Abstract
Herein we present a microfluidic-multiplexed platform that integrates electrochemical sensors based on carbon nanotubes associated with ferrocene as redox marker (carbon nanotube (CNT)/ferrocene) for direct detection of pathogenic viral DNA from Hepatitis C and genomic DNA from Mycobacterium tuberculosis in clinical isolates. By operating the fluidic device under high flow (150 μl/min), the formation of a very thin depletion layer at the sensor surface (δS = 230 nm) enhances the capture rate up to one DNA strand per second. By comparison, this capture rate is only 0.02 molecule/s in a static regime without flow. This fluidic protocol allows thus enhancing the limit of detection of the electrochemical biosensor from picomolar in bulk solution to femtomolar with a large dynamic range from 0.1 fM to 1 pM. Kinetics analysis also demonstrates an enhancement of the rate constant of electron transfer (kS) of the electrochemical process from 1 s−1 up to 6 s−1 thanks to the geometry of the miniaturized fluidic electrochemical cell. This microfluidic device working under high flow allows selective direct detection of a Mycobacterium tuberculosis (H37Rv) rpoB allele from clinical isolate extracted DNA. We envision that a microfluidic approach under high flow associated with a multiwall CNT/ferrocene sensor could find useful applications as the point-of-care for multi-target diagnostics of biomarkers in real samples.
I. INTRODUCTION
A rapid, accurate, cost-effective, and certainty microbiological diagnostics of pathogenic DNA biomarkers stills remains a challenge in the XXIst century. The development of extremely sensitive, highly selective, simple, robust, and yet inexpensive biosensing platforms appears thus essential for a wide range of applications, including not only clinical diagnostics1 but also environmental monitoring2 and food safety testing.3 Direct detection of a genomic DNA sample, without pre-analytical and amplification steps, may lead to reduced analysis times, system complexity, quantification accuracy, and other ambiguities and errors issued from contamination.4,5 Since accurate diagnostics need specific detection of very small amounts of DNA, there is a need to develop simple label-free DNA hybridization platforms able to perform highly sensitive and specific direct analyses.
Recent advances in electrochemical sensing techniques, in conjunction with microfluidics and nanomaterials, have already shown tremendous potential, offering more reliable and affordable solutions than conventional optical detection methods, which involve a complex instrumental environment around the sampling chip.4 In this regard, electrochemical detection is fully compatible with cost-effective handheld microfluidics platforms5,6 that bring the possibility of manipulating tiny amount of samples (<μl) for fast analysis.
Most often, conventional point-of-care (PoC) devices integrate several stages including a polymerase chain reaction (PCR) chamber for genomic amplification located in front of the molecular electrochemical detection cell. For example, in 2004, Liu et al.7 developed a fully integrated biochip device with an asymmetric PCR step, subsequently coupled with a two-step electrochemical hybridization sensor that succeeded to achieve ∼2 fM detection of bacterial Escherichia coli K12 DNA. Yeung et al.39 developed an electrochemical real-time PCR (ERT-PCR) technique on a silicon glass microchip for simultaneous DNA amplification and detection within a limit of detection (LoD) of 5 fM. More recently, Ferguson et al.40 reported a limit of detection of ∼0.01 fM for genomic DNA from Salmonella Enterica Serovar Typhimurium LT2 inside a monolithic disposable chip featuring asymmetric PCR, enzymatic single-stranded DNA digestion, and sequence-specific electrochemical detection elements. In such PoC devices, the on-a-chip PCR amplification reaction has to be accurately controlled during thermal cycling. The PCR protocol could be thus rather complex since the chip has to be placed inside a thermal controller8 or it has to integrate a miniaturized controller. In this context, developing simpler cost-effective microfluidic devices without a PCR unit able to detect DNA from clinical isolates appears to be of great importance.
Since the concentration of nucleic acids that are attainable from samples falls typically below 20 fM,9 new approaches for direct electrochemical detection without amplification have to be explored. A first route concerns the use of nanomaterials as ideal transducers since they possess the ability to amplify both signal transduction and molecular events. For example, in 2006, Zhang et al.41 reported the incorporation of a gold nanoparticle (NP) coupled with a nanoscale control of the self-assembly process of DNA probes. The biosensor was able to enhance significantly the sensitivity of direct electrochemical DNA sensing at the femtomolar range. Similarly, a conductometric/chemiresistive DNA sensor based on polypyrrole nanowires and optimized in terms of DNA probe sequence immobilization was reported by Bangar et al.42 with a LoD of 0.1 fM and a wide dynamic range of 0.1 fM–10 pM. Recent studies have shown that multiwall carbon nanotubes (MWCNTs) can provide a higher surface density of immobilized biomolecules in comparison with traditional planar surfaces10 and improve significantly the electrochemical reactivity.11 In 2003, Fang et al. have shown for the first time that multi-walled carbon nanotubes functionalized with a carboxylic acid group (MWNTs–COOH) dramatically increased DNA detection sensitivity.12 CNT hybrid materials combined with conducting polymers, metal NPs, or redox mediators have been receiving increased attention in amperometric biosensors due to various synergetic effects observed when hybrid materials are used.13 The integrated MWCNT/ferrocene has been shown to enhance charge transfer due to the notable properties of ferrocene as organometallic redox marker promoter.14 With a reversible and intense redox signal, ferrocene is an ideal “outer-sphere” redox system for which kinetics of electron transfer can be affected when electronic and ionic environments are modified. After attachment to ferrocene, the probe and target biomolecules could promote the charge transfer or disturb it. Ferrocene has thus been widely used as a model system for understanding charge transfer by tunnelling across various molecular interfaces to electrodes.15,16 Ferrocene could be employed as redox marker to gives information of macromolecular interaction between receptor and ligands or as redox mediators in enzymatic reaction. Attachment of ferrocene to conducting materials as redox markers to follow the biological recognition event such as DNA hybridization or protein capture has been demonstrated previously with polypyrrole17 and carbon nanotube.18
Independently of the intrinsic sensibility of the electrochemical sensor, the performance of the biosensor can be improved according to the optimization of the microfluidic protocol. Recently, it has be proven that flows properties have the ability to favour binding kinetics and thus largely reduce reaction time.19–22 For most of the applications, if the fluidic system works under “extreme fast” flow at a high Péclet number PeH, the number of captured molecules per unit of time could be largely enhanced. In 2008, Squires, Messinger, and Manalis proposed a theoretical approach to estimate the interplay between diffusion, convection, and reaction.20 Their complete analysis gives some guidelines to estimate some fundamental quantities, such as fluxes, collection rates, and equilibration times. Based on dimensionless parameters that are straightforward to compute, their simple approach is very useful in designing systems and fixing the volumetric flow rate Q. Working with such optimized parameters at high PeH (PeH ≫ 1) under high flow, the number of molecules trapped in the vicinity of the sensor surface can be increased by several orders of magnitude.
