISPRv2 was not completed, and thus a single biological replicate was used for analysis. For of the GeCKOv1-lentiCRISPRv1 library, we re-analzed the previously-published data and normalized against the pDNA pool rather than the DMSO treatment, to be consistent with the analysis of the Avana and GeCKOv2 data6.
Validation of Hits
Validation assays were performed in an arrayed format with infection conditions of 1e6 cells per well of a 24 well-plate. Cells were selected with puromycin for 5-7 days following infection to remove uninfected cells, and were treated according to the methods of their respective resistance screen. Sequences of sgRNAs and primers are included in Supplementary Table 23.
Vemurafenib resistance assay
A375-Cas9 cells were infected with sgRNAs cloned into lentiGuide and selected with puromycin for one week. Cells were mixed ~1:1 (Supplementary Figure 5) or 1:9 (Fig. 1c) with A375-Cas9 cells expressing EGFP but with no sgRNA, and vemurafenib was added to the cell mixture. Flow cytometry was performed in 96 well-plate format on a BDAccuri C6 Sampler system to measure the EGFP negative fraction of samples at the day of initial mixing of cells and then again 10 – 12 days after addition of vemurafenib. Gates were set on the cytometer software using EGFP positive and negative controls; the cell populations were first gated by forward and side scatter to exclude debris, and a second gate was applied to measure the EGFP fluorescence.
6-thiogunaine resistance assay
Cas9-containing cells were infected in two biological replicates with lentiGuide constructs, 2 targeting HPRT, 2 targeting NUDT5, and one control sgRNA targeting EGFP. Cells were infected in arrayed format in a 24 well plate with 1e6 cells per well. Cells were divided into small molecule and no-selection conditions, cell counts were taken at each passage to determine population doublings, and pellets were collected at day 0 and day 14 of 6-thioguanine treatment for analysis of indels using the TIDE web portal29.
Genomic DNA Preparation and Sequencing
Genomic DNA (gDNA) was isolated using Maxi (3e7 - 1e8 cells), Midi (5e6 - 3e7 cells), and Mini (<5e6 cells) kits according to the manufacturer's protocol (Qiagen).
Illumina sequencing
PCR of gDNA and pDNA was performed to attach sequencing adaptors and barcode samples, which were divided into multiple 100 μl reactions (total volume) containing a maximum of 10 μg gDNA. Per 96 well plate, a master mix consisted of 75 μL ExTaq DNA Polymerase (Clontech), 1000 μL of 10x Ex Taq buffer, 800 μL of dNTP provided with the enzyme, 50 μL of P5 stagger primer mix (stock at 100 μM concentration), and 2075 μL water. Each well consisted of 50 μL gDNA plus water, 40 μL PCR master mix, and 10 μL of a uniquely barcoded P7 primer (stock at 5 μM concentration). PCR cycling conditions: an initial 1 minute at 95°C; followed by 30 seconds at 94°C, 30 seconds at 52.5°C, 30 seconds at 72°C, for 28 cycles; and a final 10 minute extension at 72°C. P5/P7 primers were synthesized at Integrated DNA Technologies (IDT). Samples were purified with Agencourt AMPure XP SPRI beads according to manufacturer's instructions (Beckman Coulter, A63880). Samples were sequenced on a HiSeq2000 (Illumina). Reads were counted by first searching for the CACCG sequence in the primary read file that appears in the vector 5’ to all sgRNA inserts. The next 20 nts are the sgRNA insert, which was then mapped to a reference file of all possible sgRNAs present in the library. The read was then assigned to a condition (e.g. a well on the PCR plate) on the basis of the 8nt barcode included in the P7 primer. The resulting matrix of read counts was first normalized to a reads per million within each condition by the following formula: read per sgRNA / total reads per condition × 106. Reads per million was then log2-transformed by first adding one to all values, which is necessary in order to take the log of sgRNAs with zero reads.
