Abstract
The neuronal ceroid lipofuscinoses (NCLs) are a group of neurodegenerative genetic diseases that primarily affect children and have no known cure. A unified clinical rating scale for the juvenile form of NCL (JNCL) has been developed, but it has not been validated in other subtypes and does not give a true measure of the pathophysiological changes that may be occurring during disease progression. In this study, we have identified candidate biomarkers in blood plasma of NCL disease using multiple proteomic approaches, with the aim of developing a panel of biomarkers that could serve as a metric for therapeutic response. Candidate biomarkers were identified as proteins with levels that significantly differed between patients and controls in both sample sets. The seven candidates identified have previously been associated with neurodegenerative and inflammatory diseases. Multiplex immunoassay based testing was the most efficient and effective evaluation technique and could be employed on a broad scale to track patient response to treatment.
Keywords: NCL, Plasma Biomarkers, 2D-DIGE, Immunoassays
INTRODUCTION
The neuronal ceroid lipofuscinoses (NCLs), or Batten disease, are a group of neurodegenerative genetic diseases that primarily affect children. Most subtypes are caused by mutations in one of a dozen known genes, and they differ mainly in the age of onset and speed of progression [1, 2]. The juvenile form (JNCL) is the most common and presents around age five, with death in the third decade of life. Patients typically present with visual deterioration, which is followed by seizures, motor dysfunction, and cognitive decline (see [3] for an overview). The NCLs are classified as lysosomal storage disorders due to a characteristic intracellular accumulation of autofluorescent storage material, ceroid lipofuscin, although there is no evidence that storage material correlates with cellular pathology [4].
Researchers are working urgently to develop new therapies to treat NCL. A standardized tool to assess disease severity across all subtypes is needed to accurately evaluate the efficacy of new therapeutic interventions. The Unified Batten Disease Rating Scale (UBDRS) was developed to objectively quantify several domains of JNCL disease (i.e. physical, behavior, capability) [5]. Recently clinicians began testing the validity of this and other non-NCL scales in late infantile NCL (LINCL); however, this is a logistically intensive process that will take time to complete [6]. To our knowledge, scales are not currently being validated in other NCL subtypes. Moreover, scales reflect only outward manifestations of disease and might not be sensitive or accurate enough to gauge changes in molecular pathology. The next step is to develop a set of molecular biomarkers that provides a more direct measure of internal pathological processes, optimally across all genetic mutations comprising all NCL subtypes.
Biomarkers are objectively measurable physical signs that distinguish a diseased population from a non-diseased one. They can be used for diagnostic or prognostic purposes and to provide insight into the pathological mechanisms of a particular disorder. We report on a study using Luminex® based immunoassays, including MILLIPLEX® MAP magnetic immunoassay panels (MM) and the DiscoveryMAP ® 1.0 immunoassay from Myriad-RBM (MR), two dimensional difference gel electrophoresis (2D-DIGE) followed by mass spectrometry, and Western blot, to compare plasma protein profiles of patients representing a wide range of NCL genotypes with a control population of similar age. We chose this approach because the potential to develop a standardized assay for protein quantification and the minimally invasive nature of a blood draw makes translation of our findings into clinical practice technically and ethically feasible. Our primary goal was to identify proteins that directly reflect NCL disease itself and not disruptions in homeostasis elicited by secondary conditions (i.e. infection, malnutrition, immobility). Nonetheless, because the pathogenesis of NCL is poorly understood, we investigated a wide range of potential targets.
We have identified seven candidate biomarkers, many of which have previously been associated with neurodegeneration and/or brain injury. The remaining candidates reflect a systemic disease state. In general, we found the immunoassay to be a more sensitive and specific tool than 2D-DIGE for biomarker discovery. While 2D-DIGE theoretically allows for identification of a broader range of proteins, technical issues limited its utility.
RESULTS
Demographics
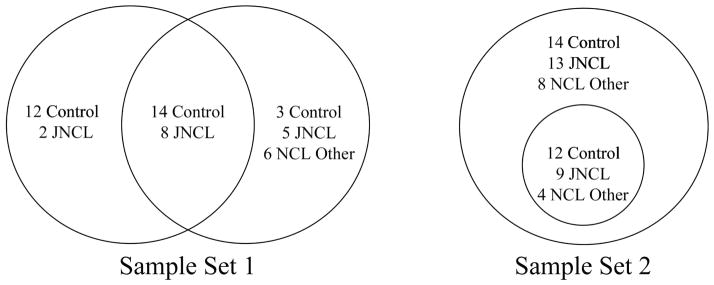
For this study we used two sets of samples collected at least two years apart (figure 1; table S1). Set 1 was used for 2D-DIGE and the MR immunoassay and set 2 for MM immunoassays and Western blot. Within set 1, 22 of the 50 samples (29 control, 21 patients) were used for both 2D-DIGE and MR assays (14 control, 8 JNCL). Within set 2, the Western blot samples were a subset of those used for MM immunoassay. Thus, analytes identified as differentially expressed by the first set required validation with the second set and vice versa. Because of the rarity of NCL, it was not possible to recruit strictly independent test populations for each set. Both sets included plasma collected from some of the same individuals but on different dates. The two years or more time between collection dates and corresponding difference in age and/ or severity of disease should offset concerns of sample redundancy between set 1 and set 2.
Figure 1.
Distribution of samples by set and proteomics technique. Sample set 1 includes samples used for Myriad RBM immunoassay on the left and 2D-DIGE on the right. In sample set 2, Western blot samples are a subset of those used for the MILLIPLEX® MAP immunoassay.
Set 1 samples used for 2D-DIGE were from 17 healthy individuals, 13 persons diagnosed with JNCL, and 6 diagnosed with a non-juvenile form of NCL [3 infantile NCL (INCL), 1 LINCL, and 2 Finnish variant late infantile NCL]. There was no significant difference in age between the control and patient groups (control versus JNCL only, P = 0.10; control versus all NCLs, P = 0.056). The ratio of males to females was similar when comparing the control group with all NCL subtypes; however, more JNCL individuals were male while those classified as “other NCL” were largely female (table 1). MR immunoassay was performed with plasma from 10 JNCL patients and 26 control individuals from set 1. The male to female ratios were similar (table 1). As there was a significant difference in age (P = 0.04) (P = 0.034), we performed post hoc analyses to rule out a confounding effect of age for candidates identified with the MR immunoassay.
Table 1.
Demographics of patient samples used in each method of analysis.
| Group | n | Mean age (SD) | Age range | M:F | |
|---|---|---|---|---|---|
| Luminex | JNCL | 22 | 19.73 (5.02) | 12 – 31 | 13:9 |
| NCL (other) | 12 | 7 (2.05) | 4 – 11 | 4:8 | |
| Control | 26 | 17.77 (7.19) | 7 – 33 | 8:18 | |
| Myriad-RBM | JNCL | 10† | 14.7 (4.22) | 9 – 23 | 5:5 |
| Control | 26§ | 19.12 (5.72) | 9 – 30 | 14:12 | |
| 2D-DIGE | JNCL | 13† | 13.91 (4.12) | 9 – 23 | 9:4 |
| NCL (other) | 6 | 12.83 (5.19) | 7 – 22 | 1:5 | |
| Control | 17§ | 16.12 (4.08) | 9 – 24 | 11:6 | |
| Western Blot | JNCL | 9 | 20.11 (4.70) | 12–26 | 6:3 |
| LINCL | 4 | 8.75 (1.71) | 7 – 11 | 3:1 | |
| Control | 12 | 16.58 (7.04) | 7 – 25 | 6:6 |
8 JNCL and
14 Control samples used in both Myriad-RBM and 2D-DIGE
MM immunoassays were performed with set 2 samples from 34 NCL patients and 26 control individuals. Subtypes were as follows: 22 persons diagnosed with JNCL, 9 with LINCL, 2 with INCL, and 1 NCL patient without a confirmed genetic diagnosis. Age did not significantly differ between the control group and JNCL patients (P = 0.29) nor between the control group and all NCL subtypes combined (P = 0.19). Females were more heavily represented in the control and other NCL groups, while males accounted for more of those diagnosed with JNCL.