Toward an universal answer for sequence-specific direct electrochemical detection, we propose herein a new microfluidic platform for direct label-free DNA detection without any amplification. The system is based on integrated electrochemical MWCNT/ferrocene sensor, which is operated under a fluidic protocol where alternative steps under fast flow and static regime are performed. This microfluidic approach is shown to improve mass transport and enhance target-probe interaction events. This simple monolithic chip integrates three independent fluidic polydimethylsiloxanes (PDMSs), each channel containing one electrochemical chamber with three gold electrodes and each working electrode being functionalized with a MWCNT/ferrocene transducing layer (Figure 1). The integrated MWCNT/ferrocene sensor is shown to enhance charge transfer due to the notable properties of ferrocene as organometallic redox marker. After attachment to ferrocene, the probe and target biomolecules could promote the charge transfer or reduce it. By choosing the nature of the immobilized probe on each MWCNT/ferrocene working electrode, this system can be configured to detect any DNA-based pathogen in clinical isolates samples. Dynamic range and LoD of this device have been first studied using model solutions of synthetic single-strand oligonucleotide from Hepatitis C virus and compared with the conventional detection in bulk solution. In a second step, we demonstrate the direct selective detection of a wild-type Mycobacterium tuberculosis (H37Rv) rpoB allele in clinical isolate extracted DNA to check the feasibility of direct sensing without amplification in real solutions.
FIG. 1.
On chip specific genetic analysis of Mycobacterium tuberculosis and Hepatitis C. (a) The PDMS/glass point-of-care device that integrates three channels: channel (1) for negative control, channel (2) for DNA detection, and channel (3) for mismatch DNA detection. (b) Schematic illustration of the chemical patterning (steps 1, 2, and 3) for the preparation of the biosensor based on modified ferrocene, as redox marker. Each step is done inside each channel of the fluidic chip before the final hybridization (step 4).
II. EXPERIMENTAL METHODS
A. Reagents
The multiwalled carbon nanotubes (purity 85%) with 20–25 nm in diameter and ∼1–5 μm in length were purchased from Strem Chemicals (France). Ethylene diamine and the lithium perchlorate were purchased from Sigma Aldrich. The modified ferrocene group Fc(NHP)2 was synthesized following a procedure described previously.23 The background electrolyte is phosphate buffer at 10 mM and pH 7.4. It has been prepared by mixing 10 mM Na2HPO4, 2.7 mM KCl, and 137 mM NaCl in ultrapure water from Millipore Milli-Q system (Billerica, USA). All other reagents are commercially available and were purchased with analytical reagent grade.
For Hepatitis C, the oligonucleotide probe with amino group at its 5′ phosphate end, abbreviated NH2-ssDNA, has the sequence as follows: NH2–(CH2)6–5′-GATACTTCTATCACC-3′. The complementary and non-complementary target oligonucleotides have, respectively, the sequences: 5′-GGTGATAGAAGTATC-3′ and 5′-CATTCCCTCTTAGG-3′. The DNA probe, specific to the Mycobacterium tuberculosis, was provided by Eurogentec Company, containing 18 bases with 5′ terminal amino group modification using a twelve carbonyl chain spacer (C12). The DNA probe sequences were wild type (5′-CCGACTGTCGGCGCTGGG-3′) and mutant (5′-CCGACTGTTGGCGCTGGG-3′).24 A single nucleotide polymorphism (SNP) in the codon 531 of rpoB gene (TCG/TTG) is associated with Mycobacterium tuberculosis rifampicin resistance and used as control experiment.
B. Sample preparation from Mycobacterium tuberculosis clinical isolates
The genomic DNA of Mycobacterium tuberculosis was extracted from a clinical sample of stump. Some pretreatments have been done off-chip, and only hybridization step has been done on-chip similarly as in other electrochemical fluidic chips.8 For the gDNA extraction, the standard CTAB (cetyltrimethylammonium bromide) method has been used.25 This approach is preferred for molecular analyses demanding high purity, whereas thermolyzates are used in other cases due to its swiftness and ease of implementation. The prepared solutions could be stored at 4 °C for two weeks or in the freezer (−20 °C) for a longer period. Finally prior to sample injection inside the chip, the gDNA solution has been exposed to thermal shock (3 min at 90 °C) for denaturation purpose. 5 μl of gDNA (20 ng of clinical isolate) was then diluted in 200 μl for the experiments inside the chip.
To compare the response of the sensor towards both wild-type DNA of Mycobacterium tuberculosis and amplified ones, PCR was also performed on several samples to amplify a 411 bp length fragment of the rpoB gene from well-characterized DNA belonging to one clinical isolate of Mycobacterium tuberculosis and Mycobacterium tuberculosis control strain H37Rv. The fragment of 411 bp of the rpoB gene includes a hot spot region of 81 bp, where the majority of mutations associated with resistance of Mycobacterium tuberculosis to rifampicin drug have been identified. To amplify a fragment of 411 bp of the rpoB gene, the primers TR1 (5′-TACGGTCGGCGAGCTGATCC-3′) and TR2b (5′-TACGGCGTTTCGATGAACC-3′) were used. 2 μl of extracted gDNA (20 ng), 1 U of Taq DNA polymerase (Go Taq flexi, Promega, Madison, USA), and 1.5 mM of MgCl2; 1 μM of each primer; 200 μM of each dNTP; and 1 X of Taq buffer were used to perform the PCR in a total volume of 25 μl. The following PCR cycle program was used: initial denaturation at 94 °C for 3 min and 25 cycles comprising 1 min at 94 °C, 1 min at 55 °C, 1 min at 72 °C, and a final extension step at 72 °C during 10 min. After amplification, we measured the concentration of PCR products by Nanodrop 1000.
