Abstract
Recent results provide evidence that cholesterol is highly accessible for removal from both cell and model membranes above a threshold concentration that varies with membrane composition. Here we measured the rate at which methyl-β-cyclodextrin depletes cholesterol from a supported lipid bilayer as a function of cholesterol mole fraction. We formed supported bilayers from two-component mixtures of cholesterol and a PC (phosphatidylcholine) lipid, and we directly visualized the rate of decrease in area of the bilayers with fluorescence microscopy. Our technique yields the accessibility of cholesterol over a wide range of concentrations (30–66 mol %) for many individual bilayers, enabling fast acquisition of replicate data. We found that the bilayers contain two populations of cholesterol, one with low surface accessibility and the other with high accessibility. A larger fraction of the total membrane cholesterol appears in the more accessible population when the acyl chains of the PC-lipid tails are more unsaturated. Our findings are most consistent with the predictions of the condensed-complex and cholesterol bilayer domain models of cholesterol-phospholipid interactions in lipid membranes.
Introduction
Mammalian cells regulate the concentration of cholesterol in their plasma membrane (∼40 mol %) and endoplasmic reticulum (∼5 mol %). When the concentration of cholesterol in the cell plasma membrane falls below a physiological set point, activation of a signaling pathway results in proteins in the endoplasmic reticulum upregulating cellular cholesterol production (1, 2, 3). The cell may maintain this set point by monitoring the chemical activity (effective concentration) of cholesterol in the plasma membrane, which can be significantly different than the total plasma membrane cholesterol concentration (1, 4, 5, 6). In humans, diets high in saturated fats correlate with hypercholesterolemia, whereas diets high in monounsaturated fats are comparatively hypocholesterolemic (7). The link between saturated fats and hypercholesterolemia may lie in the disparity between the activity and total concentration of cholesterol in the plasma membrane. Cholesterol interacts more favorably with saturated phospholipids than unsaturated phospholipids in membranes (8). These favorable interactions may reduce the activity of cholesterol with respect to total concentration, such that cells with higher concentrations of saturated lipids trigger cholesterol production even when the plasma membrane contains elevated levels of cholesterol.
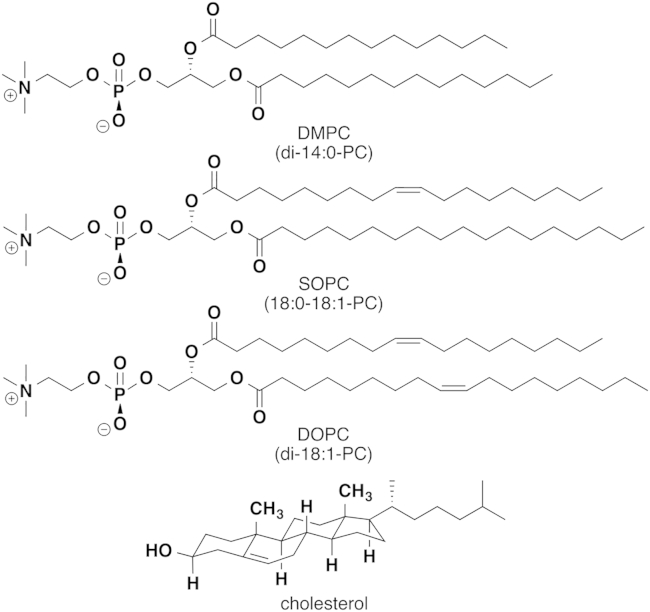
Direct measurement of the activity of cholesterol in a membrane is difficult. Determining the surface accessibility of cholesterol is more experimentally tractable, and this quantity is hypothesized to be proportional to the activity of cholesterol within the membrane (9). Here, we investigated whether all cholesterol molecules in a two-component PC (phosphatidylcholine) lipid bilayer at high concentrations of cholesterol (30–66 mol %) are equally accessible for removal by methyl-β-cyclodextrin (mβCD). Specifically, we used fluorescence microscopy to image the decrease in area of PC-lipid membranes as mβCD selectively pulled cholesterol from them. We used the rate of decrease in membrane area to determine the rate of cholesterol depletion as a function of both the mole fraction of cholesterol (χC) and the degree of unsaturation of the PC-lipid tails. Molecular structures of the three phospholipids studied (DMPC, SOPC, and DOPC in order of increasing acyl-chain unsaturation) and cholesterol are shown in Fig. 1.
Figure 1.
The molecular structures of DMPC (di-14:0-PC), SOPC (18:0-18:1-PC), DOPC (di-18:0-PC), and cholesterol. DMPC has two saturated tails. SOPC has one saturated and one monounsaturated tail. DOPC has two monounsaturated tails.
Previously, cholesterol-dependent cytolysins and cholesterol oxidase have been used to assay the accessibility of cholesterol in two-component PC-lipid membranes (10, 11, 12, 13, 14, 15, 16, 17, 18). The binding of two cytolysins, perfringolysin O and anthrolysin O, to cholesterol in the membrane increases sharply above a single χC characteristic to the PC-lipid in the bilayer (10, 11, 12, 13, 14, 15). Generally, this χC decreases with increasing lipid tail unsaturation. However, slight structural modification of the protein (19) or changes in pH (10) can cause this characteristic χC to shift independently of membrane lipid composition. These sensitivities make it difficult to isolate changes in the accessibility of cholesterol from changes in the binding behavior of the cytolysin. The use of monomeric cytolysin subunits likely alleviates this problem (15). Cholesterol oxidase converts cholesterol into cholest-4-en-3-one, altering the composition of the bilayer. This structural change should have minimal impact on the initial rate of oxidation measured, but the eventual widespread replacement of cholesterol, even by structurally similar sterols, can drastically change membrane properties (20, 21). There have been disagreements in the literature stemming from the interpretation of results from cholesterol oxidase assays. In particular, some studies claim there is only one χC above which cholesterol accessibility increases sharply (16), whereas others claim to observe several spikes in cholesterol oxidase activity at specific values of χC (17, 18). Results from previous cytolysin and cholesterol oxidase studies on two-component lipid bilayers are summarized in Table 1. Our mβCD area depletion assay avoids significant chemical modification of membrane components while giving an unambiguous readout of the accessibility of cholesterol as a function of χC.
Table 1.
Previously Published Measurements of Cholesterol Accessibility for Two-Component PC-Lipid Bilayers
| PC-Lipida | Characteristic χCb | Assay | Reference |
|---|---|---|---|
| di-16:0 | 0.63, 0.58 | COD | (18) |
| 0.50 | COD | (16) | |
| di-14:0 | 0.50 | COD | (16) |
| 18:0-18:1 | 0.50 | COD | (16) |
| 16:0-18:1 | 0.58, 0.52, 0.40, 0.25 | COD | (18) |
| 0.50 | COD | (16) | |
| 0.334, 0.250 | COD | (17)c | |
| di-18:1 | 0.62, 0.57, 0.51, 0.40, 0.25 | COD | (18) |
| 0.33 | COD | (16) | |
| di-18:0 | 0.47d | PFO | (11) |
| di-16:0 | 0.49d | PFO | (11) |
| di-14:0 | 0.51d | PFO | (11) |
| 18:0-18:1 | 0.42d | PFO | (11) |
| 16:0-18:1 | 0.47d | PFO | (14) |
| 0.45d | PFO | (14) | |
| 0.45 | PFO | (12) | |
| 0.44d | PFO | (11) | |
| 0.44d | PFO | (13) | |
| 0.28d | PFO, pH = 7.4 | (10) | |
| 0.24d | PFO, pH = 5.1 | (10) | |
| di-18:1 | 0.45 | PFO-D4e | (15) |
| 0.45 | ALO-D4e | (15) | |
| 0.41 | PFO | (15) | |
| 0.37d | PFO | (11) | |
| 0.35 | PFO | (12) | |
| 0.34d | PFO | (14) | |
| 0.26 | ALO | (15) | |
| 0.26d | PFO, pH = 7.4 | (10) | |
| 0.20d | PFO, pH = 7.0 | (10) | |
| di-(4Me-16:0) | 0.31 | PFO | (15) |
| 0.27 | PFO-D4e | (15) | |
| 0.27 | ALO-D4e | (15) | |
| 0.25 | PFO | (12) | |
COD, cholesterol oxidase; PFO, perfringolysin O; ALO, anthrolysin O.