The Western blot subset consisted of 12 control samples, 9 JNCL samples, and 4 LINCL samples. There was no significant difference in age between the control and patient groups (all NCLs, P = 0.99; JNCL only, P = 0.12). The control group had equal number of males and females, while the patient group was largely male (table1).
2D-DIGE and validation
Twenty-seven spots were identified with 2D-DIGE (tables 2 & 3). Fourteen were eliminated for the following reasons: previously uncharacterized protein with no available antibody (1), low protein concentration (2), target was a structural protein (4), target was a depleted protein (7). The seven remaining spots were comprised of seven target proteins with complement factor C4 identified on two spots, and one spot containing two different proteins (C1s and C1 esterase inhibitor) (table 2). Apolipoprotein AIV (apoAIV), zinc-α2-glycoprotein (ZAG), and high molecular weight (HMW) kininogen were significant in the male subset only. Three proteins identified via 2D-DIGE also were part of the MR immunoassay—apoAIV, C4, and CD5L. Only apoAIV was significantly different in both assays, but results stratified by gender for 2D-DIGE only. Confirmation of 2D-DIGE spots as biomarker candidates required validation with set 2 samples via MM immunoassay or Western blot. We were able to validate only HMW kininogen (table 2), for which gender was a significant factor as detailed below.
Table 2.
Identified analytes from each method of analysis. Analytes in bold were identified as validated biomarker candidates. Unless otherwise indicated, P-values were calculated with a Student’s t-test. Direction of arrows refers to protein level in patient samples compared to control samples.
| Analyte | Luminex | Myriad-RBM | 2D-DIGE | Western Blot | ||
|---|---|---|---|---|---|---|
|
| ||||||
| JNCL | all NCLs | JNCL | all NCLs | JNCL | all NCLs | |
| BDNF | ↑↑↑† | ↑↑↑† | ↑↑↑ | |||
| NrCAM | ↑† | ↑↑↑ | ↑↑ | |||
| Clusterin | ↑↑↑ | ↑↑ | ||||
| Adiponectin | ↑↑↑ | ↑ | ↑ | |||
| Apolipoprotein E | ↑ | ↑ | ↑ | |||
| VCAM-1 | N.S. | ↑ | ↑ | |||
| Myoglobin | ↑ | N.S. | ↑ | |||
| Kininogen-1 (HMW) | ↑‡ | ↑↑ | ↑ | |||
| NCAM | ↑↑↑ | ↑↑↑ | ||||
| TSH | ↑↑↑ | |||||
| Thrombopoietin | ↑↑↑ | |||||
| Apolipoprotein AIV | ↑ | ↑‡ | N.S. | N.S. | ||
| PAI-1 | ↑↑↑ | ↑↑↑ | N.S. | |||
| ICAM-1 | N.S. | N.S. | ↑↑ | |||
| CK-MB | ↑↑ | N.S. | N.S. | |||
| vWF | ↑ | N.S. | N.S. | |||
| LOX1 | ↓ | N.S. | N.S. | |||
| Complement C4/C4a | N.S. | N.S. | N.S. | ↑ | ||
| Apolipoprotein AII | ↑ | N.S. | N.S. | |||
| zinc α2-glycoprotein | ↑‡ | N.S. | ||||
| CFH | N.S. | N.S. | ↓ | |||
| CD5/CD5L | N.S. | ↑ | N.S. | N.S. | ||
| C1 inhibitor | ↓ | N.S. | ||||
| C1s | ↓ | N.S.† | ||||
↑↑↑, P < 0.0001;
↑↑, 0.0001 ≤ P ≤ 0.001;
↑, 0.002 ≤ P ≤ 0.05.
N.S., not significant.
, Mann Whitney U-test.
, P values for comparisons in males only.
Abbreviations: brain derived neurotrophic factor (BDNF), neuronal cell adhesion molecule (NrCAM), vascular cell adhesion molecule 1 (VCAM-1), high molecular weight (HMW), neural cell adhesion molecule (NCAM), thyroid stimulating hormone (TSH), platelet activating inhibitor 1 (PAI-1), immune cell adhesion molecule 1 (ICAM-1), creatine kinase MB (CK-MB), von Willebrand’s Factor (vWF), lectin-like oxidized LDL receptor 1 (LOX1), complement factor H (CFH), fatty acid binding protein (FABP)
Table 3.
Identified protein spots from 2D-DIGE.
| Spot ID | Ratio Change* | P value | Analysis Group | LC-MS/MS Identification |
|---|---|---|---|---|
| 348 | −1.67 | 0.00045 | all | depleted protein (α-2 macroglobulin) |
| 349 | −1.75 | 0.0069 | all | depleted protein (C3) |
| 355 | −1.56 | 0.0016 | all | depleted protein (α-2 macroglobulin) |
| 381 | −1.37 | 0.022 | all | low protein |
| 411 | −1.42 | 0.022 | all | depleted protein (α-2 macroglobulin) |
| 553 | −1.43 | 0.027 | all | depleted protein (α-2 macroglobulin) |
| 556 | −1.36 | 0.046 | all | depleted protein (α-2 macroglobulin) |
| 741† | 1.51 | 0.039 | all | C4 |
| 1027† | −1.5 | 0.022 | all | C1s, C1 esterase inhibitor |
| 1110† | 1.44 | 0.047 | all | C4 |
| 1354 | 1.58 | 0.02 | all | uncharacterized protein |
| 1381 | −1.65 | 0.003 | all | keratin |
| 1409 | −1.68 | 0.024 | all | low protein |
| 1467† | −1.43 | 0.42 | males | high molecular weight kininogen-1 |
| 1520 | −1.64 | 0.034 | all | depleted protein (apolipoprotein AI) |
| 1594† | 1.65 | 0.045 | all | CD5L |
| 1634 | 1.7 | 0.0044 | all | keratin |
| 1637 | 1.58 | 0.0079 | all | keratin |
| 1643 | 1.74 | 0.012 | all | actin |
| 1664† | 2.33 | 0.039 | males | apolipoprotein AIV |
| 1673† | 1.62 | 0.036 | males | zinc α2-glycoprotein |
ratio change refers to change from control conditions.
indicates spot with protein targeted for validation.
Luminex® based immunoassays and validation
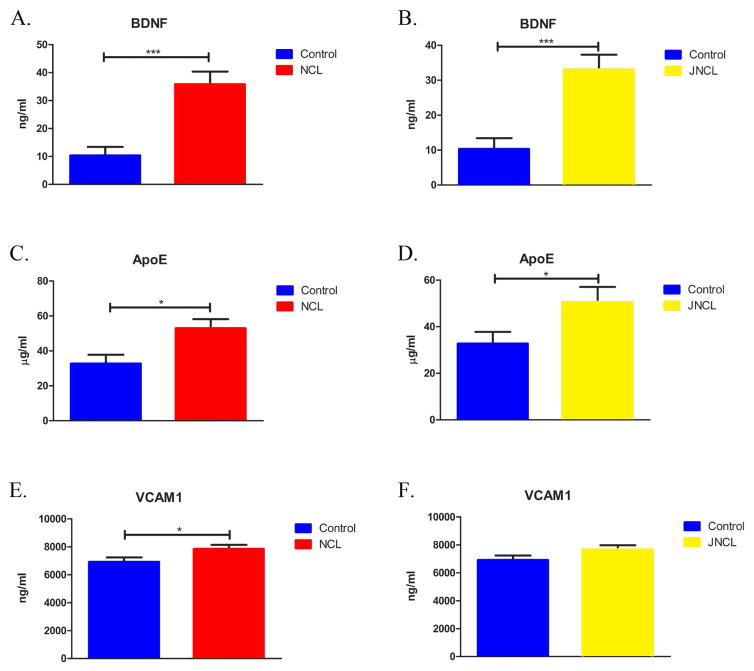
Twenty analytes were tested in both the MR immunoassay (sample set 1) and MM immunoassays (sample set 2). Analytes showing a significant change in both panels were classified as validated. Three of the 20 were significantly altered in one panel only and removed from further consideration. Three analytes significantly changed in both panels qualified as biomarker candidates: brain derived neurotrophic factor (BDNF), apolipoprotein (apoE), and vascular cell adhesion molecule 1 (VCAM-1) (table 2; figure 2; figure 3). BDNF and apoE alterations were statistically significant in comparisons of controls with all NCL patients or JNCL patients alone, while the change in VCAM-1 level was statistically significant only in the analysis including all NCL subtypes.