STARS
STARS is a gene-ranking algorithm for genetic perturbation screens. This algorithm takes a ranked list of perturbations as input. The algorithm computes a score for genes using the probability mass function of a binomial distribution:
where n is the total number of perturbations targeting a gene, k is the within-gene-rank of the perturbation, and p is the ratio of the rank of the kth perturbation over the total number of perturbations in the experiment. This calculation is performed for all sgRNAs that rank above a user-defined threshold, e.g. the top x% of sgRNAs from a ranked list. The value of the least probable perturbation for each gene is then assigned to the gene as the STARS Score. To avoid single sgRNA hits, we require that at least two perturbations rank above the user-defined threshold for a gene to receive a STARS Score. Permutation testing is also performed on the list of perturbations used in the experiment to generate the null distribution, allowing the calculation of p-values, FDR, and q-value (corrected FDR) for hit genes. STARS is written in Python, and is available on our website: http://www.broadinstitute.org/rnai/public/software/index
Development of Rule Set 2
Data processing for on-target prediction
For each sgRNA targeting a given gene, the log2-fold-change (LFC) in abundance, as judged from read-counts was computed. Next, normalized ranks were obtained for sgRNAs within each gene by assigning ranks to each guide, and then re-scaling them to lie between 0 and 1. If more than one cell type was available, these normalized ranks were averaged across cell types. Thus, a final sgRNA score for a given sgRNA and gene was in [0,1], where 1 was indicative of successful knockout. Note that normalized ranks were used instead of raw LFC because the LFC has a different maximum for genes that have more sgRNAs, suggesting that the LFC across different genes would not be comparable.
Predictive Models
We used the following statistical models in our experiments: (1) linear regression, (2) L1-regularized linear regression, (3) L2-regularized linear regression, (4) the hybrid SVM plus logistic regression approach used previously,9 (5) Random Forest, (6) Gradient-boosted regression tree, (7) L1 logistic regression (a classifier), (8) SVM Classification. Implementations for each of these used scikit-learn package in python. For (2) and (3), we set the regularization parameter range to search over100 points evenly spaced in log space, with a minimum of 10−6 and a maximum of 1.5 × 105. Random Forest used the default setting, as did the Gradient-boosted regression trees (learning rate of 0.1, and 100 base estimators each with a maximum depth of three). For the SVM, we used a linear kernel with default L2 regularization unless otherwise noted.
Featurization
A “one-hot” encoding of the nucleotide sequences refers to taking a single categorical variable and converting it to more variables each of which can take on the value 0 or 1, with at most, one of them being “hot,” or on. For example, position 1 of the 30-mer target site plus context can take on A/C/T/G, and it gets converted to four binary variables, one for each possible nucleotide. These are “order 1” features. For “order 2” features, we looked at all adjacent pairwise nucleotides as features, such as AA/AT/AG/etc. There are 4 × 4 = 16 such pairs, thus a single variable representing one such pair gets one-hot encoded into 16 binary variables. Previously, only “position-specific” nucleotide features were used in this manner, meaning that for each position on the sgRNA, a different one-hot-encoded feature was used. Here, however, taking inspiration from some of the string kernel literature, we augmented these features sets by also including “position-independent” features, where, for order 1 features for example, this would mean a feature for how many A's and how many T's, etc. were in the sgRNA, ignoring their position, and similarly for order 2.51 Therefore, for a 30mer (20mer sgRNA plus context), we obtained 80 order 1 and 320 order 2 position-specific features, and 4 order 1 and 16 order 2 position-independent features.
GC counts features were computed as before, that is, the number of Gs and Cs in the 20mer were counted, yielding one feature, and then another feature with count>10 was also used. The two nucleotides in the N and N positions relative to the PAM “NGGN” were one-hot encoded yielding 16 features, one for each NN possibility (e.g., AT).
Thermodynamic features were computed from the melting temperatures of the DNA version of the RNA guide sequence, or portions thereof, using the Biopython Tm_staluc function. 52,53 In addition to using the melting temperature of the entire 30mer target site plus context, we also separated the thermodynamic features into three further features corresponding to the melting temperature of three distinct parts of the sgRNA—in particular, the 5 nucleotides immediately proximal to the PAM, the 8 nucleotides adjacent to that (away from the PAM), and then the 5 nucleotides in turn adjacent to the 8mer (again, away from the PAM).