Lipids are listed by the number of carbons and degree of unsaturation of their tails (e.g., DMPC is di-14:0-PC).
Characteristic χC refers to the one or several mole fraction(s) of cholesterol at or above which the accessibility of cholesterol increases sharply. Only characteristic χC ≥ 0.20 are listed.
The researchers did not report on bilayers with χC > 0.345.
This value was interpreted from a graph as the half-maximum of cytolysin binding.
This version of the protein is monomeric and does not cause pores to form in the membrane.
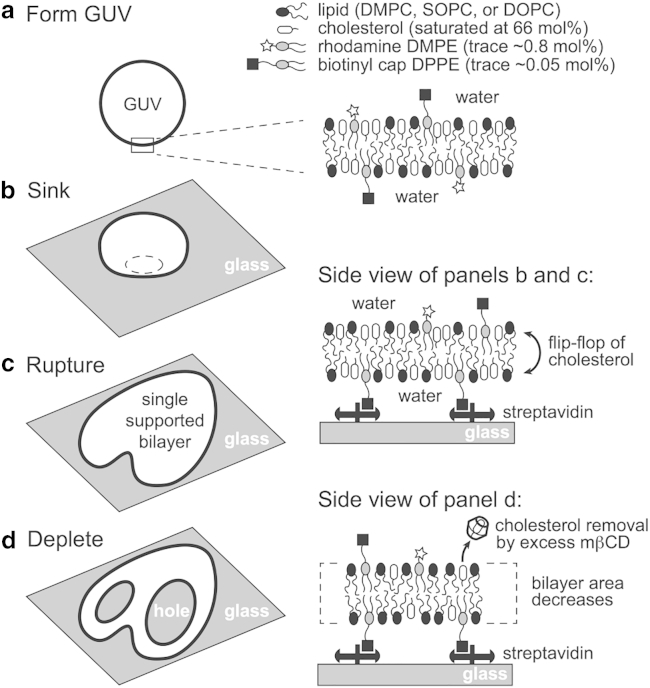
There are three significant experimental challenges to overcome in measuring mβCD-induced area depletion of bilayers to determine the accessibility of membrane cholesterol over a wide range of χC. First, membrane fluctuations must be suppressed so the decrease in area can be measured accurately. Second, membrane tension must be approximately constant throughout the depletion process so that changes in tension do not influence the measured accessibility; changes in membrane tension makes measurements involving vesicles problematic (22, 23). Third, knowledge is needed of the area per molecule of the membrane as a function of cholesterol concentration. This quantity is straightforward to measure for monolayers (24) but much more challenging to determine for bilayers (25). We addressed the first two challenges by developing a method of rupturing biotinylated giant unilamellar vesicles (GUVs) onto a glass substrate coated with streptavidin that yields supported lipid bilayers (SLBs). Upon depletion of cholesterol with mβCD, a few large holes form in the bilayer, suggesting that strong bilayer-surface pinning interactions are minimized. We tackled the third challenge by aggregating published bilayer thickness and area per unit cell data and converting those values to molecular areas. An overview of our experimental procedure is shown in Fig. 2 (full details appear in the Materials and Methods).
Figure 2.
Overview of the experimental procedure. (a) A GUV saturated with cholesterol at 66 mol % is produced via electroformation. (b) The GUV sinks to the bottom of the experimental chamber. (c) It then ruptures onto a streptavidin-functionalized glass coverslip, forming a heart-shaped SLB. Cholesterol flip-flop is rapid between the upper and lower leaflets of the fluid bilayer (39, 40, 41, 42). (d) mβCD is added to the chamber, where it selectively removes cholesterol from the SLB. Large holes form in the SLB as cholesterol is depleted, and the bilayer area decreases.
It is important that we conduct our experiments on bilayers rather than monolayers—as has been achieved previously (9)—because large differences in miscibility behavior between monolayer and bilayer systems imply that cholesterol-phospholipid interactions differ in the two systems (26). Also, our goal is to resolve disagreements about which models of cholesterol-lipid interactions best describe the accessibility of cholesterol in bilayer systems. The clearest predictions from these models apply to bilayers comprised of a single lipid and cholesterol, which led us to use binary membranes in our experiments. Membranes composed of certain ternary mixtures of lipids and cholesterol are known to phase-separate at common experimental temperatures (27). Reports of two rates of cholesterol efflux from cells and from bilayers composed of ternary lipid mixtures (22, 28, 29, 30) are challenging to interpret if the membrane demixes into coexisting liquid phases.
Here we show that the accessibility of cholesterol for removal by mβCD increases sharply above a single characteristic χC specific to the PC-lipid in the membrane. Our membranes are free of macroscopic phase separation. Our results show that cholesterol exists in multiple populations with distinct accessibilities in two-component PC-lipid membranes that appear otherwise homogeneous. Our method enables us to distinguish between different models of cholesterol-phospholipid interactions in lipid bilayers.
Materials and Methods
Chemicals
DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine, di-14:0-PC), SOPC (1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine, 18:1-18:0-PC), DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine, di-18:1-PC), biotinyl cap DPPE (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-n-(cap biotinyl), and rhodamine DMPE (1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-n-(lissamine rhodamine B sulfonyl) were from Avanti Polar Lipids (Alabaster, AL). Cholesterol was from Sigma-Aldrich (St. Louis, MO). All lipids were used without further purification and were stored at −20°C. Biotinyl cap DPPE was stored in chloroform/methanol/water (65:35:8 by volume), and all other lipids were stored in chloroform. Streptavidin and mβCD were from Sigma-Aldrich. Glucose and sucrose were from Fisher Scientific (Waltham, MA). All water was purified to 18 MΩ-cm with a Barnstead filtration system from Thermo Scientific (Waltham, MA).
Giant unilamellar vesicle formation
GUVs were generated by electroformation as in previous experiments (Fig. 2 a) (27). Electroformation was conducted using mixtures of PC-lipid/cholesterol/rhodamine DMPE/biotinyl cap DPPE (25:75:0.8:0.05 by mole) to produce GUVs saturated in cholesterol at 66 mol %; excess cholesterol precipitates as crystalline cholesterol monohydrate (21, 31). Briefly, 0.25 mg of lipids were dissolved in chloroform and spread onto two 37.5 × 25 mm glass slides coated with indium-tin-oxide (Delta Technologies, Loveland, CO). The lipid-coated slides were placed under vacuum for 30 min to remove solvent. Pairs of slides were assembled face-to-face separated by a 1-mm gap maintained by two Teflon spacers. The gap was filled with 200 mM sucrose, and the edges were sealed with vacuum grease. The two indium-tin-oxide surfaces were connected to an AC voltage of 1.5 V at 10 Hz for 1 h at 60°C. The resulting GUV-rich solution was diluted with 3.5 mL of 200 mM sucrose at 60°C, then cooled to room temperature (24–26°C). This temperature is above the gel-liquid coexistence temperature of all vesicles studied (32, 33). All experiments reported here featured vesicles that ruptured smoothly onto solid supports and that exhibited uniform distributions of dye-labeled lipids. We find that vesicles in the gel phase (e.g., 100% DMPC vesicles below 24°C) often do not rupture smoothly and that the dye-labeled lipids often accumulate at the edges of the resulting SLBs. Experiments were performed within 4 h of vesicle formation.
