Abstract
Objective:
To investigate the association of NFKB1 -94 ins/del ATTG, NFKBIA -826C>T and NFKBIA -881A>G polymorphisms with risk of lung cancer in a Chinese population.
Methods:
Genotyping of the polymorphisms were performed on 1,436 subjects (718 cases and 718 controls) by using PCR-RFLP technique, followed by DNA sequencing.
Results:
We found a significant risk reduction associated with heterozygous ins/del (OR=0.705, 95% CI=0.566-0.878, P=0.002) and variant del/del (OR=0.342, 95% CI=0.221-0.528, P<0.001) genotypes of the NFKB1 polymorphism. In contrast, the heterozygous and variantgenotypes of theNFKBIA polymorphisms showed association with increased lung cancer risk (NFKBIA -826 CT,OR=1.256, 95%CI=1.004-1.572, P=0.046; TT,OR=1.773, 95% CI=1.131-2.778, P=0.013; NFKBIA -881 AG,OR=1.277, 95% CI=1.023-1.599, P=0.031; GG,OR=1.801, 95% CI=1.169-2.775, P=0.008). Several genotypic combinations of the three polymorphisms also showed significant association with lung cancer risk. The risk association of NFKB1 polymorphism remained significant when analyses were done according to gender and smoking status (P<0.05). The significance of NFKBIA risk association was not observed when gender-specific analyses were made (P>0.05), while only NFKBIA -881 GG genotype showed significant risk association among smokers when analyzed according to smoking status (P=0.032).
Conclusions:
Polymorphisms in NFKB1 and NFKBIAgenes were associated with risk of lung cancer.
KEY WORDS: Genetic variation, Lung cancer, NF-κB, Polymorphism, Risk
INTRODUCTION
Lung cancer is the most common type of cancer worldwide and represents a leading cause of cancer-related mortality.1-3 It has been suggested that genetic variations of cancer-related genes could play a role in influencing individual susceptibility to lung cancer.4,5 Among the many candidate genes, polymorphisms within inflammatory responsegenes have received increasing attention in the past few years, since inflammation has been strongly implicated in carcinogenesis.6-8
The nuclear factor-kappa B (NF-κB) family constitutes a group of transcription factors which serve as important mediators in inflammatory responses. In addition to inflammatory responses, a wide range of signal transduction processes converge on the NF-κB pathway, including cell proliferation, apoptosis, angiogenesis and many others.9 The p105/p50 isoforms of the NF-κBfamily, denoted NF-κB1, represents the most ubiquitous form of transcription factor in the family. Given the important role of NF-κB1 as the central hub of many biological processes, it is closely regulated by its endogenous inhibitors IκBα under normal conditions.10 NF-κB1 and IκBα are encoded by the NFKB1 and NFKBIA genes respectively, and it has been shown that disrupted expressions of these genes can result in carcinogenesis.11-13
Functional polymorphisms within the promoter region of these genes could potentially influence the levels of the proteins encoded. These functional polymorphisms may therefore contribute to the inter individual differences in lung cancer risk. To test our hypothesis, we conducted a case-control study to investigate the association of NFKB1 -94 ins/del ATTG polymorphism and NFKBIA -826C>T and -881A>G polymorphisms with the risk of lung cancer in a Chinese population.
METHODS
Study Population
718 lung cancer patients and 718controls participated in this retrospective case-control study. The participants were recruited between September 2011 andAugust 2014 fromthe Zhengzhou Central Hospital Affiliated to Zhengzhou University. Controls were cancer-free individuals randomly selected from a cancer screening program, and were matched (in frequency) to cases by age, sex and smoking behavior. The study was approved by the Research Review and Ethics Board of Zhengzhou Central Hospital Affiliated to Zhengzhou University(Approval number: 21234/RESP/43.2011). Written informed consent was obtained from all the participants prior to the study.
Genotyping
Genotyping was performed by using the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method (Fig.1)described elsewhere using the Life Express thermocycler (Bioer, China), and the genotypes were confirmed by sequencing 10% of the PCR products.14,15
Fig.1.