Guide positional features such as amino acid cut position and percent peptide correspond to how far from the start of the protein coding region of the gene the cut site of the sgRNA was positioned. We also trained a version of Rule Set 2 without features that relate to protein information, as not all potential targets are protein-coding.
Not all features will appear in Supplementary Table 17, as features that did not vary at all were removed from all computations.
Stratification/CV
Unless otherwise noted, cross-validation was always performed by leaving out all sgRNAs for one gene at a time. For the inner cross-validation in the required nested cross-validation used to set the regularization weights for L1/L2, we also used leave-one-gene-out cross-validation.
When examining feature importance, we averaged either the Gini importance (for gradient boosted regression trees), or the weights (for L1-regularized regression) across genes.
Statistical significance
The only test of statistical significance used was to compare the Spearman correlation prediction measure between two approaches when using exactly the same test data and cross-validation. To do so, we used three sets of numbers: (i) the Spearman correlation performance measure for one approach, (ii) the Spearman correlation performance measure for the other approach, and (iii) the Spearman correlation between the predictions from each approach. These values were used in conjunction with a method to test dependent measures of correlations (here they are dependent because each approach uses the same ground truth)54. The precision of the implementation for this method resulted in the occasional p-value of 0.0. In such cases, we instead report p < 10−16, the smallest p-value that we observed the method reporting (strictly speaking this depends on the number of samples, and correlation between predictions, but it seemed a reasonably practical place-holder). Also of note is that this measure of statistical significance does not give equal weight to each gene, rather, each gene is represented in proportion to its number of sgRNAs.
Off-target scoring
The CCtop scoring algorithm was implemented as described42. Since the CCtop score is in the opposite direction of the Hsu-Zhang and CFD score, ranks of the CCtop scores were sorted in the opposite direction for AUC calculation, and we report the negative of the calculated correlation values. In other words, for the CFD score, a value of 0 indicate no predicted off-target activity while a value of 1 indicate a perfect match, while a low CCtop score indicates a high likelihood of off-target activity and a high CCtop score predicts no off-target activity.
The scoring algorithm that derives from data published by Hsu et al. is described on the MIT CRISPR design server (http://www.crispr.mit.edu/about)14.
The Cutting Frequency Determination (CFD) score is calculated by using the percent activity values provided in Supplementary Table 19. For example, if the interaction between the sgRNA and DNA has a single rG:dA mismatch in position 6, then that interaction receives a score of 0.67. If there are two or more mismatches, then individual mismatch values are multiplied together. For example, an rG:dA mismatch at position 7 coupled with an rC:dT mismatch at position 10 receives a CFD score of 0.57 × 0.87 = 0.50.
Evaluation of Bowtie2 performance
We initially attempted to use Bowtie2 to find off-target sites. This approach was successful and relatively straightforward for returning all perfect matches as well as those sites with only one mismatch to the query sequence. However, as the Bowtie2 algorithm is optimized for longer query sequences, we were unable to discover a set of options that would reliably find multi-mismatch sites (such as many of those identified by Guide-Seq) for our relatively short sgRNA query sequences in a single search invocation. Moreover, the runtime performance of Bowtie2 degraded significantly as we adjusted its options for greater multi-mismatch recall. One common workaround for tools with poorly performing fuzzy search is to convert the single query sequence into a list of relevant variants, perform a “less fuzzy” search on each variant, and accumulate the results. Employing this technique, we arrived at an “optimized Bowtie2” method. Specifically, we performed a systematic exploration of the Bowtie2 parameter space to find the combination of the amount and type of pre-permutation and parameter settings that yielded the highest average recall while still maintaining search time of approximately 1 second on standard computing hardware. This optimized Bowtie strategy scaled reasonably to whole-genome batches, but still did not identify all sites found by GUIDE-Seq or Cas-OFFinder.