C. Microchip fabrication
Each 300 μm-thick borosilicate slide was first cleaned by organic solvents and piranha solution (30 s in 70 vol. sulphuric acid/30 vol. peroxide water), and abundantly rinsed with deionized water before process. The slides were patterned by standard UV photolithography with image reversal AZ-5214 photosensitive resist to generate the electrode patterns. 5 nm of titanium and 200 nm of gold were then evaporated before resist removal in acetone. This conventional lift-off process allowed producing all three electrodes of each electrochemical cell, namely, the counter (CE), the reference (RE), and the working (WE) electrodes. Each slide displays three arrays of three gold electrodes (SWE = 1.2 × 10−2 cm2, SRE = 2.7 × 10−3 cm2, and SCE = 2.7 × 10−2 cm2) with the design presented in the supplementary material (see Fig. SI.1).43
First step of biosensors built-in involved oxidized carbon nanotubes coating from a diluted solution onto gold working electrode. Carbon nanotubes were oxidized in concentrated H2SO4/HNO3 (3:1) solution to produce a high concentration of carboxyl (−COOH) and C = O functions, which are of priority for dispersion purpose. The solution was prepared by dissolving 1 mg of CNTs-COOH in 1 ml of dimethylformamide (DMF) in ultrasonic bath. 1 μl of this solution was deposited onto the working electrode and the solvent was evaporated at 90 °C for 30 min.
A conventional SU–8 2050 mould containing the 60 μm-thick patterns of fluidic channels and chambers was prepared by standard UV-lithography. The PDMS and its curing agent were mixed at a 10:1 ratio and poured over the SU–8 mould that has to be degassed under vacuum and cured at 70 °C for 2 h. The PDMS replica can be then separated from the mould after ambient cooling. Inlets and outlets were formed by gently punching small needles through the PDMS layer. Then, watertight sealing of the glass-PDMS chip is achieved after exposing surfaces to UV ozone plasma for 1 min (triggering power of plasma is 400 mTorr). The microfluidic PDMS network was aligned on top of the glass with patterned electrodes and was finally annealed at 70 °C for 2 h to improve adhesion and to prevent leakages. The chamber volume of each electrochemical fluidic cell is 300 nl (see SI.1 in the supplementary material for more details).43
D. Fluidic protocol
Before use, each microfluidic device was systematically filled and rinsed gently during 1 h with phosphate-buffered saline (PBS) solution. For the construction of the biolayer on each WE electrode (steps 1–3 in Figure 1(b)), each fluidic channel was filled with the desired solution at 150 μl/min during 1 min, namely, ethylenediamine (EDA) for step 1, redox ferrocene (Fc(NHP)2) for step 2, and ssDNA solution for step 3 (in channels 2 and 3—Figure 1(a)). Between step 2 and 3, acetonitrile was used to flush the channel bearing the microelectrodes in order to remove any adsorbed ferrocene. For the step 3 of covalent immobilization of the oligonucleotides probes, the incubation time was fixed to 30 min.
For the last step (step 4 in Figure 1(b)) of DNA hybridization in channels 2 and 3, a two-sequence specific fluidic protocol was used: (1) a first dynamic step to inject the DNA target solution at 150 μl/min during t1 = 1 min followed by (2) a conventional static diffusion regime during t1 = 9 min. This sequence was repeated four times for a total hybridization time of 40 min. The high volumetric flow rate Q (Q = 150 μl/min) and the geometry of the fluidic channel (H = 60 μm) allowed the system working at high Péclet numbers and producing a very thin depletion zone δS (δS = 230 nm) at the transducer surface (see supplementary material SI.2 and Table SI.2-PeH and PeS ≫ 1).43 Since our fluidic system operates in a mass-transport-limited regime, capture of target molecules within the first dynamic step was favoured (t1 = 1 min).
E. On-a-chip electrochemical measurements
Electrochemical measurements were performed using Autolab 30 (Metrohm) equipped with software Nova by connecting one gold electrode as pseudo reference and the larger gold electrode as counter electrode. The potential of gold reference was then measured with free ferrocene in order to report all potential values versus potential of ferrocene redox couple. The electrochemical impedance spectroscopy (EIS) measurements were performed with the same potentiostat using FRA32M module and Nova as software. This was obtained in 10 mM KCl solution in the presence of redox marker 5 mM [Fe(CN)6]4−/[Fe(CN)6]3− at steady state potential and DC potential of 10 mV in the frequency range from 10 kHz to 0.5 Hz.
III. RESULTS AND DISCUSSION
A. Preparation of the biolayer on the working electrode
The covalent immobilization of EDA at 7.4 mM on CNTs/Au was performed by electro-chemical patterning of functional groups bearing amine functions (step 1) on each WE covered with the physically adsorbed CNT layer (supplementary Figure SI.1(b)).43 This reaction was monitored by cyclic voltammetry (CV) by scanning the potential from −0.2 V to 0.75 V (vs. Au) at a scan rate 200 mV s−1 during 8 cycles in 7.4 mM EDA solution containing 0.5 M LiClO4/H2O. This electrochemical oxidation reaction produced cationic radicals allowing covalent binding between ethylene diamine and carbon nanotubes.