Supported lipid bilayer formation and cholesterol depletion
A 25 × 25 mm glass coverslip (Fisher Scientific) was plasma-etched (Harrick, Ithaca, NY) for 50 s. A quantity of 1.5 mL of 0.1 mg/mL streptavidin in water was deposited on the coverslip. After 20 min, the water was poured off, and the coverslip was adhered with vacuum grease to form the base of a cylindrical well with a radius of 10 mm and a height of 12 mm. The chamber was gently rinsed with water five times and placed on the stage of an inverted epifluorescence microscope.
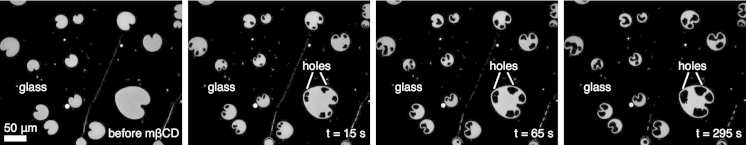
One milliliter of 200 mM glucose was added to the chamber, followed by 20–50 μL of GUV-rich solution. GUVs sank to the bottom of the chamber (Fig. 2 b). Within 1–2 min, hundreds of vesicles ruptured onto the coverslip to create a field of heart-shaped SLBs spatially separated by bare glass (Figs. 2 c and 3). The asymmetric rupture mechanism that produces heart-shaped SLBs has been explored previously (34), and we found our protocol predominantly yields this rupturing pattern. The chamber was washed with 1 mL of 200 mM glucose 10 times. Care was taken to ensure that SLBs were never directly exposed to air. After an initial image was collected, 2 mL of 3.75 mM (for membranes with DMPC or SOPC) or 1.5 mM (for membranes with DOPC) mβCD in 200 mM aqueous glucose was added to the experimental chamber, yielding 3 mL of 2.5 mM (for membranes with DMPC or SOPC) or 1 mM (for membranes with DOPC) mβCD in 200 mM aqueous glucose. Approximately 5 s later, images were collected every 2 s with 500 ms exposures for a total of 300 s (DMPC or DOPC) or 100 s (SOPC) (Figs. 2 d and 3). All experiments were performed at 24–26°C. Two different concentrations of mβCD were used because 2.5 mM mβCD removes cholesterol from DOPC bilayers too quickly to accurately measure the rate of area depletion, whereas 1 mM mβCD depletes cholesterol prohibitively slowly from SOPC and DMPC bilayers. In total, we obtained data from 82 distinct bilayers of cholesterol with DMPC, 95 with SOPC, and 49 with DOPC.
Figure 3.
Micrographs of the cholesterol depletion process. Heart-shaped regions (gray) are individual supported lipid bilayers formed from the rupture of giant unilamellar vesicles onto a glass coverslip (black). mβCD is added at t = 0 s. mβCD removes cholesterol from the supported lipid bilayers. Large holes form in the bilayers as their areas decrease, further revealing the glass coverslip beneath them. DMPC/cholesterol bilayers are shown here, [mβCD] = 2.5 mM.
At concentrations <5–20 mM, mβCD selectively removes cholesterol from PC-lipid bilayers, leaving the phospholipids behind and the membrane intact (35, 36, 37, 38). As a control to ensure that our procedure does not remove significant amounts of PC-lipids from the bilayer, we performed our assay using SLBs of pure DMPC, SOPC, or DOPC. No area depletion was observed on the timescale of our experiment.
It is likely that mβCD pulls cholesterol from only the upper leaflet of the SLBs. Because the rate of cholesterol flip-flop is on the order of milliseconds in fluid phase lipid bilayers (39, 40, 41, 42) and our experiment occurs on the order of seconds, we expect that cholesterol equilibrates across the leaflet of our bilayers faster than we could detect. If cholesterol flip-flop were hindered and cholesterol could only move between the two leaflets by edge diffusion, we would expect to see depletion rates vary with SLB size. We found no correlation between SLB size and cholesterol depletion rate (see Fig. S1 in the Supporting Material), nor did we find evidence of an asymmetric distribution of cholesterol across the two leaflets in our observations.
Images and image processing
Epifluorescence microscopy was performed with a 10× objective on an inverted microscope (Nikon, Melville, NY) with a Coolsnap fx charge-coupled device camera (Photometrics, Tucson, AZ). All micrographs were analyzed using the open-source software package Fiji (43). Each time series of images was converted to a series of binary images using Fiji’s isodata algorithm. The number of pixels in each SLB as a function of time was determined using Fiji’s Analyze Particles function. The fraction of bilayer area remaining is determined by dividing the area remaining of each bilayer by its initial area. We imaged 3–15 separate SLBs per experiment. Data for any bilayer with an initial area of fewer than 250 μm2 was discarded to limit sensitivity to noise in the area measurement. Occasionally, over the course of an experiment, a free-floating GUV or other lipid debris drifted over one of the bilayers being imaged, briefly obfuscating measurement of the bilayer area. In this case, the affected data points were discarded.
Curve fitting
We fit all data using the freely available Python module PyMC (44). PyMC implements Markov chain Monte Carlo algorithms to provide a computationally tractable approach to Bayesian curve fitting. For each fit, we started with a uniform prior probability distribution and generated 100,000 samples of the posterior distribution using a likelihood function that assumes independent, unbiased, and normally distributed errors in the data. The first 20,000 of these samples were discarded as burn-in to avoid sampling from the Markov chain before it reached its equilibrium distribution. We determined the means and standard deviations of fitting parameters and derived values in the results directly from these sets of 80,000 samples (see Fig. S2 for plots of the sampled posterior distribution).
Bilayer area per molecule
We aggregated published bilayer electron density peak-to-peak distances, dpp, determined by x-ray diffraction (45, 46, 47) and area per unit cell data, AUC, determined by neutron scattering (48, 49) or jointly analyzed neutron and x-ray scattering (50, 51) for bilayers comprised of cholesterol and either DMPC, SOPC, or DOPC at 30°C (see Tables S1–S5 in the Supporting Material for all data used).
From x-ray experiments, we determined the thickness of the approximately incompressible hydrocarbon region of the bilayer as a function of mole fraction of cholesterol, dHC(χC), as
| (1) |
where dH1 is the distance from the peak of the x-ray scattering electron density profile to the interface of the hydrocarbon region of the bilayer, as determined by the joint analysis of x-ray and neutron scattering data for single-component bilayers consisting of the appropriate PC-lipid (50, 51). It is likely that dH1 varies with χC, but there are no published values of this relationship for the systems we studied. We therefore approximated dH1 as constant with respect to χC.
From dHC, we determined the average area per molecule in the bilayer, aavg, as
| (2) |
where VHC is the average volume per molecule in the hydrophobic portion of the bilayer, computed as
| (3) |
and where VL, VH, and VC are the reported volumes of the PC-lipid (52, 53), the PC-headgroup (54, 55), and a single cholesterol molecule in a PC-lipid bilayer (53), respectively. It is likely that cholesterol does not lie entirely within the hydrocarbon portion of the bilayer (14, 46), but no value for the fraction of excluded volume has been reported. We therefore approximated cholesterol as being completely located within the hydrocarbon region of the bilayer.