GUIDE-Seq analysis
The sites identified by Guide-Seq were available as a supplementary table in that publication43. Cas-OFFinder was used to find all off-target sites with up to 6 mismatches in the human genome for the 10 sgRNAs assayed with Guide-Seq46. To make the fairest comparison between off-target scoring algorithms, we only scored off-target sites with NRG PAMs, as required by CCtop and the Hsu et al. algorithms, which meant excluding 38 sites.
Design of Brunello and Brie libraries
To design these libraries, we targeted protein-coding transcripts annotated by the GENCODE database (http://www.gencodegenes.org). GENCODE annotation is created by merging the Havana manual annotation and Ensembl automated gene annotation. We picked a transcript for each gene based on three tags defined by GENCODE: a) APPRIS (http://appris.bioinfo.cnio.es) annotations, which prioritize transcripts based on protein structural information, functionally important residues and evidence from cross-species alignments; b) level of confidence on the transcript, such as whether the transcript was a verified loci, manually annotated loci or automatically annotated loci; and c) transcript level support in GENCODE, which is based on how well mRNA and EST alignments match over the full length of the transcript.
We annotated all possible sgRNAs targeting each transcript for its Rule Set 2 score, and used these scores to rank, per transcript, the on-target activity of sgRNAs. In this implementation, we used the version of Rule Set 2 that does not incorporate protein target site information, as that criterion was used later in picking sgRNAs.
Likewise, we annotated each sgRNA by the number of potential off-target sites within various tiers of CFD scores: (i) 1.0; (ii) 0.2 – 1.0; (iii) 0.05 – 0.2; (iv) < 0.05. We then ranked, per transcript, each sgRNA by the fewest number of off-target sites in tier (i), using the additional tiers as tiebreakers. To break ties within a tier, we prioritized sgRNAs with the fewest off-target sites in protein-coding regions.
The on-target and off-target ranks of each sgRNA were then combined at equal weight to provide a final rank for each sgRNA targeting a particular transcript.
To pick sgRNAs for each transcript, we first chose the best-ranked sgRNA that targeted within the 5 – 65% of the protein-coding region of the target gene. We then selected additional sgRNAs per transcript, requiring that the next-picked sgRNA targets a site at least 5% away, from a protein-coding standpoint, from previously-picked sgRNAs. This ensures diversity in target space, especially useful due to the potential for exons that are present in the reference transcript not to be included in any particular cellular model to which the library is applied. We selected up to 4 sgRNAs per gene using this heuristic. In order to meet quota for some genes, we eliminated the 5 – 65% protein-coding region and 5% spacing criteria.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Tomko, M. Greene, A. Brown, D. Alan, and T. Green for software engineering support, and T. Nguyen, N. Tran, and X. Yang for library production support (Broad Institute). Z.T. is funded by NIH 5K12CA087723-12, ASCO Young Investigator Award, LLS Special Fellow Award. J.G.D. is a Merkin Institute Fellow and is supported by the Next Generation Fund at the Broad Institute of MIT and Harvard.
Footnotes
AUTHOR CONTRIBUTIONS
J.G.D., M.S., E.W.V., Z.T., C.W., and R.O. designed experiments; M.S., E.W.V., K.F.D., Z.T., C.W., and R.O. performed experiments; J.G.D., M.H., and I.S. analyzed experiments; N.F. and J.L. performed the computational modeling; J.G.D., N.F., J.L., and D.E.R. wrote the manuscript with assistance from other authors; J.G.D., H.W.V., and D.E.R. supervised the research.