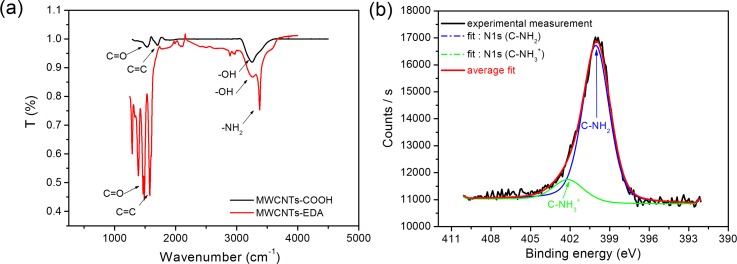
Functionalization of Au-MWCNTs and their conjugation with EDA were investigated by Fourier Transform Infra-Red spectroscopy (FT-IR). The FT-IR spectrum (Figure 2(b)) of the MWCNT-COOH layer revealed the presence of three peaks corresponding to the vibration modes of C=C, −OH, and C=O at 1500 cm−1, 3400 cm−1, and 1710 cm−1, respectively, which confirms the modification of MWCNTs with carboxylic bonds. The spectrum measured after the amine functionalization (Figure 2(a), red curve) shows the appearance of a new peak, which corresponds to the vibration mode of the −NH2 bond at 3375 cm−1. FT-IR spectroscopy experiments validated this first step of electrochemical grafting of the amine. High resolution X-Ray Photoelectron (XPS) spectroscopy confirmed this functionalization since the intensity of the N1s characteristic peak after electrochemical deposition of the EDA enhanced (Fig. 2(b)). Deconvolution of the corresponding nitrogen peak has been performed using the Shirley-type baseline and Lorentzian-Doniac-Sunsic curves, with Gaussian profile. The fit revealed the appearance of two nitrogen peaks at 399.90 eV and 402.11 eV which could be, respectively, attributed to C-N link and free amine NH2. This chemical modification of CNTs by electrochemical patterning allowed adding amine as functional group in the desired surface layer of CNTs, which will be employed for further chemical functionalization.
FIG. 2.
Spectroscopy measurements performed before and after the covalent immobilization of ethylenediamine (EDA) (step 1 in Figure 1(b)) with (a) FT-IR and (b) XPS at the N1s peak.
Subsequently, the redox marker ferrocene was covalently attached through the aminated surface of MWCNT-EDA electrode (step 2 in Figure 1(b)). Here, the ferrocenyl group has been modified by two phthalimido ester groups in order to promote covalent attachment with MWCNT-EDA groups without addition of any reagent.25 All the channels and chambers were rinsed with water and acetonitrile before injection at 150 μl/min during 1 min of the Fc(NHP)2 (1 mM). The redox ferrocene (Fc(NHP)2) served also for the covalent attachment of DNA probe bearing amine group in 5′ terminal position. This last chemical step (step 3 in Figure 1(b)) consisted in preparing the sensing layer based on the attachment of single-strand DNA (ssDNA) modified by amine group in end position to activate ester of immobilized ferrocenyl group following amide link. After a flush of acetonitrile to remove the adsorbed ferrocene, ssDNA probes (1 μM) were injected similarly under high flow (150 μl/min) during 1 min and incubated for immobilization during 30 min inside the microfluidic channel. After a final rinse of the channel with PBS, the chip was ready for the hybridization reaction (step 4).
B. Biosensor electrochemical characterization
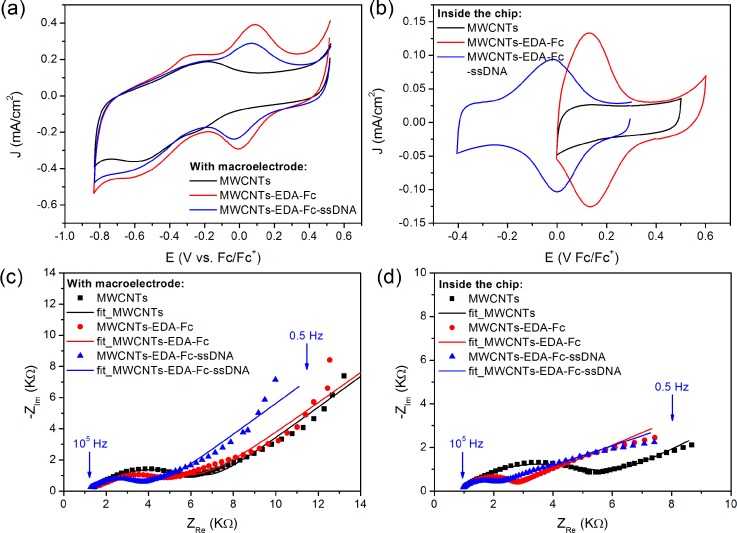
At the molecular scale, the presence of both contributions at 720.8 eV and 708 eV in the high resolution XPS spectrum can be attributed to Fe2+ (Fe 2p 1/2) and Fe3+ (Fe 2p 3/2), respectively, which proved the covalent attachment of ferrocene on the CNT surface. The grafting of ssDNA was also confirmed by the appearance of a peak at 133.6 eV, which can be attributed to phosphate ions (see supplementary material SI.3).43 The electrical properties of each patterned layer were first examined by CV in free PBS solution. High current values of several μA and reversible redox signal of ferrocene were observed either in bulk solution (Fig. 3(a)) or in the fluidic cell (Fig. 3(b)), which demonstrated the potential of this redox marker to follow the biological immobilization and interaction in the microfluidic system. The reversibility was observed in the microfluidic device with ΔE0 = Epc-Epa = 0.004 V ∼ 0 V vs. Fc/Fc+, as the signature of a high electron transfer compared to the macroelectrode for which ΔE0 ∼ 0.1 V vs. Fc/Fc+ where redox signal displayed quasi-reversible behavior. The reaction of the covalent immobilization of the ssDNA probe to the ferrocenyl was followed similarly through the redox signal of ferrocene (blue curves in Figure 3). In the macrocell, the redox potential remained the same and decrease in the current density was observed. However, after immobilization of single strand DNA on the surface of electrode in microfluidic cell, the redox potential of ferrocene shifted to lower value. This could be related to the large electronic effect due to the negative charge of DNA, which affected directly the redox process of the thin layer redox ferrocene.
FIG. 3.