From neutron scattering experiments, we determined aavg as
| (4) |
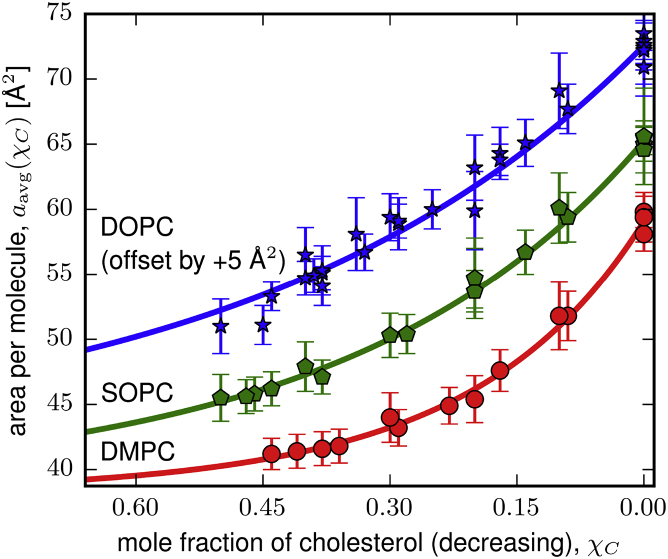
We found that aavg is well described as exponentially increasing with decreasing χC,
| (5) |
where p1, p2, and p3 are the fitting parameters (see Fig. 4). The fitting parameters are summarized in Table 2. We do not assign a physical meaning to these fitting parameters or relationship, and we use this analytic form only as an approximation of aavg to enable further analysis.
Figure 4.
The average area per molecule as a function of mole fraction of cholesterol, aavg (χC), for two-component bilayers of cholesterol with DMPC (circles, red line), SOPC (pentagons, green line), or DOPC (stars, blue line). All data (symbols) shown here are determined from previously reported measurements of bilayer thickness or area per unit cell conducted at 30°C (45, 46, 47, 48, 49, 50, 51). The average area per molecule is well approximated as exponentially increasing with decreasing cholesterol mole fraction (colored lines). DOPC data is offset (increased) by 5 Å2 to avoid overlap with the SOPC data. To see this figure in color, go online.
Table 2.
Fitting Parameters for the Average Area per Molecule in a Bilayer as a Function of Mole Fraction of Cholesterol, aavg(χC)
| Lipid | p1 (Å2) | p2 (Å2) | p3 |
|---|---|---|---|
| DMPC | 38.1 ± 2.4 | 21.0 ± 2.4 | 5.1 ± 1.4 |
| SOPC | 38.5 ± 3.0 | 26.8 ± 2.9 | 2.9 ± 0.7 |
| DOPC | 36.5 ± 2.1 | 31.1 ± 2.1 | 2.2 ± 0.3 |
The method of generating uncertainties in the fitting parameters is described in the Curve Fitting subsection.
Results
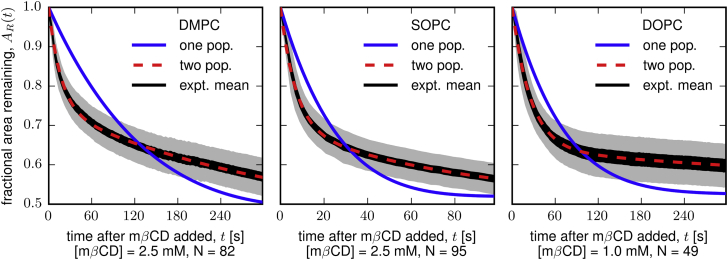
We added mβCD to an aqueous solution above a field of SLBs and recorded a time-series of micrographs to determine the rate at which cholesterol is depleted from the lipid bilayers. Full experimental details are compiled in the Materials and Methods. A representative time-series of images for bilayers of DMPC and cholesterol appears in Fig. 3. We collected similar time-series for bilayers of SOPC and cholesterol and for bilayers of DOPC and cholesterol. As cholesterol is depleted from each bilayer, a few holes form and grow. We plot the fraction of each bilayer area remaining as a function of time after mβCD is added, AR(t). In Fig. 5 the mean of the experimental data ± two standard errors is shown in black, and the standard deviation of the data is shown in gray. Our goal is to use AR(t) to quantitatively determine how the rate of cholesterol depletion depends on χC.
Figure 5.
The fraction of bilayer area remaining as a function of time, AR(t), after addition of mβCD. AR(t) decreases as mβCD depletes cholesterol from the bilayer. (Black region) Our experimental mean ± 2 SE; (gray region) standard deviation of our data. The model with two cholesterol populations describes the data well (red dotted line), whereas the model with only one cholesterol population underfits the data (blue line). N is the number of replicate bilayers studied. To see this figure in color, go online.
Our approach to analyzing AR(t) is as follows. We define
| (6) |
| (7) |
where nL is the number of PC-lipid molecules initially in the bilayer, χC is the mole fraction of cholesterol in the bilayer, nC is the monotonically decreasing number of cholesterol molecules in the bilayer, and aavg is the composition-dependent average area per molecule in the bilayer.
Because we deplete with a gross excess of mβCD, the concentration of mβCD remains approximately constant throughout the experiment. Therefore, the rate of depletion for a given SLB varies with nC only. If all cholesterol in the bilayer were equally accessible by mβCD and this accessibility were independent of χC, then the rate law governing the depletion process would be pseudo-first order in nC with
| (8) |
| (9) |
where k1 is the depletion rate constant. Then by Eq. 7,
| (10) |
| (11) |
where k1 is the single fitting parameter used to generate the one-population fit shown in Fig. 5 (blue line). This model underfits our data, implying that a model in which all cholesterol in the membrane is in a single population is inadequate to describe our system.
If instead there were two independent populations of cholesterol, one with high accessibility and one with low accessibility, then the depletion would be the sum of two pseudo-first-order processes with
| (12) |
| (13) |
where nf and ns are the numbers of cholesterol molecules in the two populations with corresponding rate constants kf and ks. We choose kf > ks so that kf (ks) represents the fast (slow) cholesterol population. By Eq. 7,
| (14) |
We then use the fact that nf(0) = nC(0) – ns(0), and define as the initial mole fraction of the slow population of cholesterol, ≡ ns(0)/(nL + nC(0)), to obtain
| (15) |
where , ks, and kf are the three fitting parameters used to generate the two-population fit shown in Fig. 5 (red dotted line). This model describes our data well, and a summary of the fitting parameters is given in Table 3. This model is the simplest description of a membrane with more than one population of cholesterol. It is possible that there are more than two populations of cholesterol or that the two populations readily interconvert, but adding further complexity to the model is unwarranted by our data (see Fig. S3 for a three-population model).
Table 3.
Experimental Parameters and Fitting Results for the Two-Population Model of Cholesterol
| Lipid | Na | [mβCD] (mM)b | c | ks × 103 (s−1)d | kf × 103 (s−1)e |
|---|---|---|---|---|---|
| DMPC | 82 | 2.5 | 0.391 ± 0.002 | 2.38 ± 0.03 | 57.4 ± 1.2 |
| SOPC | 95 | 2.5 | 0.312 ± 0.004 | 8.2 ± 0.2 | 138.2 ± 3.9 |
| DOPC | 49 | 1.0 | 0.226 ± 0.004 | 0.98 ± 0.10 | 36.7 ± 0.8 |
The method of generating uncertainties in the fitting parameters is described in the Curve Fitting subsection.
The number of replicate bilayers measured.
The concentration of mβCD used (see the Materials and Methods).
Fitting parameter corresponding to the initial mole fraction of the slow population of cholesterol.
Fitting parameter corresponding to the rate constant for the depletion of the slow population of cholesterol.
Fitting parameter corresponding to the rate constant for the depletion of the fast population of cholesterol.
Discussion
We found evidence for two populations of cholesterol in bilayers containing cholesterol and either DMPC, SOPC, or DOPC. The fraction of cholesterol that is more highly accessible increases with the degree of unsaturation of the PC-lipid’s acyl chains. These results, in conjunction with the previous literature results in Table 1, enable us to assess current models of cholesterol-phospholipid interactions.
Models of cholesterol-phospholipid interactions
There are four commonly referenced descriptions of cholesterol-phospholipid interactions in membranes that yield distinct predictions about how the accessibility of cholesterol should vary with composition. These are the umbrella, superlattice, condensed-complex, and cholesterol bilayer domain models (Table 4).