Figure 2.
Identified and validated analytes by Luminex (MM). In comparison to control samples, BDNF, ApoE and VCAM-1 were significantly elevated in all NCLs samples (a,c,e). When comparing only JNCL and control samples, BDNF and ApoE were significantly elevated; VCAM-1 was not significantly different (b,d,f). Bars indicate mean ± SEM. Statistical significance determined using Student’s t test for all analytes except BDNF, determined by Mann-Whitney U test (***, P < 0.0001; *, 0.002 ≤ P ≤ 0.05).
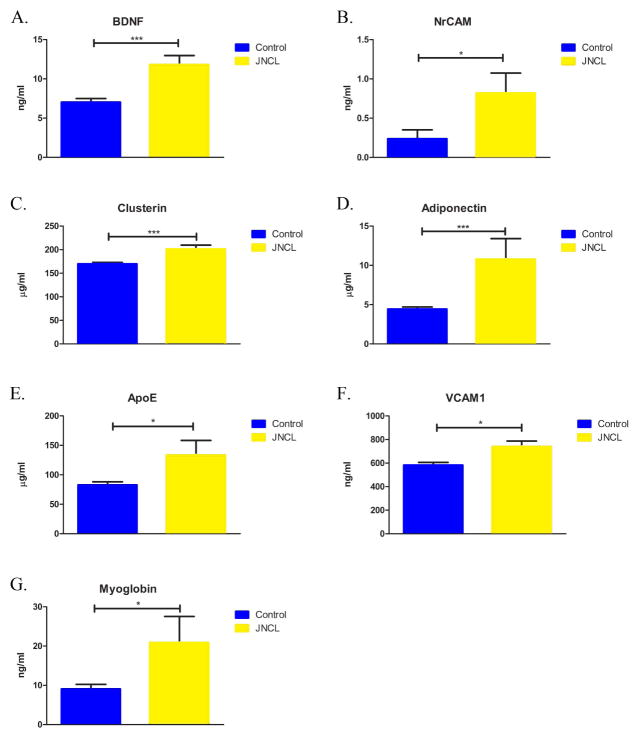
Figure 3.
Identified and validated analytes by Myriad-RBM (MR). In comparison to control samples, BDNF, NrCAM, Clusterin, Adiponectin, ApoE, VCAM-1, and Myoglobin were significantly elevated in JNCL samples. Bars indicate mean ± SEM. Statistical significance determined using Student’s t test for all analytes except NrCAM, determined by Mann-Whitney U test (***, P < 0.0001; *, 0.002 ≤ P ≤ 0.05).
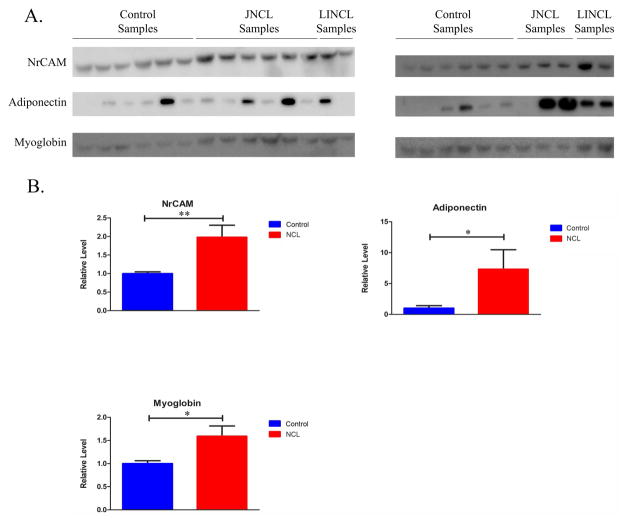
Western blots were run for 12 of the significant remaining analytes, P ≤ 0.005, identified by the MR immunoassay. Neuronal cell adhesion molecule (NrCAM), adiponectin, myoglobin, and clusterin, were validated (table 2; figure 4). We were unable to optimize blot conditions for cardiac fatty acid binding protein (FABP), thyroid stimulating hormone (TSH), and thrombopoietin, despite trying several different antibodies. Interestingly, the MR data indicated that several patients but none of the control individuals had values falling above the physiological plasma range for TSH (four of ten), myoglobin (two of ten), and adiponectin (four of eleven). For other analytes, values were either within the normal plasma range or normal parameters were not available for reference.
Figure 4.
Validation of analytes identified by Myriad-RBM. (a) Western blots were used to confirm and determine the expression profiles of identified analytes. (b) Densitometric analyses were used to quantify relative expression levels of MR identified analytes. Relative levels were normalized to total protein. Bars indicate mean ± SEM. Statistical significance for NrCAM and Adiponectin was determined using Mann Whitney Test (**, 0.0001 ≤ P ≤ 0.001; *, 0.002 ≤ P ≤ 0.05); Myoglobin was determined using Student’s T-test (*, 0.002 ≤ P ≤ 0.05).
We could not pursue validation of the significant MM immunoassay analytes cathepsin D and neural cell adhesion molecule (NCAM) as additional plasma was not available. Cathepsin D was of potential interest because mutation of the cathepsin D gene (CTSD, aka CLN10) causes a very rare autosomal recessive form of NCL [7, 8]. No patients with this NCL subtype were included in the study. NCAM levels were significantly higher in patient samples (all NCLs and JNCL only, table 2), with P values lower than the alpha value required by Bonferonni correction for multiple testing (< .002 for 26 analytes.) Interestingly, serum levels of S100B, a calcium binding protein found in glia and other neural crest derived cells, did not differ in our sample (P = 0.912). S100B has been broadly identified as a serum biomarker for a variety of neuropathological conditions, including psychiatric disorders, brain injury, neuroinflammation, and neurodegenerative disease (see [9] for review).
Effect of gender and age in candidate proteins
In light of a more severe disease course in females and disparate gender proportions in several of our sample subsets, candidate biomarkers were interrogated for an effect of gender [10, 11]. Gender already was considered in 2D-DIGE by analyzing females and males separately and together, with results mentioned above. For other techniques, a significant effect of gender was found only for NCAM and HMW kininogen (Western blot), with post hoc pairwise analyses revealing similar trends. For NCAM, analysis of variance (ANOVA) showed significant main effects of gender (all NCLs: F1,40 = 7.21, P = 0.011; JNCL: F1,36 = 6.433, P = 0.016) and sample group (all NCLs: F1,40 = 12.322, P = 0.001; JNCL: F1,36 = 11.936, P = 0.001), and a significant gender by group interaction (all NCLs: F1,40 = 8.108, P = 0.007; JNCL: F1,36 = 6.85, P = 0.013). HMW kininogen was significant for an effect of gender in the JNCL group (all NCLs: F1,21 = 3.971, P = 0.059; JNCL: F1,17 = 5.099, P = 0.037). There was a main effect of group (all NCLs: F1,21 = 4.630, P = 0.059; JNCL: F1,17 = 11.940, P = 0.003), but no significant interaction of gender and group (all NCLs: F1,21 = 0.624, P = 0.439; JNCL: F1,17 = 0.567, P = 0.462).
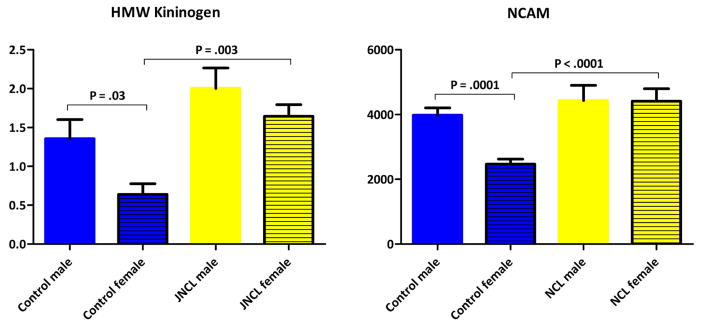
For both NCAM and HMW kininogen, post hoc pairwise analyses revealed that the male control population had significantly higher serum plasma levels than female controls, a trend not present in the patient population (figure 5). Accordingly, NCAM levels differed between patient and control groups in females only. This indicates that the utility of NCAM as a biomarker could be gender dependent. These findings call into question the validity of HMW kininogen as a biomarker, because we identified differences via 2D-DIGE in the male subset only, opposite of what was found with Western blot. Further investigation is needed to resolve gender stratification issues. The paucity of female samples used for HMW kininogen testing could have been an issue and should be addressed.