Cyclic voltammograms recorded in a PBS phosphate buffer solution (10 mM and pH = 7.4) with a scan rate of 50 mV/s during steps 1 and 2 (a) for a macroelectrode and (b) inside the chip; Nyquist plots obtained in the frequency range from 10 kHz to 0.5 Hz by applying 10 mV (AC mode) in 10 mM [Fe(CN)6]3−/4−: (c) for the macroelectrode at the fixed potential 0.25 V vs. Ag/AgCl and (d) for the electrode integrated in the fluidic channel at 0.25 V vs. Fc/Fc+. The symbols are the experimental data and the solid lines are the fitted curves using the equivalent circuit showed in inset. EIS data have been satisfactorily fitted by the equivalent circuits that are reported in supplementary material SI.3.43
To analyse kinetics of electron transfer in the microfluidic system compared to macro-cell, the heterogeneous electron transfer rate kS has been determined following Laviron's model.26 kS values were extracted from cyclic voltammograms obtained at various scan rates after DNA hybridization at saturation (see Fig. SI.5 in the supplementary material).43 For the macro-cell, kS ranges between 1 and 2 s−1, which is in agreement with previous reported values for ferrocene modified carbon nanofibers27 or CNTs.28 For the fluidic device, an enhanced kS value of 5.8 s−1 was obtained, which suggests that the miniaturization of the electrochemical chamber and the immobilization of redox molecules by reducing the diffusion layer lead to high distribution of redox ferrocene on the surface. The electron hoping between adjacent ferrocene centers is thus favored and kinetics of electron transfer increases.
To confirm this increase of kS and this hypothesis, electrochemical impedance spectroscopy was also used to compare the two electrochemical systems, i.e., the macrocell and the microfluidic chamber. The corresponding EIS Nyquist plots show that the semi-circle of impedance decreases after ferrocene immobilization (Figs. 3(c) and 3(d)). This variation was related to decrease of charge transfer resistance which demonstrated the efficiency of electron transfer of the MWCNT/Fc layer formed in the microfluidic cell. The decrease of diffusion layer of the microfluidic system should favor the chemical attachment of the ferrocene leading to the high distribution of redox center on the MWCNT surface as demonstrated by cyclic voltammetry. This demonstrates that the dynamic flow used for the formation of biosensor improved also the reactivity of the surface by increasing the distribution and the accessibility of the redox marker.
C. Hepatitis C 15 base pair DNA detection in the microfluidic device
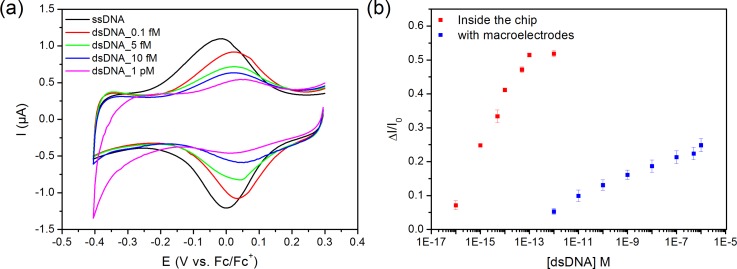
Model solutions of PBS containing different concentrations from 0.1 fM to 1 pM of complementary Hepatitis C oligonucleotide were prepared and used. The hybridization reaction was realized inside the fluidic biochip, based on the specific two-step protocol at 38 °C. The CVs obtained after DNA hybridization in a buffer free solution are presented in Figure 4(a). The hybridization reaction with the DNA target induced the formation of double strand dsDNA in a more rigid conformation leading to a decrease of electron transfer due to blocking effect.28,29 This was confirmed by the decrease in the redox signal and EIS experiments. Comparing the calibration curve deduced from our experimental results in the microfluidic PoC cell to those measured in a conventional macro-cell (see Figure 4(b)), a large enhancement of sensitivity associated to a decrease of LoD was obtained. LoD was calculated in both systems as being 3 times the standard deviation of the smallest concentration divided by the slope of the linear part of the calibration curve. LoD values are 7 fM in the microfluidic system and 2 pM with the same biosensor measured in a standard macro-cell (Figure 4(b)). Such three orders magnitude improvement is mainly related to the enhanced kinetics through the proposed pulsed flow microfluidic approach. Analogue behaviours were previously reported in the case of enzymatic biosensors integrated in microfluidic devices for glucose sensing.30
FIG. 4.
(a) Electrochemical response of the biosensor measured in free PBS solution of biosensors after the hybridization reaction (step 4 in Figure 1) with different solutions of complementary DNA with concentrations ranging from 0.1 fM to 1 pM. (b) Calibration curves of the relative changes of the current peak ΔI = (I0 − I) at a fixed potential versus concentration of complementary DNA, where I0 and I are, respectively, the peak current prior to DNA hybridization and after incubation of complementary DNA, which were measured either in microfluidic cell (red) or in conventional macro-cell (blue).
The microfluidic biosensor exhibits a dynamic range of detection from 0.1 fM to 1 pM (Figure 4(b), red curve), whereas the dynamic range measured for conventional macro-cell is 1 pM–100 nM (Figure 4(b), blue curve). A good reproducibility is observed with standard deviation of 1.5% for microfluidic and 2% for macrocell. As negative control, to check the specificity of the biosensing layer, similar experiments with non-complementary DNA strands have been performed. A decrease of around 7% in signal intensity compared to decrease in current related to complementary target was observed, which can be attributed to non-specific adsorption of DNA strands on the surface of the microelectrode. This could be improved by the covalent attachment on the surface layers of polyethylene glycol (PEG) to prevent this phenomenon.19
To explain such enhancement of sensitivity of the biosensors in the microfluidic device compared to macrocell system, the mass-transport from the two dynamic and static regimes applied during hybridization reaction has been analysed. The first dynamic step of the fluidic protocol at high flow (t1 = 1 min with Q = 150 μl/min)20 leads to very large values of both Péclet numbers (PeH and PeS ≫ 1), producing a very thin depletion zone δS (δS = 230 nm) at the transducer surface. The collection rate of target molecules in this depletion zone can be easily estimated from the theoretical approach of Squires et al.20,21 Details of calculations made to extract values of all the pertinent parameters are reported in supplementary material SI.2.43 Since this microfluidic system operates in a mass-transport-limited regime, a significant high number of target molecules can thus be captured within the first dynamic step under high flow enhancing the kinetics of hybridization reaction. In other words, a high number of DNA targets can be trapped in the thin depletion layer where they can reach rapidly the sensor surface to fast hybridize on the probe (supplementary Fig. SI.2).43 The calculation following this model was performed in the case of lowest DNA target concentration detected, i.e., 7 fM for DNA target of Hepatitis C. We show here that the first step of our protocol allows capture in the diffusion zone of around 1 molecule/s, whereas the rate of DNA capture is only 0.02 molecule/s for the static regime (supplementary material SI.2).43
For the static mode, the volume where DNA has time to diffuse during 9 min was estimated to be around δ* = 2.3 μm from diffusion equation.30 Note that the relative fraction of DNA molecules (around 270 molecules) collected during the whole experiment of 40 min corresponds to 0.01% of the DNA molecules in the 7 fM sample. The strong confinement of the reactive species, first in the depletion layer δS and second in the diffusion layer δ*, explains the improvement in sensitivity of our direct DNA sensing measurements.