Table 4.
Summary of Models of Cholesterol-Phospholipid Interactions in Lipid Membranes
| Model | Number of Characteristic χCa (0.66 > χC > 0.30) | Dependent on PC-Lipid Head or Tail?b |
|---|---|---|
| Umbrella (at solubility limit) | 0 | head |
| Condensed-complex | 1 | tail |
| Cholesterol bilayer domain | 1 | both |
| Umbrella (regular distributions) | ≥3 | head |
| Superlattice | ≥3 | head |
The number of characteristic χC at which the accessibility of cholesterol is predicted to change sharply.
Statement of whether the predictions of the model vary with the structure of the phospholipid head, tail, or both.
At the solubility limit of cholesterol in the bilayer, the umbrella model predicts nonspecific cholesterol-phospholipid interactions that are independent of the PC-lipid tail (56). The large hydrophilic PC-headgroups form a protective canopy under which cholesterol diffuses freely. Each PC-umbrella shields up to two molecules of cholesterol from water. This model successfully predicts that the solubility limit of cholesterol in PC-lipid bilayers is largely independent of PC-lipid tail structure (31), with notable exceptions being polyunsaturated (57, 58) and methylated (59) PC-lipids. However, by the shielding mechanism alone, all cholesterol in the bilayer should be equally accessible for removal by mβCD. Our results provide evidence that there are at least two populations of cholesterol with distinct accessibilities, and treating all cholesterol as equally accessible underfits our data (Fig. 5, blue line).
Both the umbrella and superlattice models predict that bilayers contain latticelike, regular distributions of cholesterol and phospholipids at several characteristic values of χC. By the umbrella model, lattice formation is driven by unfavorable multibody cholesterol-phospholipid interactions; the energy of these interactions increases nonlinearly as the number of cholesterol molecules per phospholipid increases (56). By the superlattice model, long-range cholesterol-cholesterol repulsions drive lattice formation (60). An essential similarity between these models is that both claim that the characteristic χC at which regular distributions form are independent of the degree of unsaturation of the PC-lipid tail (18, 56). The results reported in Wang et al. (17) and Ali et al. (18) are consistent with some of the predictions of these models. However, our results clearly show that the accessibility of cholesterol depends on the PC-lipid tail, with starkly different values of , ks, and kf for the three PC-lipids studied (Table 3). These models also predict a change in cholesterol accessibility at several values of χC. Our results show that a two-population model of cholesterol with a single characteristic χC is sufficient to describe our systems.
The third model, the condensed-complex model, predicts that cholesterol and phospholipids react reversibly to form thermodynamically stable complexes with a well-defined stoichiometry characteristic to the phospholipid tail (61). These complexes behave as a third chemical species in the two-component bilayer. Cholesterol accessibility increases once χC exceeds the complex stoichiometry. At this point, the population of cholesterol that is more accessible exists in excess of the amount of cholesterol needed to fully partner with each PC-lipid. Our results, as well as many results reported in the literature (10, 11, 12, 13, 14, 15, 16), are consistent with this model. We found our data are well described with two populations of cholesterol and that the characteristic χC at which cholesterol accessibility begins to increase varies with the degree of unsaturation of the PC-lipid tail.
The fourth model, the cholesterol bilayer domain model, states that cholesterol in excess of a threshold mole fraction, χCBD, forms submicroscopic domains of bilayer-thick, tail-to-tail cholesterol monohydrate that are soluble in the membrane (62, 63, 64, 65). Signatures of these domains in two-component PC-lipid bilayers have been interpreted from x-ray diffraction, neutron scattering, and electron paramagnetic resonance (Table 5) (45, 66, 67, 68, 69, 70). Our results are also consistent with this model. The characteristic χC at which cholesterol accessibility begins to increase would correspond to the formation of cholesterol bilayer domains made of cholesterol in excess of χCBD; cholesterol in these domains would be less shielded from the aqueous phase than those in PC-lipid rich regions.
Table 5.
Experimental Reports of Cholesterol Bilayer Domains in Two-Component PC-Lipid Bilayers
| PC-Lipida | χCBDb | Technique | Reference |
|---|---|---|---|
| di-14:0 | 0.50 | electron paramagnetic resonance | (69) |
| 16:0-18:1 | 0.50 | (67) | |
| di-16:0 | 0.325 | neutron scattering | (70) |
| di-16:0 | 0.54 ± 0.02 | x-ray diffraction | (66) |
| di-14:0 | 0.44 | (45) | |
| 0.40 | (68) | ||
| 18:0-18:1 | 0.47 | (45) | |
| di-18:1 | 0.40 | (45) |
Lipids are listed by the number of carbons and degree of unsaturation of their tails (e.g., DMPC is di-14:0-PC).
The mole fraction of cholesterol above which signatures of cholesterol bilayer domains are reported.
These four models need not be mutually exclusive. The cholesterol bilayer domain model makes predictions only about cholesterol concentrations above χCBD; it does not preclude the predictions of the condensed-complex model or the regular distributions predicted by the superlattice or umbrella models below χCBD. Attempts have been made to resolve the condensed-complex and superlattice models by treating the condensed-complexes as subunits that aggregate to form larger superlattice clusters over time (71). Also, for χC < 0.50, the umbrella and superlattice models yield similar characteristic values of χC (56).
Condensed-complex stoichiometry and χCBD
From our two-cholesterol population model, we derived values pertinent to the condensed-complex and cholesterol bilayer domain models (Table 6). We determined the values of the phospholipid-cholesterol complex stoichiometries that result from applying the condensed-complex model to our data. This stoichiometry is equivalent to the ratio of the number of cholesterol molecules in the less accessible (slow) pool to the total number of PC-lipid molecules in the bilayer. We compute this ratio as
| (16) |
Table 6.
Values Derived from a Two-Population Model of Cholesterol
| Lipid | Complex Stoichiometrya | χCBDb | kf/ksc |
|---|---|---|---|
| DMPC | 1.150 ± 0.006 | 0.535 ± 0.001 | 24.1 ± 0.4 |
| SOPC | 0.92 ± 0.01 | 0.479 ± 0.004 | 16.8 ± 0.4 |
| DOPC | 0.67 ± 0.01 | 0.399 ± 0.005 | 37.9 ± 3.3 |
The method of generating uncertainties is described in the Curve Fitting subsection.
The average number of cholesterol molecules complexed with a single PC-lipid molecule in the context of the condensed complex-model.
The mole fraction of cholesterol above which cholesterol bilayer domains would be expected to exist in the context of the cholesterol bilayer domain model. This value is most directly comparable to the characteristic χC values in Table 1.
The ratio of depletion rate constants between the fast and slow populations of cholesterol.
In the context of the cholesterol bilayer domain model, we determine the mole fraction of cholesterol above which cholesterol bilayer domains would form in the bilayer, χCBD. This quantity is approximated by assuming that all cholesterol in the more accessible population is located in cholesterol bilayer domains. This value is most directly comparable to the characteristic χC values reported in Table 1. We determine
| (17) |
We also compute the ratio of the depletion rate constants of the two populations of cholesterol (kf/ks).
Depletion rate coefficient
Because the two-component membranes we investigate appear homogeneous, it is natural to think of the depletion process as pseudo-first order in nC with a composition-dependent rate coefficient of depletion, kD:
| (18) |
| (19) |
| (20) |
Our depletion rate coefficient is proportional to the average accessibility of all cholesterol in the bilayer. Encapsulated within kD is the effect of the concentration of mβCD on the rate of depletion. For purposes of comparison with other literature values, we treat the depletion as second order in mβCD (35) and therefore scale our results for kD of DOPC by a factor of 6.25 (2.52) to account for differences in the concentration of mβCD used (Table 3).