Figure 5.
Gender as a significant factor in plasma protein levels. High molecular weight (HMW) kininogen and neural cell adhesion molecule (NCAM) significantly differed between patient and control populations in females only as measured by Western blot and MILLIPLEX® MAP immunoassay, respectively.
Age was a significant factor in the MR immunoassay for apoE only. The ANOVA model indicated a main effect of age (F1,32 = 5.517, P = 0.025) but no main effect of group (F1,32 = 2.09, P = 0.39) nor a significant age by group interaction (F1,32 = 1.589, P = 0.22). These data might suggest that the significant effect of group was, in fact, due to differences in age. It also is possible that the small number of patient samples (n = 10) lowered statistical power to the extent that detection of moderate age, group, and age x group effects was not possible (figure 1). Age was not a significant factor for apoE in the MM data set, which included a larger patient sample size. Future studies should be cognizant of age and that it might affect levels of apoE.
DISCUSSION
Our results suggest that in the later stages of disease, the central nervous system (CNS) of NCL patients appears to affect protective and reparative processes. BDNF is a neurotrophin involved in synaptic plasticity and adult neurogenesis. It also has a neuroprotective role, increasing with neuroinflammation and after brain injury, and crossing the blood brain barrier in pathological conditions [12, 13]. It has been widely explored as a biomarker for common neurodegenerative diseases including Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease [14–18]. In general, these conditions have been associated with a decrease in circulating BDNF, the conclusion being that a loss of trophic support permits progression of disease. In NCL, BDNF might be elevated as a compensatory response to brain injury and/or as part of an inflammatory response. It is a promising protein to follow during disease as it seems to directly reflect NCL associated neural pathology and can be easily tested via immunoassay. The mood stabilizers lithium and valproic acid (the latter also used as an anti-epileptic) might increase BDNF levels [19, 20]. We did not have complete information about which drugs participants were taking, something that must be accounted for in future studies.
We detected increased levels of the immunoglobulin gene superfamily adhesion molecules NrCAM and VCAM-1 in patients compared to non-diseased individuals. NrCAM is expressed primarily in the brain where it helps to mediate adhesion between neurons, glia, and the extracellular matrix (see [21] for review). The adrenal glands and pancreas are physiological peripheral sources [22]. Increased NrCAM levels have been reported in the plasma of females with Asperger’s syndrome [23], while decreased cerebrospinal fluid (CSF) concentrations are associated with cognitive impairment and Alzheimer’s disease [24, 25].
VCAM-1 is expressed by endothelial cells. Upregulation by inflammatory mediators such as TNF-α, IL-1β, and IFN-γ, increases permeability of the microvasculature to leukocytes. To our knowledge, VCAM-1 is the only candidate biomarker reported in a previous NCL study. Plasma VCAM-1 levels were increased in the INCL mouse model and in several non-NCL mouse models of lysosomal storage disease compared to wild type animals [26]. VCAM-1 has been associated with multiple auto-immune conditions as well as neurodegeneration and Alzheimer’s disease [27–32]. Identification of VCAM-1 is interesting in the context of JNCL-associated autoantibody production.
We identified several candidates potentially associated with lipid metabolism—adiponectin and apoE. Adiponectin is an adipocytokine that plays an anti-inflammatory role in the cardiovascular system. Its regulation is not well understood, but blood levels change with nutritional status, pharmaceuticals, horomones, cytokines, and inflammation [33–36]. Increased levels of adiponectin were reported in association with mild cognitive impairment (plasma and CSF) and Alzheimer’s disease (CSF) and also in multiple sclerosis (CSF) [37, 38]. There has been speculation that adiponectin might be promoting neuronal apoptosis or that it could be associated with the reduced body mass index that often precedes Alzheimer’s disease in elderly patients [38, 39]. NCL patients experience both. In our study, 4 of 11 JNCL patients included in the MR panel registered adiponectin levels above the normal plasma range.
Mutations in APOE predispose to late onset Alzheimer’s disease [40]. Peripherally, apoE is produced mainly in the liver and serves as one of the protein components of chylomicron remnants and intermediate density lipoproteins (IDLs). In the brain, apoE transports cholesterol from astrocytes to neurons and also is involved in synaptic plasticity and maintenance of dendritic spines [41]. Astrocytes and microglia produce and secrete apoE into cerebrospinal fluid [42, 43]. A healthy blood brain barrier should prevent flux of ApoE between these two compartments [44]. Increased apoE could reflect the expansion of glial cell populations observed in NCL; however, even in conditions of increased blood brain barrier permeability, central apoE is present in pg/ml concentrations versus systemic levels of ug/ml [45–47].
Clusterin is atypical of the apolipoproteins in that it is a multi-functional protein that serves as an extracellular protein chaperone and is not involved in lipid transport. It is non-specifically increased with inflammation and disease stress and is thought to play a protective role in the brain [48, 49]. Levels were increased in patient samples; although clusterin’s involvement in primary mechanisms of NCL pathology is unlikely as it is one of the most broadly identified biomarkers across a wide range of diseases [50–54].
Myoglobin and cardiac FABP were elevated in patient plasma. They are released into the circulation after damage to striated muscle fibers, including the myocardium or skeletal muscle [55, 56]. We could not validate cardiac FABP as we were unable to optimize antibody binding conditions for Western blot. Cardiac involvement in JNCL, presents as bradycardia with sinus arrests, sometimes necessitating placement of a pacemaker [57].
In this study we used three distinct proteomics techniques. All were largely successful; however, the accuracy, precision, and ease of use varied greatly. 2D-DIGE was most problematic overall. Complement factors, the apolipoproteins, and CD5L, have been identified as potential biomarkers with the same or a similar technique across a wide range of seemingly unrelated conditions [58–64]. These targets are reflective of disease in general such that non-specific differential expression might be expected. It is also possible that 2D-DIGE biases toward detection of moderate to high concentration plasma proteins (e.g. complement and apolipoproteins), which can have excessive inherent inter- and intra- individual variability in plasma levels in a normal population, sometimes in excess of 50% [65]. HMW kininogen, a coagulation and kinin system protein, was the only validated 2D-DIGE target. This result is still uncertain, however, since changes in HMW kininogen was specific to males in the first sample set and females in the second sample set.
Western blots are frequently used to investigate biomarkers where targets are few and known or to validate results from other, more high-throughput methods such as 2D-DIGE or multiplex immunoassay. In general, we found Western blot to be an effective validation tool. Occasionally, the fact that most antibodies are not intended for use with plasma became an issue. Plasma-tested antibodies for cardiac FABP, TSH, and thrombopoiten, were not available, and the antibodies we tried produced extensive non-specific binding without a discernible band for the target protein. Thrombopoiten and TSH both remained significant after Bonferroni correction (a significance level of P<0.0004 for 129 MR immunoassay analyzed); platelet activating factor 1 did as well but failed validation. TSH is particularly noteworthy as the MR immunoassay revealed that TSH was elevated above the clinically normal range in almost half of the patients and none of the control individuals. Clinicians have not reported changes in TSH or other thyroid associated hormones, but our results should elicit awareness of the potential for thyroid dysfunction.
Multiplex immunoassay using Luminex® based or other technologies was our method of choice. The kits were easy to use, required little to no optimization, and could assess multiple plasma proteins simultaneously. Samples also might be sent to an ever increasing number of commercial labs with Luminex® capabilities for assessment.
