Abstract
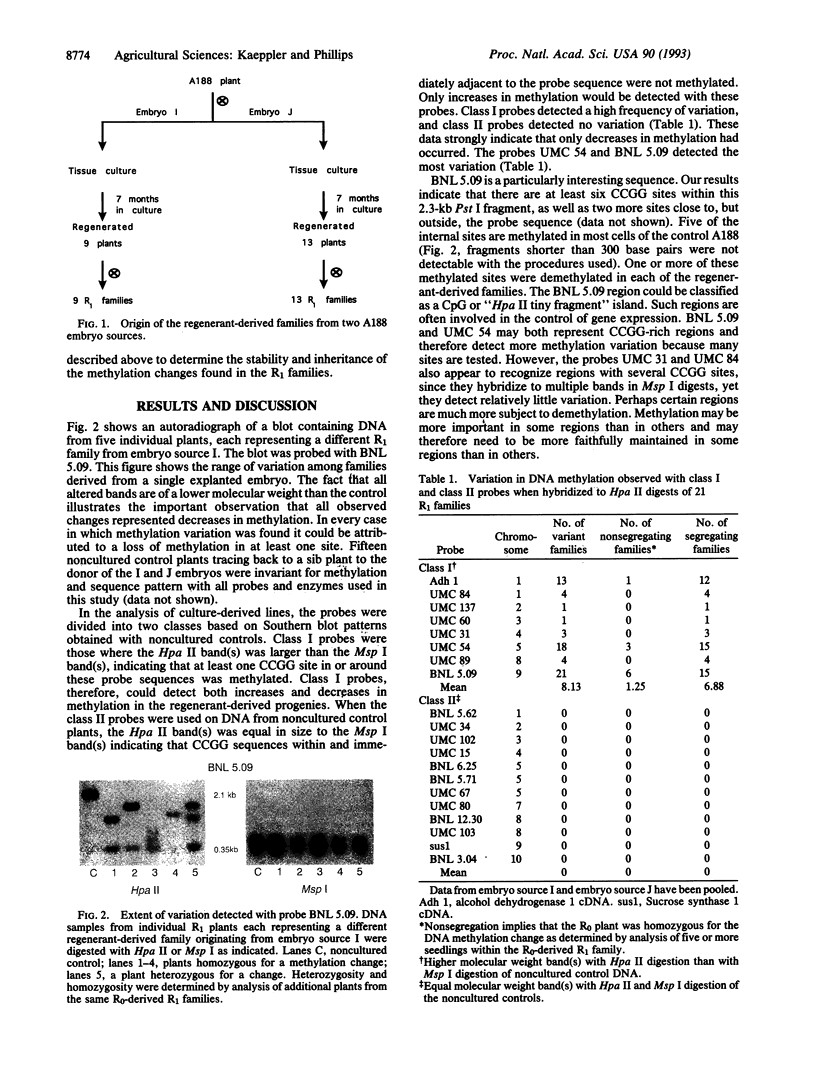
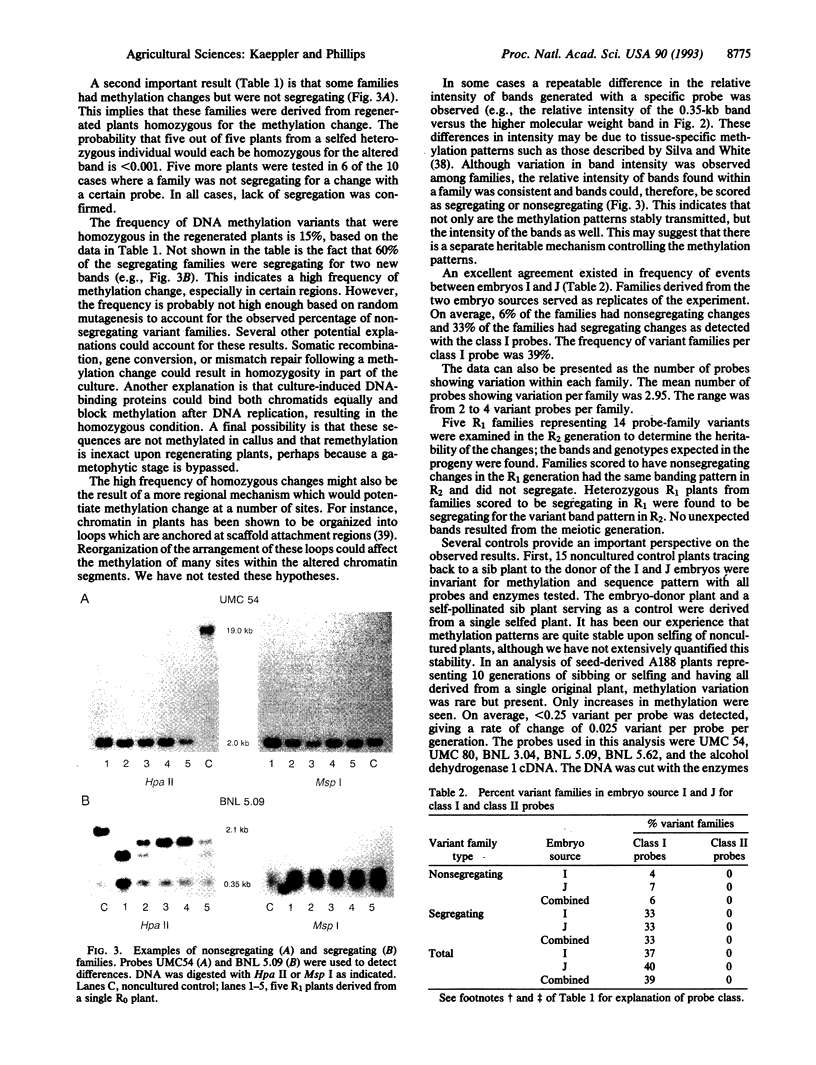
Twenty-one progeny lines derived from tissue cultures of two embryo sources of maize inbred strain A188 were examined for DNA methylation changes. Total DNA was cut with the isoschizomers Hpa II and Msp I and probed with 18 single-copy Pst I genomic clones and two cDNA clones. Eight of these probes could detect both increases and decreases in methylation. With these probes 39% of the families were found to contain an altered methylation pattern. All changes represented a decrease in methylation. The other 12 probes could detect only increases in methylation; no methylation variation was seen with these probes. Fifteen percent of the methylation changes were homozygous in the original regenerated plant. Changes were stably inherited upon two generations of self-pollination. No sequence variation was observed in Msp I-digested DNA from the same 21 progeny lines. Certain probes detected methylation changes much more often than others. Our study provides evidence that demethylation occurs at a high frequency and could be an important cause of tissue culture-induced variation. Occurrence of the frequent homozygous alterations in original regenerated plants implies a non-random mutational mechanism.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown P. T., Kyozuka J., Sukekiyo Y., Kimura Y., Shimamoto K., Lörz H. Molecular changes in protoplast-derived rice plants. Mol Gen Genet. 1990 Sep;223(2):324–328. doi: 10.1007/BF00265070. [DOI] [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Hall G., Jr, Allen G. C., Loer D. S., Thompson W. F., Spiker S. Nuclear scaffolds and scaffold-attachment regions in higher plants. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9320–9324. doi: 10.1073/pnas.88.20.9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. The inheritance of epigenetic defects. Science. 1987 Oct 9;238(4824):163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- Klaas M., Amasino R. M. DNA Methylation is Reduced in DNasel-Sensitive Regions of Plant Chromatin. Plant Physiol. 1989 Oct;91(2):451–454. doi: 10.1104/pp.91.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M. Changes in DNA methylation during mouse embryonic development in relation to X-chromosome activity and imprinting. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1235):299–312. doi: 10.1098/rstb.1990.0013. [DOI] [PubMed] [Google Scholar]
- Peschke V. M., Phillips R. L., Gengenbach B. G. Discovery of transposable element activity among progeny of tissue culture--derived maize plants. Science. 1987 Nov 6;238(4828):804–807. doi: 10.1126/science.238.4828.804. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A., Allard R. W. Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A. 1984 Dec;81(24):8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. Gene-controlled cytosine demethylation in the promoter region of the Ac transposable element in maize. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2789–2793. doi: 10.1073/pnas.86.8.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. J., White R. Inheritance of allelic blueprints for methylation patterns. Cell. 1988 Jul 15;54(2):145–152. doi: 10.1016/0092-8674(88)90546-6. [DOI] [PubMed] [Google Scholar]