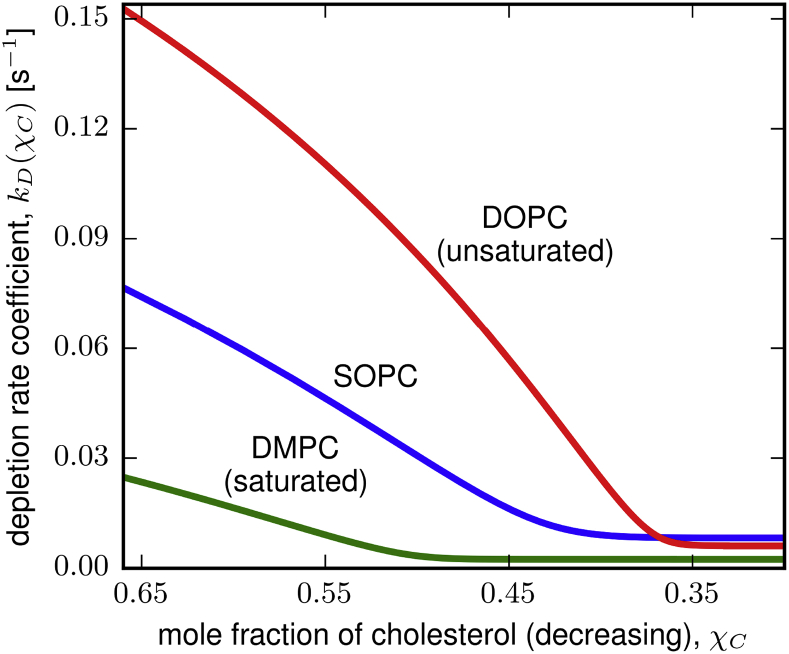
In Fig. 6, we show that, at a given χC, kD increases with the degree of unsaturation of the PC-lipid tails. This is consistent with cholesterol interacting more favorably with saturated lipids than unsaturated lipids (8). The value of kD decreases sharply as the more readily accessible population of cholesterol is depleted. The proportion of cholesterol in this population at high χC increases with the degree of lipid tail unsaturation.
Figure 6.
The depletion rate coefficient as a function of the mole fraction of cholesterol in the bilayer, kD(χC). The rate coefficient decreases sharply as the more accessible population of cholesterol is depleted. The fraction of cholesterol in this population increases with increasing PC-lipid tail unsaturation from DMPC (green) to SOPC (blue) to DOPC (red). At a given χC, the rate coefficient increases with increasing lipid tail unsaturation. We treat the depletion process as second order in mβCD (35) and scale kD for DOPC by a factor of 6.25. To see this figure in color, go online.
Advantages and disadvantages of the mβCD area depletion assay
With our area depletion assay, a single experiment yields data for up to 15 individual SLBs over a wide range of cholesterol concentrations, enabling rapid acquisition of replicate data. The cholesterol-dependent cytolysin and cholesterol oxidase assays also report on the accessibility of membrane cholesterol but require a separate experiment for every χC tested. In addition, the experimental observable in our assay, namely membrane area visualized by fluorescence microscopy, enables us to directly witness any evidence of large-scale structural changes of the membrane during the course of our experiment. A disadvantage of our assay is that the signal/noise diminishes in the low-cholesterol regime (≤30 mol %) because at these χC values, aavg increases sharply as cholesterol is removed, meaning that large changes in χC yield very small changes in the total bilayer area remaining. Our discussion focuses on the accessibility of cholesterol. A vast literature of complementary techniques exists to assess cholesterol-lipid interactions and has been reviewed by others (72). As only one example, isothermal titration calorimetry detects nonideal lipid-cholesterol mixing in bilayers at χC ≥ 0.3, although the membrane partition coefficient of cholesterol appears to increase with χC (35).
Conclusions
We found that there are two populations of cholesterol with distinct accessibilities in lipid bilayers composed of DMPC, SOPC, or DOPC at high concentrations of cholesterol. The accessibility of cholesterol decreases sharply as the more accessible population is depleted from the bilayer, yielding a single abrupt change in slope in all three traces of kD versus χC in Fig. 6. The proportion of cholesterol initially in the more accessible population increases with the degree of unsaturation of the PC-lipid tail. Treating the depletion rate coefficient as a measure of accessibility that is proportional to chemical activity (9), our results are consistent with cholesterol activity being higher in membranes comprised of monounsaturated lipids as compared to those comprised of saturated lipids. Our results are consistent with the condensed-complex and cholesterol bilayer domain models of lipid membranes and complement previous experiments using different systems to build a body of work elucidating how the chemical potential of cholesterol changes with its mole fraction in membranes (10, 11, 12, 13, 14, 15, 16, 17, 18). We converted our results into predictions about these two models (Table 6). Our assay enables robust determination of the accessibility of cholesterol in lipid bilayers as a function of both phospholipid structure and cholesterol concentration.
Author Contributions
J.P.L., T.P., and S.L.K. designed the research; J.P.L. performed the research; J.P.L. and N.T. analyzed the data; and J.P.L. and S.L.K. wrote the article.
Acknowledgments
The authors thank Matthew C. Blosser for comments on the article.
This research was funded by the National Science Foundation (grant Nos. MCB-07444852 and MCB-1402059). J.P.L. and N.T. were supported by National Science Foundation Graduate Research Fellowships Nos. DGE-0718124 and DGE-1256082, respectively. T.P. was supported by the Raymond and Beverly Sackler Foundation and the Fondation Bettencourt Schueller.
Editor: David Cafiso.
Footnotes
Supporting Materials and Methods, four figures, and five tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)01178-9.