Because we anticipate that any therapeutic application will require samples to be collected and potentially processed at a number of clinics, we emphasize the importance of standardization of collection, processing, and storage conditions. In our study, we were unable to maintain consistency in time of day and nutritional status at sample collection. This could have been a factor in our ability to detect and verify differentially expressed proteins and should be considered when planning future protocols. On the other hand, the biomarkers we identified, and especially those that were verified, could be particularly robust for detection under nonstandardized conditions. Care must also be taken in determining how samples are handled at collection through storage and beyond as this will affect protein concentrations. Most important is the duration between sample collection and storage and the temperature at which samples are handled during that time. Many proteins degrade the older and warmer they are. Some proteins, such as BDNF, begin to break down with long term storage even at −80°C [66]. All samples should be treated as uniformly as possible to ensure reliable results. In our study, samples were processed similarly, though not identically. We compensated for slight differences in processing as well as varying lengths of time in storage by ensuring that equal proportions of samples were collected and processed at the same time across patient and control groups.
While questions remain about what methods are best used for identifying blood biomarkers of disease, we identified seven candidate biomarkers that clearly differed between NCL patients and control participants—BDNF, NrCAM, clusterin, adiponectin, apoE, VCAM-1, and myoglobin. We postulate that the biomarkers are best suited for assessing effectiveness of therapy by measuring baseline protein levels of these identified biomarkers and then tracking the levels through therapy as related to disease progression.
Materials and methods
Participants
Study participants included patients with a diagnosis of any NCL subtype (all except one with confirmed genetic mutation) and non-affected control individuals. All were enrolled between July of 2008 and October of 2012. Each participant was fully informed of the nature of the study and voluntarily signed an informed consent approved by the Sanford Independent Review Board (IRB) for “Biochemical Analysis of Batten Disease” and the University of Rochester IRB, for “Biochemical Analysis of Batten Disease”. Parents signed informed consents for persons younger than 18 years of age. Assent was obtained when possible. For logistical reasons, we were unable to control for time of day or nutritional status. Blood was collected by venipuncture in heparin or EDTA and then centrifuged for 10 minutes at 2000 x g. Plasma was aliquoted and stored at −80. Storage tubes were coded such that they contained no personal identifying information.
2D-DIGE
Samples were thawed and then filtered to remove particulates. Protein was quantified with the Pierce 660nm Protein assay (Thermo Fisher Scientific, Rockford, IL). A volume containing 500 μg of protein was diluted to 1μg/μl in Seppro® dilution buffer (Sigma-Aldrich Corp, St. Louis, MO). To prevent masking of less abundant proteins, samples were depleted of the 14 most abundant plasma proteins with Seppro® IgY14 columns (Sigma-Aldrich Corp) according to the manufacturer’s instructions. The IgY polyclonal hen egg yolk antibody based system prevents cross reactivity encountered with mammalian antibody based immunodepletion systems and is the current method of choice [67]. Depleted samples were checked for albumin with gel electrophoresis followed by total protein staining with SYPRO® Ruby Protein Gel Stain (Sigma-Aldrich Corp). Depletion was repeated where excessive albumin was detected, delineated by a large smear in the 70 kDa region of the gel. Protein concentrations again were measured after depletions and the sample pH was adjusted to 8.5.
2D-DIGE was performed according to the manufacturer’s instructions (GE Healthcare, Piscataway, NJ). Briefly, samples were labeled with CyDye DIGE Fluor saturation dyes (GE Healthcare)—50 μg protein from patient and control samples, one labeled with Cy3 and the other with Cy5, and a Cy2 labeled pooled standard. Reciprocal labeling within experimental groups was used to control for dye bias. Pick gels required 100 μg protein of unlabeled patient and control samples and 50ug protein of Cy2 labeled pooled standard. Protein preparations were loaded onto Ready Strip IPG strips (pH 3 – 10 or pH 5 – 8) (Bio-Rad, Hercules, CA) for first dimension focusing on a Protean IEF Cell (Bio-Rad). Strips then were incubated in equilibration buffer (ingredients). For second dimension focusing, strips were applied to 10% SDS-PAGE gels and run 8 to 16 hours in an Ettan DALT electrophoresis chamber (GE Healthcare). Gels then were scanned with a Typhoon Trio+ variable mode imager (GE Healthcare). Pick gels were stained with Lava Purple total protein stain (Gel Company, San Francisco, CA) and scanned a second time. All images were uploaded into DeCyder Differential Analysis v7 Software (GE Healthcare).
Following analyses, pick gels were placed on the Ettan Spot Picker (GE Healthcare). Coordinates for spots were uploaded from a picklist generated by the DeCyder software, and spots were picked according to reference markers. Picked spots were sent to the Proteomics Core Facility at the Sanford Burnham in La Jolla, CA for protein identification by liquid chromatography followed by tandem mass spectrometry (LC-MS/MS). Proteins identified by LC-MS/MS were validated by Western blot or immunoassay when possible.
Myriad-RBM Luminex® multiplex immunoassay
Heparinized plasma samples were sent to Myriad-RBM (MR) (Austin, TX) for quantification of the 190 serum proteins comprising the Human DiscoveryMAP ® 1.0 immunoassay. Myriad-RBM is a CLIA certified laboratory that specializes in the identification of serum biomarkers using Luminex xMAP ® technology.
EMD-Millipore Luminex® multiplex immunoassay
Plasma samples collected in EDTA were assessed for 26 serum proteins with MILLIPLEX ® MAP magnetic immunoassay kits (MM) (EMD-Millipore, Billerica, MA) according to the manufacturer’s instructions. Briefly, samples were vortexed and then centrifuged to settle particulates before aliquotting. Samples were diluted with assay buffer as follows: Human Neurodegenerative Disease Panel 1, 1:40,000; Human Neurodegenerative Disease Panel 2, 1:2000; Human Neurodegenerative Disease Panel 3, 1:100; and Human Neurological Disorders Panel 2, 1:25,000. In general, samples were first incubated with beads conjugated to antibodies against proteins of interest. After a washing step, secondary, biotinylated antibodies were added for an hour followed by streptavidin-phycoerythrin for 30 minutes. After another washing step, samples were read on a Luminex ®. Absolute protein concentrations were derived from a standard curve generated from eight serial dilutions of a standard sample.
Western blot
Western blot was used to verify select 2D-DIGE spots and MR immunoassay results for analytes with P ≤ 0.005. Samples first were filtered to remove excess lipid and then quantified using the Pierce 660nm Protein assay (Thermo Fisher Scientific, Rockford, IL). Twenty to 40 μg of protein in reducing sample buffer was resolved with sodium dodecyl sulfate – polyacrylamide gel electrophoresis and then transferred to PVDF membrane. Membranes were stained with a Ponceau S (Sigma-Aldrich Corp) solution (0.1% w/v in 5% acetic acid) and imaged for total protein with a Biospectrum 500 Imaging System (UVP, Upland, CA). Proteins then were detected with the following primary antibodies—from Abcam: anti-adiponectin (ab22554, 1:1000), anti-apolipoprotein AIV (ab81616, 1:500), anti-C1s (ab92706), anti-cardiac FABP (ab16915), anti-CD5L (ab45408, 1:500), anti-creatine kinase MB (ab72002, 1:2000), anti-HMW kininogen (ab79653, 1:60), anti-LOX1 (ab93423, 1:100), anti-myoglobin (ab77232, 1:500), anti-TSH beta (ab93151), anti-von Willebrand Factor (ab9378, 1:1000), anti-zinc α2-glycoprotein (ab167573, 1:1000); from Sigma: anti-apolipoprotein AII (SAB1403558, 1:1000), anti-NrCAM (AV44832, 1:2000); from Fitzgerald International (Acton, MA): anti-C1 esterase inhibitor (60R-1076, 1:1000); from EMD-Millipore: anti-apolipoprotein J/clusterin (AB825, 1:5000). Membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibody (all 1:10,000 except myoglobin at 1:1000) followed by development with chemiluminescence (Luminata Forte, EMD-Millipore). Membranes were imaged with a Biospectrum 500 Imaging System (UVP). Band densities were quantified and then normalized to total protein stain with ImageJ (NIH, Bethesda, MD).
Statistical analysis
Data from 2D-DIGE was potentially analyzed three ways—healthy males versus male patients, healthy females versus female patients, and all healthy participants versus all patients. DeCyder Differential Analysis Software was used to detect differential protein expression on 2D-DIGE scanned images. Parameters were adjusted for best spot identification and to minimize non-specific background noise. Differential In-gel Analysis (DIA) was used to identify potential spots on each gel, with manual adjustment. Selection criteria was a ratio change >1.5 or < −1.5, P value ≤ 0.05. DIA results were grouped for Biological Variance Analysis, which identified targets across all samples.