Table I compares the performances of our microfluidic system that works without PCR amplification stage, with other reported LoD values for electrochemical DNA detection in microfluidic cells. The DNA biosensor based on MWCNT/Fc as transducer presented here demonstrates high sensitivity compared to the performances of other electrochemical affinity biosensing systems in microfluidic cells. This could be related to both the transducer formed with MWCNT/Fc and the dynamic flow mode applied to introduce the reagents which both improves the chemical reactivity with the surface and then the response of the biosensor. Here, we show that the majority of DNA strands are captured under high flow during 4 min (4 steps of 1 min under dynamic flow). This improvement in time of capture has to be compared with the very recent work of Taller et al.31 that is based on a membrane sensor able to detect the target at a concentration of 13 pM in a SAW-lysed sample with a total analysis time of ∼1.5 h. Our device offers a real reduction of time analysis.
TABLE I.
Comparison of affinity biosensors in microfluidic cells.
| Electrodes | Biosensor | Method | LoD (mol. l−1) | Reference |
|---|---|---|---|---|
| Au | DNA | EIS | 3.8 × 10−9 | 32 |
| Au | DNA | DPV | 4 × 10−7 | 33 |
| Al-Au | Immunoassay | CV | 2.7 × 10−13 | 34 |
| MWCNT_NiO | Immunoassay | CV, CA | 3 × 10−5 | 35 |
| Au (PCR on chip) | DNA | ACV | <10−18 | 11 |
| Au (PCR on chip) | DNA | CA | 2 × 10−13 | 18 |
| CNTs | DNA | EIS | 1 × 10−7 | 36 and 37 |
| Au-MWCNT—Fc | DNA | CV | 7 × 10−16 | This work |
D. Direct genetic detection of Mycobacterium tuberculosis
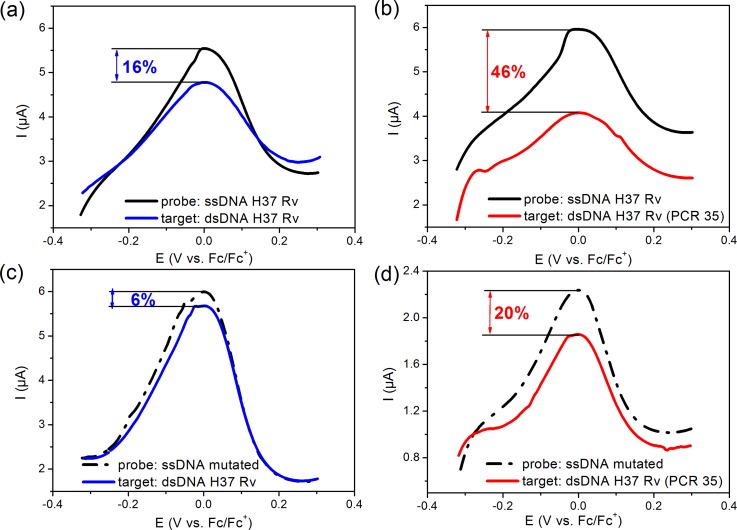
To validate our direct electrochemical detection for more relevant samples for diagnosis, a similar study has been done with this microfluidic device for the detection of rpob gene from gDNA of M. tuberculosis. The control was therefore realized with the DNA probe SNP T (TCG/TTG) in the codon position 531 of rpoB gene conferring resistance to rifampicin drug. The wild-type codon is TCG and the targeted mutant codon by the probe is TTG. Cyclic voltammograms recorded after hybridization with DNA H37Rv flowing under similar quasi-dynamic protocol in the fluidic device are shown, respectively, in Figures 5(a) and 5(b).
FIG. 5.
Electrochemical response of the biosensor of M. tuberculosis H37Rv genomic matrix DNA from clinical isolate before (black curve) and after the hybridization reaction (blue and red curve): (a) and (b) with the DNA probe corresponding to the M. tuberculosis H37Rv, (c) and (d) with the DNA probe specific to mutated (rifampicin RIF resistant) with only one mismatch. In figure (a) and (c), the hybridization reaction occurs in clinical isolates without PCR amplification (blue curves), whereas in (b) and (d) the analysis was done with the same sample after 35 cycles of PCR (red lines curves)—the sample was diluted to have the same concentration of DNA target for all experiments.