Supporting Material
References
- 1.Steck T.L., Lange Y. Cell cholesterol homeostasis: mediation by active cholesterol. Trends Cell Biol. 2010;20:680–687. doi: 10.1016/j.tcb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Das A., Goldstein J.L., Radhakrishnan A. Use of mutant 125I-perfringolysin O to probe transport and organization of cholesterol in membranes of animal cells. Proc. Natl. Acad. Sci. USA. 2013;110:10580–10585. doi: 10.1073/pnas.1309273110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Das A., Brown M.S., Radhakrishnan A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. eLife. 2014;3:e02882. doi: 10.7554/eLife.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lange Y., Steck T.L. The role of intracellular cholesterol transport in cholesterol homeostasis. Trends Cell Biol. 1996;6:205–208. doi: 10.1016/0962-8924(96)20016-9. [DOI] [PubMed] [Google Scholar]
- 5.Lange Y., Ye J., Steck T.L. How cholesterol homeostasis is regulated by plasma membrane cholesterol in excess of phospholipids. Proc. Natl. Acad. Sci. USA. 2004;101:11664–11667. doi: 10.1073/pnas.0404766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lange Y., Ye J., Steck T.L. Essentially all excess fibroblast cholesterol moves from plasma membranes to intracellular compartments. PLoS One. 2014;9:e98482. doi: 10.1371/journal.pone.0098482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kris-Etherton P.M., Yu S. Individual fatty acid effects on plasma lipids and lipoproteins: human studies. Am. J. Clin. Nutr. 1997;65(Suppl):1628S–1644S. doi: 10.1093/ajcn/65.5.1628S. [DOI] [PubMed] [Google Scholar]
- 8.Silvius J.R. Role of cholesterol in lipid raft formation: lessons from lipid model systems. Biochim. Biophys. Acta. 2003;1610:174–183. doi: 10.1016/s0005-2736(03)00016-6. [DOI] [PubMed] [Google Scholar]
- 9.Radhakrishnan A., McConnell H.M. Chemical activity of cholesterol in membranes. Biochemistry. 2000;39:8119–8124. doi: 10.1021/bi0005097. [DOI] [PubMed] [Google Scholar]
- 10.Nelson L.D., Johnson A.E., London E. How interaction of perfringolysin O with membranes is controlled by sterol structure, lipid structure, and physiological low pH: insights into the origin of perfringolysin O-lipid raft interaction. J. Biol. Chem. 2008;283:4632–4642. doi: 10.1074/jbc.M709483200. [DOI] [PubMed] [Google Scholar]
- 11.Flanagan J.J., Tweten R.K., Heuck A.P. Cholesterol exposure at the membrane surface is necessary and sufficient to trigger perfringolysin O binding. Biochemistry. 2009;48:3977–3987. doi: 10.1021/bi9002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sokolov A., Radhakrishnan A. Accessibility of cholesterol in endoplasmic reticulum membranes and activation of SREBP-2 switch abruptly at a common cholesterol threshold. J. Biol. Chem. 2010;285:29480–29490. doi: 10.1074/jbc.M110.148254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moe P.C., Heuck A.P. Phospholipid hydrolysis caused by Clostridium perfringens α-toxin facilitates the targeting of perfringolysin O to membrane bilayers. Biochemistry. 2010;49:9498–9507. doi: 10.1021/bi1013886. [DOI] [PubMed] [Google Scholar]
- 14.Olsen B.N., Bielska A.A., Ory D.S. The structural basis of cholesterol accessibility in membranes. Biophys. J. 2013;105:1838–1847. doi: 10.1016/j.bpj.2013.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gay A., Rye D., Radhakrishnan A. Switch-like responses of two cholesterol sensors do not require protein oligomerization in membranes. Biophys. J. 2015;108:1459–1469. doi: 10.1016/j.bpj.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange Y., Tabei S.M.A., Steck T.L. Stability and stoichiometry of bilayer phospholipid-cholesterol complexes: relationship to cellular sterol distribution and homeostasis. Biochemistry. 2013;52:6950–6959. doi: 10.1021/bi400862q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M.M., Olsher M., Chong P.L.G. Cholesterol superlattice modulates the activity of cholesterol oxidase in lipid membranes. Biochemistry. 2004;43:2159–2166. doi: 10.1021/bi035982+. [DOI] [PubMed] [Google Scholar]
- 18.Ali M.R., Cheng K.H., Huang J. Assess the nature of cholesterol-lipid interactions through the chemical potential of cholesterol in phosphatidylcholine bilayers. Proc. Natl. Acad. Sci. USA. 2007;104:5372–5377. doi: 10.1073/pnas.0611450104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson B.B., Moe P.C., Heuck A.P. Modifications in perfringolysin O domain 4 alter the cholesterol concentration threshold required for binding. Biochemistry. 2012;51:3373–3382. doi: 10.1021/bi3003132. [DOI] [PubMed] [Google Scholar]
- 20.Beattie M.E., Veatch S.L., Keller S.L. Sterol structure determines miscibility versus melting transitions in lipid vesicles. Biophys. J. 2005;89:1760–1768. doi: 10.1529/biophysj.104.049635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevens M.M., Honerkamp-Smith A.R., Keller S.L. Solubility limits of cholesterol, lanosterol, ergosterol, stigmasterol, and β-sitosterol in electroformed lipid vesicles. Soft Matter. 2010;6:5882–5890. doi: 10.1039/c0sm00373e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yancey P.G., Rodrigueza W.V., Rothblat G.H. Cellular cholesterol efflux mediated by cyclodextrins. Demonstration Of kinetic pools and mechanism of efflux. J. Biol. Chem. 1996;271:16026–16034. doi: 10.1074/jbc.271.27.16026. [DOI] [PubMed] [Google Scholar]
- 23.Melzak K.A., Melzak S.A., Toca-Herrera J.L. Cholesterol organization in phosphatidylcholine liposomes: a surface plasmon resonance study. Materials (Basel) 2012;5:2306–2325. [Google Scholar]
- 24.Mohwald H. In: Structure and Dynamics of Membranes. Lipowsky R., Sackmann E., editors. Elsevier; Amsterdam, the Netherlands: 1995. pp. 161–211. [Google Scholar]
- 25.Kučerka N., Nagle J.F., Katsaras J. Lipid bilayer structure determined by the simultaneous analysis of neutron and x-ray scattering data. Biophys. J. 2008;95:2356–2367. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stottrup B.L., Stevens D.S., Keller S.L. Miscibility of ternary mixtures of phospholipids and cholesterol in monolayers, and application to bilayer systems. Biophys. J. 2005;88:269–276. doi: 10.1529/biophysj.104.048439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veatch S.L., Keller S.L. Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys. J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kilsdonk E.P.C., Yancey P.G., Rothblat G.H. Cellular cholesterol efflux mediated by cyclodextrins. J. Biol. Chem. 1995;270:17250–17256. doi: 10.1074/jbc.270.29.17250. [DOI] [PubMed] [Google Scholar]
- 29.Haynes M.P., Phillips M.C., Rothblat G.H. Efflux of cholesterol from different cellular pools. Biochemistry. 2000;39:4508–4517. doi: 10.1021/bi992125q. [DOI] [PubMed] [Google Scholar]
- 30.Beseničar M.P., Bavdek A., Anderluh G. Kinetics of cholesterol extraction from lipid membranes by methyl-β-cyclodextrin—a surface plasmon resonance approach. Biochim. Biophys. Acta. 2008;1778:175–184. doi: 10.1016/j.bbamem.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 31.Huang J., Buboltz J.T., Feigenson G.W. Maximum solubility of cholesterol in phosphatidylcholine and phosphatidylethanolamine bilayers. Biochim. Biophys. Acta. 1999;1417:89–100. doi: 10.1016/s0005-2736(98)00260-0. [DOI] [PubMed] [Google Scholar]
- 32.Silvius J.R. In: Lipid-Protein Interactions. Jost P.C., Griffith O.H., editors. Wiley; New York: 1982. pp. 239–281. [Google Scholar]
- 33.Marsh D. Liquid-ordered phases induced by cholesterol: a compendium of binary phase diagrams. Biochim. Biophys. Acta. 2010;1798:688–699. doi: 10.1016/j.bbamem.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 34.Hamai C., Cremer P.S., Musser S.M. Single giant vesicle rupture events reveal multiple mechanisms of glass-supported bilayer formation. Biophys. J. 2007;92:1988–1999. doi: 10.1529/biophysj.106.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsamaloukas A., Szadkowska H., Heerklotz H. Interactions of cholesterol with lipid membranes and cyclodextrin characterized by calorimetry. Biophys. J. 2005;89:1109–1119. doi: 10.1529/biophysj.105.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niu S.-L., Litman B.J. Determination of membrane cholesterol partition coefficient using a lipid vesicle-cyclodextrin binary system: effect of phospholipid acyl chain unsaturation and headgroup composition. Biophys. J. 2002;83:3408–3415. doi: 10.1016/S0006-3495(02)75340-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson T.G., Tan A., Seelig J. Calorimetric measurement of phospholipid interaction with methyl-β-cyclodextrin. Biochemistry. 2004;43:2251–2261. doi: 10.1021/bi0358869. [DOI] [PubMed] [Google Scholar]