Immunoassay and Western blot data were analyzed with SYSTAT 13 version 13.1 (Systat Software Inc, Chicago, IL). Cytokines and chemokines were eliminated from the MR data set to decrease confounding effects of acute infectious processes and to reduce potential interference of heparin. Reproductive hormones also were removed as were analytes with excessive (>75% of total) undetectable values. Within each sample, analytes present at undetectably low levels were set at zero, while analytes present at unquantifiably high levels were set at the maximum detectable value. Values were log transformed when data was non normally distributed. Solo outliers (a single value falling more than 2.5 standard deviations away from the mean) were discarded. Normally distributed data was analyzed with the unpaired Student’s t test. For non-normally distributed, non-transformable variables, the non-parametric Mann Whitney U-test was used. MM immunoassay and Western blot data were analyzed two ways—controls versus all NCL subtypes and controls versus JNCL only. Candidate biomarkers were checked for an effect of age (MR immunoassay only) and gender (all approaches) with analysis of variance (ANOVA) followed by post-hoc analyses with the Student’s t test where appropriate.
Supplementary Material
Complete list of patient samples with their associated demographics and proteomic technique.
Acknowledgments
The authors would like to thank Attila Kovacs for technical assistance and reviewing the manuscript. This work was supported in part by NIH grants R21 NS60185 and R01 NS44310.
Abbreviations
- 2D-DIGE
two dimensional difference gel electrophoresis
- ANOVA
analysis of variance
- apoAIV
apolipoprotein AIV
- apoE
apolipoprotein E
- BDNF
brain derived neurotrophic factor
- CFH
complement factor H
- CK-MB
creatine kinase MB
- CNS
central nervous system
- CSF
cerebrospinal fluid
- FABP
fatty acid binding protein
- HMW
high molecular weight
- ICAM-1
immune cell adhesion molecule 1
- IFN-γ
interferon gamma
- INCL
infantile neuronal ceroid lipofuscinosis
- IRB
institutional review board
- LOX1
lectin-like oxidized LDL receptor 1
- MM
MILLIPLEX MAP
- MR
Myriad-RBM
- JNCL
juvenile neuronal ceroid lipofuscinosis
- LINCL
late infantile neuronal ceroid lipofuscinosis
- NCL
neuronal ceroid lipofuscinosis
- NCAM
neural cell adhesion molecule
- NrCAM
neuronal cell adhesion molecule
- PAI-1
platelet activating inhibitor 1
- TNF-α
tumor necrosis factor alpha
- TSH
thyroid stimulating hormone
- UBDRS
Unified Batten Disease Rating Scale
- VCAM-1
vascular cell adhesion molecule 1
- vWF
von Willebrand’s Factor
- ZAG
zinc-α2-glycoprotein
Footnotes
Conflict of Interest Statement:
The authors declare no conflicts of interest.
Author Contribution: All authors contributed in planning of experiments and drafting of the manuscript. SLH, KLW, and RDG conducted experiments and provided data.
References
- 1.Jalanko A, Braulke T. Neuronal ceroid lipofuscinoses. Biochim Biophys Acta. 2009;1793:697–709. doi: 10.1016/j.bbamcr.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Mole SE. The genetic spectrum of human neuronal ceroid-lipofuscinoses. Brain Pathol. 2004;14:70–76. doi: 10.1111/j.1750-3639.2004.tb00500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gardiner RM. In: Myoclonus and Paroxysmal Dyskinesias. Fahn S, Hallett M, Frucht SJ, Truong DD, editors. Vol. 89. 2002. pp. 211–215. [Google Scholar]
- 4.Cooper JD, Russell C, Mitchison HM. Progress towards understanding disease mechanisms in small vertebrate models of neuronal ceroid lipofuscinosis. Biochim Biophys Acta. 2006;1762:873–889. doi: 10.1016/j.bbadis.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Marshall FJ, de Blieck EA, Mink JW, Dure L, Adams H, Messing S, Rothberg PG, Levy E, McDonough T, DeYoung J, et al. A clinical rating scale for Batten disease: reliable and relevant for clinical trials. Neurology. 2005;65:275–279. doi: 10.1212/01.wnl.0000169019.41332.8a. [DOI] [PubMed] [Google Scholar]
- 6.Williams R. 13th International Conference on Neuronal Ceroid Lipofusinoses (Batten Disease) & Patient Organisation Meeting; London, England. 2012. in press. [Google Scholar]
- 7.Siintola E, Partanen S, Stromme P, Haapanen A, Haltia M, Maehlen J, Lehesjoki AE, Tyynela J. Cathepsin D deficiency underlies congenital human neuronal ceroid-lipofuscinosis. Brain. 2006;129:1438–1445. doi: 10.1093/brain/awl107. [DOI] [PubMed] [Google Scholar]
- 8.Steinfeld R, Reinhardt K, Schreiber K, Hillebrand M, Kraetzner R, Bruck W, Saftig P, Gartner J. Cathepsin D deficiency is associated with a human neurodegenerative disorder. Am J Hum Genet. 2006;78:988–998. doi: 10.1086/504159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michetti F, Corvino V, Geloso MC, Lattanzi W, Bernardini C, Serpero L, Gazzolo D. The S100B protein in biological fluids: more than a lifelong biomarker of brain distress. J Neurochem. 2012;120:644–659. doi: 10.1111/j.1471-4159.2011.07612.x. [DOI] [PubMed] [Google Scholar]
- 10.Cialone J, Adams H, Augustine EF, Marshall FJ, Kwon JM, Newhouse N, Vierhile A, Levy E, Dure LS, Rose KR, et al. Females experience a more severe disease course in Batten disease. J Inherit Metab Dis. 2012;35:549–555. doi: 10.1007/s10545-011-9421-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen AK, Ostergaard JR. Do females with juvenile ceroid lipofuscinosis (Batten disease) have a more severe disease course? The Danish experience. Eur J Paediatr Neurol. 2013;17:265–268. doi: 10.1016/j.ejpn.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Di Lazzaro V, Profice R, Pilato F, Dileone M, Florio L, Tonali PA, Angelucci F. BDNF plasma levels in acute stroke. Neurosci Lett. 2007;422:128–130. doi: 10.1016/j.neulet.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Sharma HS, Johanson CE. In: Slikker W, Andrew RJ, Trembly B, editors. Neuroprotective Agents: Eighth International Neuroprotection Society Meeting; 2007. pp. 112–129. [Google Scholar]
- 14.Laske C, Stransky E, Leyhe T, Eschweiler GW, Maetzler W, Wittorf A, Soekadar S, Richartz E, Koehler N, Bartels M, et al. BDNF serum and CSF concentrations in Alzheimer’s disease, normal pressure hydrocephalus and healthy controls. J Psychiatr Res. 2007;41:387–394. doi: 10.1016/j.jpsychires.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Laske C, Stransky E, Leyhe T, Eschweiler GW, Wittorf A, Richartz E, Bartels M, Buchkremer G, Schott K. Stage-dependent BDNF serum concentrations in Alzheimer’s disease. J Neural Transm. 2006;113:1217–1224. doi: 10.1007/s00702-005-0397-y. [DOI] [PubMed] [Google Scholar]
- 16.Lee JG, Shin BS, You YS, Kim JE, Yoon SW, Jeon DW, Baek JH, Park SW, Kim YH. Decreased Serum Brain-Derived Neurotrophic Factor Levels in Elderly Korean with Dementia. Psychiatry Investig. 2009;6:299–305. doi: 10.4306/pi.2009.6.4.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciammola A, Sassone J, Cannella M, Calza S, Poletti B, Frati L, Squitieri F, Silani V. Low brain-derived neurotrophic factor (BDNF) levels in serum of Huntington’s disease patients. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:574–577. doi: 10.1002/ajmg.b.30501. [DOI] [PubMed] [Google Scholar]