If the PCR35 sample produced the maximum response (ΔI/I0 = 46%), the high flow protocol allowed detection of the non-amplified sample (Fig. 5(a)) with a significant response (ΔI/I0 = 16%). To check the reliability of our biosensor, we also tested in the same conditions the detection of DNA H37Rv on a probe that differs from only one mismatch, namely, the mutated rifampicin resistant probe. Figures 5(c) and 5(d) present the corresponding CV curves with a small but reproducible response (ΔI/I0 = 6% for the native target and 20% for the PCR35 target). All these results confirm that the dynamic regime under high flow leads to high sensitivity of our microfluidic platform for DNA sensing from clinical isolates without any amplification step. Improvement of the sensitivity and selectivity could be reached by modification of CNT with nanomaterials such dendrimer polyamidoamine (PAMAM) as we demonstrated recently.38
IV. CONCLUSION
We have shown that a DNA grafted MWCNT biosensor integrated in a microfluidic device in conjunction with enhanced flow protocol for target mass transport improvement features very promising sensitivity and selective perspectives. Indeed, for Hepatitis C amplicon model solutions, we demonstrated enhanced dynamic range compared to macroscale systems associated to LoD of 0.7 fM. Although many POC systems have demonstrated tremendously impressive advances since the inception of microfluidics, little work focused on exclusive optimization of mass transport issues has been attempted. Therefore, through this work, we aimed to establish how such a strategy could bring higher performance to microfluidic systems. Indeed, the extremely high flow protocol generates such high values of Péclet numbers that the height of the depletion layer at the sensor surface can be reduced to hundreds of nanometers. Therefore, those extreme microfluidic conditions enhanced target-probes interaction events. Beyond this purely microfluidic enhancement, pursing the same thought, we believe dedicated efforts geared toward reagent selection, channel geometries, and thermal tuning optimization among other approaches could significantly contribute to further kinetics and mass transport optimization in confined environments.
In addition, our patterned receptor modified with the ferrocene redox marker with aminated linker on carbon nanotubes demonstrates strong electrochemical transduction upon DNA hybridization through charge transfer variation of the redox marker. The electrochemical microfluidic biosensor developed in this study demonstrates high potential for the detection of DNA samples from clinical isolates such as the rpob gene of M. tuberculosis without PCR amplification. The perspective of direct genomic pathogenic identification without an amplification step very positively promotes two major concerns. First, it implicitly induces lower microfluidic system complexity, thus indirectly causing performance and robustness improvements; and second, it offers a tremendously important simplified and accurate pathogenic load quantification perspective.
ACKNOWLEDGMENTS
The authors would like to thank CNRS for funding this research within the Fultrace project and LPN and ICMMO engineers' platforms for measurements of XPS and SEMs images.
References
- 1. Gubala V., Harris L. F., Ricco A. J., Tan M. X., and Williams D. E., Anal. Chem. 84, 487 (2012). 10.1021/ac2030199 [DOI] [PubMed] [Google Scholar]
- 2. Jokerst J. C., Emory J. M., and Henry C. S., Analyst 137, 24 (2012). 10.1039/C1AN15368D [DOI] [PubMed] [Google Scholar]
- 3. Atalay Y. T., Vermeir S., Witters D., Vergauwe N., Verbruggen B., Verboven P., Nicolaï B. M., and Lammertyn J., Trends Food Sci. Technol. 22, 386 (2011). 10.1016/j.tifs.2011.05.001 [DOI] [Google Scholar]
- 4. Falconnet D., She J., Tornay R., Leimgruber E., Bernasconi D., Lagopoulos L., Renaud P., Demierre N., and van den Bogaard P., Anal. Chem. 87, 1582 (2015). 10.1021/ac502741c [DOI] [PubMed] [Google Scholar]
- 5. Floris A., Staal S., Lenk S., Staijen E., Kohlheyer D., Eijkel J., and van den Berg A., Lab Chip 10, 1799 (2010). 10.1039/c003899g [DOI] [PubMed] [Google Scholar]
- 6. Gorkin R., Park J., Siegrist J., Amasia M., Lee B. S., Park J.-M., Kim J., Kim H., Madou M., and Cho Y.-K., Lab Chip 10, 1758 (2010). 10.1039/b924109d [DOI] [PubMed] [Google Scholar]
- 7. Liu R. H., Yang J., Lenigk R., Bonanno J., and Grodzinski P., Anal. Chem. 76, 1824 (2004). 10.1021/ac0353029 [DOI] [PubMed] [Google Scholar]
- 8. Ferguson B. S., Buchsbaum S. F., Wu T. T., Hsieh K., Xiao Y., Sun R., and Soh H. T., J. Am. Chem. Soc. 133, 9129 (2011). 10.1021/ja203981w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goodrich T. T., Lee H. J., and Corn R. M., Anal. Chem. 76, 6173 (2004). 10.1021/ac0490898 [DOI] [PubMed] [Google Scholar]
- 10. Wang J., Analyst 130, 421 (2005). 10.1039/b414248a [DOI] [PubMed] [Google Scholar]
- 11. Musameh M., Wang J., Merkoci A., and Lin Y., Electrochem. Commun. 4, 743 (2002). 10.1016/S1388-2481(02)00451-4 [DOI] [Google Scholar]
- 12. Cai H., Cao X., Jiang Y., He P., and Fang Y., Anal. Bioanal. Chem. 375, 287 (2003). 10.1007/s00216-002-1652-9 [DOI] [PubMed] [Google Scholar]
- 13. Tiwari J. N., Vij V., Kemp K. C., and Kim K. S., ACS Nano 10, 46 (2016). 10.1021/acsnano.5b05690 [DOI] [PubMed] [Google Scholar]