ACCESSION CODES
SRA: SRP063278
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests
REFERENCES
- 1.Jinek M, et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mali P, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartenian E, Doench JG. Genetic screens and functional genomics using CRISPR/Cas9 technology. FEBS J. 2015;282:1383–1393. doi: 10.1111/febs.13248. [DOI] [PubMed] [Google Scholar]
- 6.Shalem O, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2013;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 9.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Veres A, et al. Low Incidence of Off-Target Mutations in Individual CRISPR-Cas9 and TALEN Targeted Human Stem Cell Clones Detected by Whole-Genome Sequencing. Cell Stem Cell. 2014;15:27–30. doi: 10.1016/j.stem.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ran FA, et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whittaker SR, et al. A Genome-Scale RNA Interference Screen Implicates NF1 Loss in Resistance to RAF Inhibition. Cancer Discovery. 2013;3:350–362. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies BR, et al. AZD6244 (ARRY-142886), a potent inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1/2 kinases: mechanism of action in vivo, pharmacokinetic/pharmacodynamic relationship, and potential for combination in preclinical models. Molecular Cancer Therapeutics. 2007;6:2209–2219. doi: 10.1158/1535-7163.MCT-07-0231. [DOI] [PubMed] [Google Scholar]
- 20.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae S, et al. TRIAD1 inhibits MDM2-mediated p53 ubiquitination and degradation. FEBS Letters. 2012;586:3057–3063. doi: 10.1016/j.febslet.2012.07.022. [DOI] [PubMed] [Google Scholar]
- 24.Gamper AM, Roeder RG. Multivalent binding of p53 to the STAGA complex mediates coactivator recruitment after UV damage. Molecular and Cellular Biology. 2008;28:2517–2527. doi: 10.1128/MCB.01461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Molecular Systems Biology. 2014;10:733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015 doi: 10.1126/science.aac7041. doi:10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caskey CT, Kruh GD. The HPRT locus. Cell. 1979;16:1–9. doi: 10.1016/0092-8674(79)90182-x. [DOI] [PubMed] [Google Scholar]
- 29.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Research. 2014;42:e168–e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zha M, et al. Molecular mechanism of ADP-ribose hydrolysis by human NUDT5 from structural and kinetic studies. J. Mol. Biol. 2008;379:568–578. doi: 10.1016/j.jmb.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Cheok MH, Evans WE. Acute lymphoblastic leukaemia: a model for the pharmacogenomics of cancer therapy. Nat Rev Cancer. 2006;6:117–129. doi: 10.1038/nrc1800. [DOI] [PubMed] [Google Scholar]
- 32.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi J, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chari R, Mali P, Moosburner M, Church GM. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nature Methods. 2015;12:823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu H, et al. Sequence determinants of improved CRISPR sgRNA design. Genome Research. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jinek M, et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science. 2014;343:1247997–1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bae S, Kweon J, Kim HS, Kim J-S. Microhomology-based choice of Cas9 nuclease target sites. Nature Methods. 2014;11:705–706. doi: 10.1038/nmeth.3015. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Research. 2014;42:7473–7485. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stemmer M, Thumberger T, del Sol Keyer M, Wittbrodt J, Mateo JL. CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS ONE. 2015;10:e0124633–11. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2014;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature Publishing Group. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nature Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 46.Bae S, Park J, Park J, Kim JS. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kampmann M, et al. Next-generation libraries for robust RNA interference-based genome-wide screens. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E3384–E3391. doi: 10.1073/pnas.1508821112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steiger JH. Tests for comparing elements of a correlation matrix. Psychological Bulletin. 1980;87:245–251. [Google Scholar]
- 49.Blasi E, Radzioch D, Durum SK, Varesio L. A murine macrophage cell line, immortalized by v-raf and v-myc oncogenes, exhibits normal macrophage functions. Eur. J. Immunol. 1987;17:1491–1498. doi: 10.1002/eji.1830171016. [DOI] [PubMed] [Google Scholar]
- 50.Stansley B, Post J, Hensley K. A comparative review of cell culture systems for the study of microglial biology in Alzheimer's disease. J Neuroinflammation. 2012;9:115. doi: 10.1186/1742-2094-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ben-Hur A, Ong CS, Sonnenburg S, Schölkopf B, Rätsch G. Support vector machines and kernels for computational biology. PLoS Comput. Biol. 2008;4:e1000173. doi: 10.1371/journal.pcbi.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cock PJA, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le Novère N. MELTING, computing the melting temperature of nucleic acid duplex. Bioinformatics. 2001;17:1226–1227. doi: 10.1093/bioinformatics/17.12.1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.