- 18.Scalzo P, Kuemmer A, Bretas TL, Cardoso F, Teixeira AL. Serum levels of brain-derived neurotrophic factor correlate with motor impairment in Parkinson’s disease. J Neurol. 2010;257:540–545. doi: 10.1007/s00415-009-5357-2. [DOI] [PubMed] [Google Scholar]
- 19.Leyhe T, Eschweiler GW, Stransky E, Gasser T, Annas P, Basun H, Laske C. Increase of BDNF Serum Concentration in Lithium Treated Patients with Early Alzheimer’s Disease. J Alzheimers Dis. 2009;16:649–656. doi: 10.3233/JAD-2009-1004. [DOI] [PubMed] [Google Scholar]
- 20.Pillai A, Terry AV, Mahadik SP. Differential effects of long-term treatment with typical and atypical antipsychotics on NGF and BDNF levels in rat striatum and hippocampus. Schizophr Res. 2006;82:95–106. doi: 10.1016/j.schres.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 21.Sakurai T. The role of NrCAM in neural development and disorders--beyond a simple glue in the brain. Mol Cell Neurosci. 2012;49:351–363. doi: 10.1016/j.mcn.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Wang B, Williams H, Du JS, Terrett J, Kenwrick S. Alternative Splicing of Human NrCAM in Neural and Nonneural Tissues. Mol Cell Neurosci. 1998;10:287–295. doi: 10.1006/mcne.1997.0658. [DOI] [PubMed] [Google Scholar]
- 23.Schwarz E, Guest PC, Rahmoune H, Wang L, Levin Y, Ingudomnukul E, Ruta L, Kent L, Spain M, Baron-Cohen S, et al. Sex-specific serum biomarker patterns in adults with Asperger’s syndrome. Mol Psychiatry. 2011;16:1213–1220. doi: 10.1038/mp.2010.102. [DOI] [PubMed] [Google Scholar]
- 24.Hu WT, Chen-Plotkin A, Arnold SE, Grossman M, Clark CM, Shaw LM, Pickering E, Kuhn M, Chen Y, McCluskey L, et al. Novel CSF biomarkers for Alzheimer’s disease and mild cognitive impairment. Acta Neuropathol. 2010;119:669–678. doi: 10.1007/s00401-010-0667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perrin RJ, Craig-Schapiro R, Malone JP, Shah AR, Gilmore P, Davis AE, Roe CM, Peskind ER, Li G, Galasko DR, et al. Identification and validation of novel cerebrospinal fluid biomarkers for staging early Alzheimer’s disease. PloS one. 2011;6:e16032. doi: 10.1371/journal.pone.0016032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woloszynek JC, Coleman T, Semenkovich CF, Sands MS. Lysosomal dysfunction results in altered energy balance. J Biol Chem. 2007;282:35765–35771. doi: 10.1074/jbc.M705124200. [DOI] [PubMed] [Google Scholar]
- 27.Gimenez MAT, Sim JE, Russell JH. TNFR1-dependent VCAM-1 expression by astrocytes exposes the CNS to destructive inflammation. J Neuroimmunol. 2004;151:116–125. doi: 10.1016/j.jneuroim.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 28.James WG, Bullard DC, Hickey MJ. Critical role of the alpha(4) integrin/VCAM-1 pathway in cerebral leukocyte trafficking in lupus-prone MRL/fas(lpr) mice. J Immunol. 2003;170:520–527. doi: 10.4049/jimmunol.170.1.520. [DOI] [PubMed] [Google Scholar]
- 29.Jublanc C, Beaudeux JL, Aubart F, Raphael M, Chadarevian R, Chapman MJ, Bonnefont-Rousselot D, Bruckert E. Serum levels of adhesion molecules ICAM-1 and VCAM-1 and tissue inhibitor of metalloproteinases, TIMP-1, are elevated in patients with autoimmune thyroid disorders: Relevance to vascular inflammation. Nutr Metab Cardiovasc Dis. 2011;21:817–822. doi: 10.1016/j.numecd.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 30.Vercellino M, Votta B, Condello C, Piacentino C, Romagnolo A, Merola A, Capello E, Mancardi GL, Mutani R, Giordana MT, et al. Involvement of the choroid plexus in multiple sclerosis autoimmune inflammation: a neuropathological study. J Neuroimmunol. 2008;199:133–141. doi: 10.1016/j.jneuroim.2008.04.035. [DOI] [PubMed] [Google Scholar]
- 31.Yusuf-Makagiansar H, Anderson ME, Yakovleva TV, Murray JS, Siahaan TJ. Inhibition of LFA-1/ICAM-1 and VLA-4/VCAM-1 as a therapeutic approach to inflammation and autoimmune diseases. Med Res Rev. 2002;22:146–167. doi: 10.1002/med.10001. [DOI] [PubMed] [Google Scholar]
- 32.Ewers M, Mielke MM, Hampel H. Blood-based biomarkers of microvascular pathology in Alzheimer’s disease. Exp Gerontol. 2010;45:75–79. doi: 10.1016/j.exger.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brochu-Gaudreau K, Rehfeldt C, Blouin R, Bordignon V, Murphy BD, Palin MF. Adiponectin action from head to toe. Endocrine. 2010;37:11–32. doi: 10.1007/s12020-009-9278-8. [DOI] [PubMed] [Google Scholar]
- 34.Combs TP, Berg AH, Rajala MW, Klebanov S, Iyengar P, Jimenez-Chillaron JC, Patti ME, Klein SL, Weinstein RS, Scherer PE. Sexual differentiation, pregnancy, calorie restriction, and aging affect the adipocyte-specific secretory protein adiponectin. Diabetes. 2003;52:268–276. doi: 10.2337/diabetes.52.2.268. [DOI] [PubMed] [Google Scholar]
- 35.Fasshauer M, Klein J, Neumann S, Eszlinger M, Paschke R. Hormonal regulation of adiponectin gene expression in 3T3-L1 adipocytes. Biochem Biophys Res Comm. 2002;290:1084–1089. doi: 10.1006/bbrc.2001.6307. [DOI] [PubMed] [Google Scholar]
- 36.Maeda N, Shimomura I, Kishida K, Nishizawa H, Matsuda M, Nagaretani H, Furuyama N, Kondo H, Takahashi M, Arita Y, et al. Diet-induced insulin resistance in mice lacking adiponectin/ACRP30. Nat Med. 2002;8:731–737. doi: 10.1038/nm724. [DOI] [PubMed] [Google Scholar]
- 37.Hietaharju A, Kuusisto H, Nieminen R, Vuolteenaho K, Elovaara I, Moilanen E. Elevated cerebrospinal fluid adiponectin and adipsin levels in patients with multiple sclerosis: a Finnish co-twin study. Eur J Neurol. 2010;17:332–334. doi: 10.1111/j.1468-1331.2009.02701.x. [DOI] [PubMed] [Google Scholar]
- 38.Une K, Takei YA, Tomita N, Asamura T, Ohrui T, Furukawa K, Arai H. Adiponectin in plasma and cerebrospinal fluid in MCI and Alzheimer’s disease. Eur J Neurol. 2011;18:1006–1009. doi: 10.1111/j.1468-1331.2010.03194.x. [DOI] [PubMed] [Google Scholar]
- 39.Thundyil J, Pavlovski D, Sobey CG, Arumugam TV. Adiponectin receptor signalling in the brain. Br J Pharmacol. 2012;165:313–327. doi: 10.1111/j.1476-5381.2011.01560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadigh-Eteghad S, Talebi M, Farhoudi M. Association of apolipoprotein E epsilon 4 allele with sporadic late onset Alzheimer’s disease A meta-analysis. Neurosciences. 2012;17:321–326. [PubMed] [Google Scholar]
- 41.Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10:333–344. doi: 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pitas RE, Boyles JK, Lee SH, Foss D, Mahley RW. Astrocytes synthesize apolipoprotein E and metabolize apolipoprotein E-containing lipoproteins. Biochim Biophys Acta. 1987;917:148–161. doi: 10.1016/0005-2760(87)90295-5. [DOI] [PubMed] [Google Scholar]
- 43.Uchihara T, Duyckaerts C, He Y, Kobayashi K, Seilhean D, Amouyel P, Hauw JJ. ApoE immunoreactivity and microglial cells in Alzheimer’s disease brain. Neurosci Lett. 1995;195:5–8. doi: 10.1016/0304-3940(95)11763-m. [DOI] [PubMed] [Google Scholar]