- 14. Gooding J. J., Chou A., Liu J., Losic D., Shapter J. G., and Hibbert D. B., Electrochem. Commun. 9, 1677 (2007). 10.1016/j.elecom.2007.03.023 [DOI] [Google Scholar]
- 15. Ruther R. E., Cui Q., and Hamers R. J., J. Am. Chem. Soc. 135, 5751 (2013). 10.1021/ja312680p [DOI] [PubMed] [Google Scholar]
- 16. Adams D. M., Brus L., Chidsey C. E. D., Creager S., Creutz C., Kagan C. R., Kamat P. V., Lieberman M., Lindsay S., Marcus R. A., Metzger R. M., Michel-Beyerle M. E., Miller J. R., Newton M. D., Rolison D. R., Sankey O., Schanze K. S., Yardley J., and Zhu X., J. Phys. Chem. B 107, 6668 (2003). 10.1021/jp0268462 [DOI] [Google Scholar]
- 17. Bouchet A., Chaix C., Marquette C. A., Blum L. J., and Mandrand B., Biosens. Bioelectron. 23, 735 (2007). 10.1016/j.bios.2007.06.018 [DOI] [PubMed] [Google Scholar]
- 18. Du Y., Chen C., Li B., Zhou M., Wang E., and Dong S., Biosens. Bioelectron. 25, 1902 (2010). 10.1016/j.bios.2010.01.003 [DOI] [PubMed] [Google Scholar]
- 19. Kim T.-H., Abi-Samra K., Sunkara V., Park D.-K., Amasia M., Kim N., Kim J., Kim H., Madou M., and Cho Y.-K., Lab Chip 13, 3747 (2013). 10.1039/c3lc50374g [DOI] [PubMed] [Google Scholar]
- 20. Squires T. M., Messinger R. J., and Manalis S. R., Nat. Biotechnol. 26, 417 (2008). 10.1038/nbt1388 [DOI] [PubMed] [Google Scholar]
- 21. Leyden M. R., Messinger R. J., Schuman C., Sharf T., Remcho V. T., Squires T. M., and Minot E. D., Lab Chip 12, 954 (2012). 10.1039/c2lc21020g [DOI] [PubMed] [Google Scholar]
- 22. Parsa H., Chin C. D., Mongkolwisetwara P., Lee B. W., Wang J. J., and Sia S. K., Lab Chip 8, 2062 (2008). 10.1039/b813350f [DOI] [PubMed] [Google Scholar]
- 23. Korri-Youssoufi H. and Makrouf B., Anal. Chim. Acta 469, 85 (2002). 10.1016/S0003-2670(02)00539-1 [DOI] [Google Scholar]
- 24. Gomgnimbou M. K., Abadia E., Zhang J., Refregier G., Panaiotov S., Bachiyska E., and Sola C., J. Clin. Microbiol. 50, 3172 (2012). 10.1128/JCM.00976-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murray M. G., Thompson W. F., and City S. L., Nucleic Acids Res. 8, 4321 (1980). 10.1093/nar/8.19.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laviron E., J. Electroanal. Chem. 101, 19 (1979). 10.1016/S0022-0728(79)80075-3 [DOI] [Google Scholar]
- 27. Landis E. C. and Hamers R. J., J. Phys. Chem. C 112, 16910 (2008). 10.1021/jp806173d [DOI] [Google Scholar]
- 28. Miodek A., Castillo G., Hianik T., and Korri-Youssoufi H., Biosens. Bioelectron. 56, 104 (2014). 10.1016/j.bios.2013.12.051 [DOI] [PubMed] [Google Scholar]
- 29. Rahman M. M., Kim Y. J., and Lee J.-J., J. Electrochem. Soc. 162, B159 (2015). 10.1149/2.0291507jes [DOI] [Google Scholar]
- 30. Nie Z., Nijhuis C. A., Gong J., Chen X., Kumachev A., Martinez A. W., Narovlyansky M., and Whitesides G. M., Lab Chip 10, 477 (2010). 10.1039/B917150A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taller D., Richards K., Slouka Z., Senapati S., Hill R., Go D. B., and Chang H.-C., Lab Chip 15, 1656 (2015). 10.1039/C5LC00036J [DOI] [PubMed] [Google Scholar]
- 32. Ben-Yoav H., Dykstra P. H., Bentley W. E., and Ghodssi R., Biosens. Bioelectron. 38, 114 (2012). 10.1016/j.bios.2012.05.009 [DOI] [PubMed] [Google Scholar]
- 33. Pavlovic E., Lai R. Y., Wu T. T., Ferguson B. S., Sun R., Plaxco K. W., and Soh H. T., Langmuir 24, 1102 (2008). 10.1021/la702681c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Triroj N., Jaroenapibal P., Shi H., Yeh J. I., and Beresford R., Biosens. Bioelectron. 26, 2927 (2011). 10.1016/j.bios.2010.11.039 [DOI] [PubMed] [Google Scholar]
- 35. Ali M. A., Srivastava S., Solanki P. R., Reddy V., Agrawal V. V., Kim C., John R., and Malhotra B. D., Sci. Rep. 3, 2661 (2013). 10.1038/srep02661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Basuray S., Senapati S., Aijian A., Mahon A. R., and Chang H., ACS Nano 3, 1823 (2009). 10.1021/nn9004632 [DOI] [PubMed] [Google Scholar]
- 37. Mahon A. R., Barnes M. A., Senapati S., Feder J. L., Darling J. A., Chang H. C., and Lodge D. M., PLoS One 6, e17280 (2011). 10.1371/journal.pone.0017280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miodek A., Mejri N., Gomgnimbou M., Sola C., and Korri-Youssoufi H., Anal. Chem. 87, 9257 (2015). 10.1021/acs.analchem.5b01761 [DOI] [PubMed] [Google Scholar]
- 39. Yeung S. S. W., Lee T. M. H., and Hsing I., Anal. Chem. 80, 363 (2008). 10.1021/ac071198+ [DOI] [PubMed] [Google Scholar]
- 40. Ferguson B. S., Buchsbaum S. F., Swensen J. S., Hsieh K., Lou X., and Soh H. T., Anal. Chem. 81, 6503 (2009). 10.1021/ac900923e [DOI] [PubMed] [Google Scholar]
- 41. Zhang J., Song S., Zhang L., Wang L., Wu H., Pan D., and Fan C., J. Am. Chem. Soc. 128, 8575 (2006). 10.1021/ja061521a [DOI] [PubMed] [Google Scholar]
- 42. Bangar M. A., Shirale D. J., Purohit H. J., Chen W., Myung N. V., and Mulchandani A., Electroanalysis 23, 371 (2011). 10.1002/elan.201000388 [DOI] [Google Scholar]
- 43.See supplementary material at http://dx.doi.org/10.1063/1.4940887E-BIOMGB-10-014601 for a description of the microfluidic chamber, fluidic parameters, XPS analyses, impedance measurements and characterization of the sensor's kinetics.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4940887E-BIOMGB-10-014601 for a description of the microfluidic chamber, fluidic parameters, XPS analyses, impedance measurements and characterization of the sensor's kinetics.