- 38.Huang Z., London E. Effect of cyclodextrin and membrane lipid structure upon cyclodextrin-lipid interaction. Langmuir. 2013;29:14631–14638. doi: 10.1021/la4031427. [DOI] [PubMed] [Google Scholar]
- 39.Steck T.L., Ye J., Lange Y. Probing red cell membrane cholesterol movement with cyclodextrin. Biophys. J. 2002;83:2118–2125. doi: 10.1016/S0006-3495(02)73972-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamilton J.A. Fast flip-flop of cholesterol and fatty acids in membranes: implications for membrane transport proteins. Curr. Opin. Lipidol. 2003;14:263–271. doi: 10.1097/00041433-200306000-00006. [DOI] [PubMed] [Google Scholar]
- 41.Bruckner R.J., Mansy S.S., Szostak J.W. Flip-flop-induced relaxation of bending energy: implications for membrane remodeling. Biophys. J. 2009;97:3113–3122. doi: 10.1016/j.bpj.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jo S., Rui H., Im W. Cholesterol flip-flop: insights from free energy simulation studies. J. Phys. Chem. B. 2010;114:13342–13348. doi: 10.1021/jp108166k. [DOI] [PubMed] [Google Scholar]
- 43.Schindelin J., Arganda-Carreras I., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patil A., Huard D., Fonnesbeck C.J. PyMC: Bayesian stochastic modelling in Python. J. Stat. Softw. 2010;35:1–81. [PMC free article] [PubMed] [Google Scholar]
- 45.Hung W.-C., Lee M.-T., Huang H.W. The condensing effect of cholesterol in lipid bilayers. Biophys. J. 2007;92:3960–3967. doi: 10.1529/biophysj.106.099234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan J., Tristram-Nagle S., Nagle J.F. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2009;80:021931. doi: 10.1103/PhysRevE.80.021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heftberger P., Kollmitzer B., Pabst G. Global small-angle x-ray scattering data analysis for multilamellar vesicles: the evolution of the scattering density profile model. J. Appl. Cryst. 2014;47:173–180. doi: 10.1107/S1600576713029798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallová J., Uhríková D., Balgavý P. Partial area of cholesterol in monounsaturated diacylphosphatidylcholine bilayers. Chem. Phys. Lipids. 2010;163:765–770. doi: 10.1016/j.chemphyslip.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 49.Kučerka N., Pencer J., Katsaras J. Influence of cholesterol on the bilayer properties of monounsaturated phosphatidylcholine unilamellar vesicles. Eur. Phys. J. E Soft Matter. 2007;23:247–254. doi: 10.1140/epje/i2007-10202-8. [DOI] [PubMed] [Google Scholar]
- 50.Kučerka N., Gallová J., Katsaras J. Areas of monounsaturated diacylphosphatidylcholines. Biophys. J. 2009;97:1926–1932. doi: 10.1016/j.bpj.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kučerka N., Nieh M.-P., Katsaras J. Fluid phase lipid areas and bilayer thicknesses of commonly used phosphatidylcholines as a function of temperature. 2011;1808:2761–2771. doi: 10.1016/j.bbamem.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 52.Koenig B.W., Gawrisch K. Specific volume of unsaturated phosphatidylcholines in the liquid crystalline phase. Biochim. Biophys. Acta. 2005;1715:65–70. doi: 10.1016/j.bbamem.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 53.Greenwood A.I., Tristram-Nagle S., Nagle J.F. Partial molecular volumes of lipids and cholesterol. Chem. Phys. Lipids. 2006;143:1–10. doi: 10.1016/j.chemphyslip.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tristram-Nagle S., Liu Y., Nagle J.F. Structure of gel phase DMPC determined by x-ray diffraction. Biophys. J. 2002;83:3324–3335. doi: 10.1016/S0006-3495(02)75333-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uhríková D., Rybár P., Balgavý P. Component volumes of unsaturated phosphatidylcholines in fluid bilayers: a densitometric study. Chem. Phys. Lipids. 2007;145:97–105. doi: 10.1016/j.chemphyslip.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 56.Huang J., Feigenson G.W. A microscopic interaction model of maximum solubility of cholesterol in lipid bilayers. Biophys. J. 1999;76:2142–2157. doi: 10.1016/S0006-3495(99)77369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brzustowicz M.R., Cherezov V., Wassall S.R. Controlling membrane cholesterol content. A role for polyunsaturated (docosahexaenoate) phospholipids. Biochemistry. 2002;41:12509–12519. doi: 10.1021/bi0262808. [DOI] [PubMed] [Google Scholar]
- 58.Brzustowicz M.R., Cherezov V., Wassall S.R. Molecular organization of cholesterol in polyunsaturated membranes: microdomain formation. Biophys. J. 2002;82:285–298. doi: 10.1016/S0006-3495(02)75394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baykal-Caglar E., Hassan-Zadeh E., Huang J. Preparation of giant unilamellar vesicles from damp lipid film for better lipid compositional uniformity. Biochim. Biophys. Acta. 2012;1818:2598–2604. doi: 10.1016/j.bbamem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 60.Chong P.L.-G. Evidence for regular distribution of sterols in liquid crystalline phosphatidylcholine bilayers. Proc. Natl. Acad. Sci. USA. 1994;91:10069–10073. doi: 10.1073/pnas.91.21.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McConnell H.M., Radhakrishnan A. Condensed complexes of cholesterol and phospholipids. Biochim. Biophys. Acta. 2003;1610:159–173. doi: 10.1016/s0005-2736(03)00015-4. [DOI] [PubMed] [Google Scholar]
- 62.Preston Mason R., Tulenko T.N., Jacob R.F. Direct evidence for cholesterol crystalline domains in biological membranes: role in human pathobiology. Biochim. Biophys. Acta. 2003;1610:198–207. doi: 10.1016/s0005-2736(03)00018-x. [DOI] [PubMed] [Google Scholar]
- 63.Ziblat R., Kjaer K., Addadi L. Structure of cholesterol/lipid ordered domains in monolayers and single hydrated bilayers. Angew. Chem. Int. Ed. 2009;48:8958–8961. doi: 10.1002/anie.200903847. [DOI] [PubMed] [Google Scholar]
- 64.Raguz M., Mainali L., Subczynski W.K. The immiscible cholesterol bilayer domain exists as an integral part of phospholipid bilayer membranes. Biochim. Biophys. Acta. 2011;1808:1072–1080. doi: 10.1016/j.bbamem.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Varsano N., Fargion I., Addadi L. Formation of 3D cholesterol crystals from 2D nucleation sites in lipid bilayer membranes: implications for atherosclerosis. J. Am. Chem. Soc. 2015;137:1601–1607. doi: 10.1021/ja511642t. [DOI] [PubMed] [Google Scholar]
- 66.Ziblat R., Leiserowitz L., Addadi L. Crystalline domain structure and cholesterol crystal nucleation in single hydrated DPPC:Cholesterol:POPC bilayers. J. Am. Chem. Soc. 2010;132:9920–9927. doi: 10.1021/ja103975g. [DOI] [PubMed] [Google Scholar]
- 67.Raguz M., Mainali L., Subczynski W.K. Using spin-label electron paramagnetic resonance (EPR) to discriminate and characterize the cholesterol bilayer domain. Chem. Phys. Lipids. 2011;164:819–829. doi: 10.1016/j.chemphyslip.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barrett M.A., Zheng S., Rheinstädter M.C. Solubility of cholesterol in lipid membranes and the formation of immiscible cholesterol plaques at high cholesterol concentrations. Soft Matter. 2013;9:9342–9351. [Google Scholar]
- 69.Mainali L., Raguz M., Subczynski W.K. Formation of cholesterol bilayer domains precedes formation of cholesterol crystals in cholesterol/dimyristoylphosphatidylcholine membranes: EPR and DSC studies. J. Phys. Chem. B. 2013;117:8994–9003. doi: 10.1021/jp402394m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toppozini L., Meinhardt S., Rheinstädter M.C. Structure of cholesterol in lipid rafts. Phys. Rev. Lett. 2014;113:228101. doi: 10.1103/PhysRevLett.113.228101. [DOI] [PubMed] [Google Scholar]
- 71.Sugár I.P., Chong P.L.-G. A statistical mechanical model of cholesterol/phospholipid mixtures: linking condensed complexes, superlattices, and the phase diagram. J. Am. Chem. Soc. 2012;134:1164–1171. doi: 10.1021/ja2092322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ohvo-Rekilä H., Ramstedt B., Slotte J.P. Cholesterol interactions with phospholipids in membranes. Prog. Lipid Res. 2002;41:66–97. doi: 10.1016/s0163-7827(01)00020-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.