- 44.Linton MF, Gish R, Hubl ST, Butler E, Esquivel C, Bry WI, Boyles JK, Wardell MR, Young SG. Phenotypes of apolipoprotein B and apolipoprotein E after liver transplantation. J Clin Invest. 1991;88:270–281. doi: 10.1172/JCI115288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fagan AM, Younkin LH, Morris JC, Fryer JD, Cole TG, Younkin SG, Holtzman DM. Differences in the Abeta40/Abeta42 ratio associated with cerebrospinal fluid lipoproteins as a function of apolipoprotein E genotype. Ann Neurol. 2000;48:201–210. [PubMed] [Google Scholar]
- 46.Autti T, Raininko R, Santavuori P, Vanhanen SL, Poutanen VP, Haltia M. MRI of neuronal ceroid lipofuscinosis. II. Postmortem MRI and histopathological study of the brain in 16 cases of neuronal ceroid lipofuscinosis of juvenile or late infantile type. Neuroradiology. 1997;39:371–377. doi: 10.1007/s002340050427. [DOI] [PubMed] [Google Scholar]
- 47.Tyynela J, Cooper JD, Khan MN, Shemilts SJ, Haltia M. Hippocampal pathology in the human neuronal ceroid-lipofuscinoses: distinct patterns of storage deposition, neurodegeneration and glial activation. Brain Pathol. 2004;14:349–357. doi: 10.1111/j.1750-3639.2004.tb00077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Charnay Y, Imhof A, Vallet PG, Kovari E, Bouras C, Giannakopoulos P. Clusterin in neurological disorders: Molecular perspectives and clinical relevance. Brain Res Bull. 2012;88:434–443. doi: 10.1016/j.brainresbull.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 49.Falgarone G, Chiocchia G. In: Adv Cancer Res. Bettuzzi S, Pucci S, editors. Vol. 104. Vol. 104. Elsevier Academic Press Inc; San Diego: 2009. pp. 139–170. [Google Scholar]
- 50.Antonelou MH, Kriebardis AG, Stamoulis KE, Trougakos IP, Papassideri IS. Apolipoprotein J/Clusterin Is a Novel Structural Component of Human Erythrocytes and a Biomarker of Cellular Stress and Senescence. PloS one. 2011:6. doi: 10.1371/journal.pone.0026032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li JY, Xie DD, Jiang JY, Zhou T. Clusterin is a potential biomarker for the renal tubular epithelial cell injury in kidney stone formers. J Urol. 2013;189:E850–E851. [Google Scholar]
- 52.Panico F, Casali C, Rossi G, Rizzi F, Morandi U, Bettuzzi S, Davalli P, Corbetta L, Storelli ES, Corti A, et al. Prognostic role of clusterin in resected adenocarcinomas of the lung. Lung Cancer. 2013;79:294–299. doi: 10.1016/j.lungcan.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 53.Thambisetty M, An Y, Kinsey A, Koka D, Saleem M, Guntert A, Kraut M, Ferrucci L, Davatzikos C, Lovestone S, et al. Plasma clusterin concentration is associated with longitudinal brain atrophy in mild cognitive impairment. Neuroimage. 2012;59:212–217. doi: 10.1016/j.neuroimage.2011.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu HR, Kuo HC, Huang EY, Liang CD, Hwang KP, Lin IC, Sheen JM, Wang TJ, Wang CL, Yang KD. Plasma Clusterin Levels in Predicting the Occurrence of Coronary Artery Lesions in Patients With Kawasaki Disease. Pediatr Cardiol. 2010;31:1151–1156. doi: 10.1007/s00246-010-9769-7. [DOI] [PubMed] [Google Scholar]
- 55.Zschiesche W, Kleine AH, Spitzer E, Veerkamp JH, Glatz JF. Histochemical localization of heart-type fatty-acid binding protein in human and murine tissues. Histochem Cell Biol. 1995;103:147–156. doi: 10.1007/BF01454012. [DOI] [PubMed] [Google Scholar]
- 56.Van Nieuwenhoven FA, Kleine AH, Wodzig WH, Hermens WT, Kragten HA, Maessen JG, Punt CD, Van Dieijen MP, Van der Vusse GJ, Glatz JF. Discrimination between myocardial and skeletal muscle injury by assessment of the plasma ratio of myoglobin over fatty acid-binding protein. Circulation. 1995;92:2848–2854. doi: 10.1161/01.cir.92.10.2848. [DOI] [PubMed] [Google Scholar]
- 57.Ostergaard JR, Rasmussen TB, Molgaard H. Cardiac involvement in juvenile neuronal ceroid lipofuscinosis (Batten disease) Neurology. 2011;76:1245–1251. doi: 10.1212/WNL.0b013e31821435bd. [DOI] [PubMed] [Google Scholar]
- 58.Heywood WE, Madgett TE, Wang D, Wallington A, Hogg J, Mills K, Avent ND. 2D DIGE analysis of maternal plasma for potential biomarkers of Down Syndrome. Proteome Sci. 2011;9:56. doi: 10.1186/1477-5956-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu HR, Kuo HC, Sheen JM, Wang L, Lin IC, Wang CL, Yang KD. A unique plasma proteomic profiling with imbalanced fibrinogen cascade in patients with Kawasaki disease. Pediatr Allergy Immunol. 2009;20:699–707. doi: 10.1111/j.1399-3038.2008.00844.x. [DOI] [PubMed] [Google Scholar]
- 60.Gangadharan B, Antrobus R, Dwek RA, Zitzmann N. Novel serum biomarker candidates for liver fibrosis in hepatitis C patients. Clin Chem. 2007;53:1792–1799. doi: 10.1373/clinchem.2007.089144. [DOI] [PubMed] [Google Scholar]
- 61.Ando T, Nagai K, Chikada M, Okamoto K, Kurokawa MS, Kobayashi T, Kato T, Makuuchi H. Proteomic analyses of aortic wall in patients with abdominal aortic aneurysm. J Cardiovasc Surg. 2011;52:545–555. [PubMed] [Google Scholar]
- 62.Michlmayr A, Bachleitner-Hofmann T, Baumann S, Marchetti-Deschmann M, Rech-Weichselbraun I, Burghuber C, Pluschnig U, Bartsch R, Graf A, Greil R, et al. Modulation of plasma complement by the initial dose of epirubicin/docetaxel therapy in breast cancer and its predictive value. Br J Cancer. 2010;103:1201–1208. doi: 10.1038/sj.bjc.6605909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Domenico M, Scumaci D, Grasso S, Gaspari M, Curcio A, Oliva A, Ausania F, Di Nunzio C, Ricciardi C, Santini AC, et al. Biomarker discovery by plasma proteomics in familial Brugada Syndrome. Front Biosci. 2013;18:564–571. doi: 10.2741/4120. [DOI] [PubMed] [Google Scholar]
- 64.Liu WT, Liu BY, Cai Q, Li JF, Chen XH, Zhu ZG. Proteomic identification of serum biomarkers for gastric cancer using multi-dimensional liquid chromatography and 2D differential gel electrophoresis. Clin Chim Acta. 2012;413:1098–1106. doi: 10.1016/j.cca.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 65.Corzett TH, Fodor IK, Choi MW, Walsworth VL, Turteltaub KW, McCutchen-Maloney SL, Chromy BA. Statistical analysis of variation in the human plasma proteome. J Biomed Biotechnol. 2010;2010:258494. doi: 10.1155/2010/258494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trajkovska V, Marcussen AB, Vinberg M, Hartvig P, Aznar S, Knudsen GM. Measurements of brain-derived neurotrophic factor: methodological aspects and demographical data. Brain Res Bull. 2007;73:143–149. doi: 10.1016/j.brainresbull.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 67.Boschetti E, Chung MC, Righetti PG. “The quest for biomarkers”: are we on the right technical track? Proteomics Clin Appl. 2012;6:22–41. doi: 10.1002/prca.201100039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete list of patient samples with their associated demographics and proteomic technique.