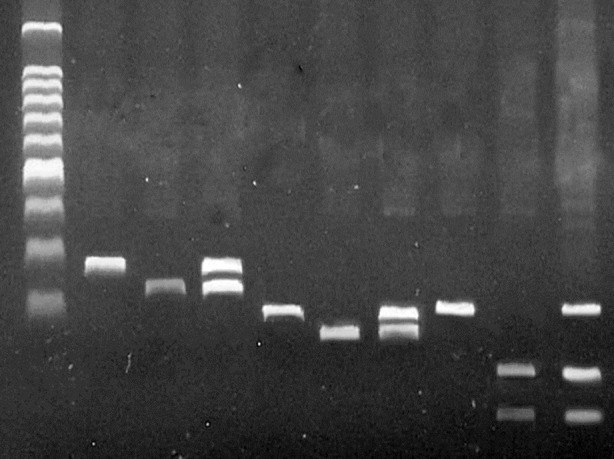
Gel image of PCR-RFLP. From left to right, NFKB1 del/del, NFKB1 ins/ins, NFKB1 ins/del, NFKBIA -826TT, NFKBIA -826CC, NFKBIA -826CT, NFKBIA -881AA, NFKBIA -881GG, NFKBIA -881AG.
Statistical Analysis
Differences in age, smoking status and gender between cases and controls were evaluated using the chi-square test. The association between the polymorphisms and lung cancer risk was determined using the logistic regression method to assess the odds ratio (OR). P values less than 0.05 were considered statistically significant.
RESULTS
Association of NFKB1 and NFKBIA polymorphisms with lung cancer risk
The association between NFKB1 and NFKBIA polymorphisms and lung cancer risk is shown in Table-I. By using ins/ins genotype as the reference, the heterozygous and homozygous variant genotypes of the NFKB1 polymorphism were found to be significantly associated with the reduced risk of lung cancer(P=0.002 for heterozygous, P<0.001 for variant). On the other hand, the heterozygous and homozygous variant genotypes of the NFKBIA polymorphisms appeared to be significantly associated with increased lung cancer risk(P=0.046 for -826CT, P=0.013 for -826TT, P=0.031 for -881AG, P=0.008 for -881GG,).
Table-I.
Association between NFKB1 -94 ins/del ATTG polymorphism, NFKBIA -826C>Tpolymorphism and NFKBIA -881A>G polymorphism and lung cancer risk.
| Genotype | Case, N=718 | Control, N=718 | OR (95% CI) | P |
|---|---|---|---|---|
| -94 ins/del ATTG | ||||
| Ins/ins | 434(60.4%) | 352(49.0%) | 1.000 | - |
| Ins/del | 252(35.1%) | 290(40.4%) | 0.705 (0.566-0.878) | 0.002* |
| Del/del | 32(4.5%) | 76(10.6%) | 0.342 (0.221-0.528) | <0.001* |
| -826C>T | ||||
| CC | 413(57.5%) | 461(64.2%) | 1.000 | - |
| CT | 251(35.0%) | 223(31.1%) | 1.256 (1.004-1.572) | 0.046* |
| TT | 54(7.5%) | 34(4.7%) | 1.773 (1.131-2.778) | 0.013* |
| -881A>G | ||||
| AA | 402(56.0%) | 454(63.2%) | 1.000 | - |
| AG | 257(35.8%) | 227(31.6%) | 1.277 (1.023-1.599) | 0.031* |
| GG | 59(8.2%) | 37(5.2%) | 1.801 (1.169-2.775) | 0.008* |
significant.
Combinations of polymorphisms and lung cancer risk
The association of combinations of NFKB1 -94 ins/del ATTG polymorphism and NFKBIA -826C>T polymorphism with lung cancer risk is shown in Table-II. Of the nine possible combinations, significant risk association was observed only for ins/del-CC(P=0.006), del/del-CC(P<0.001), and del/del-CT (P=0.003)combination genotypes. All the three combination genotypes showed decreased risk association.
Table-II.
Association between combinations of NFKB1 -94 ins/del ATTG polymorphism and NFKBIA -826C>T polymorphism and lung cancer risk.
| Genotype | Case | Control | OR (95% CI) | P | |
|---|---|---|---|---|---|
| -94 ins/del ATTG | -826C>T | ||||
| Ins/ins | CC | 259 (36.1%) | 232 (32.3%) | 1.000 | -- |
| Ins/del | CC | 134 (18.7%) | 179 (24.9%) | 0.671 (0.504-0.892) | 0.006* |
| Del/del | CC | 20 (2.8%) | 50 (7.0%) | 0.358 (0.207-0.620) | <0.001* |
| Ins/ins | CT | 144 (20.1%) | 103 (14.3%) | 1.252 (0.920-1.705) | 0.153 |
| Ins/del | CT | 99 (13.8%) | 95 (13.2%) | 0.934 (0.669-1.302) | 0.685 |
| Del/del | CT | 8 (1.1%) | 25 (3.5%) | 0.287 (0.127-0.648) | 0.003* |
| Ins/ins | TT | 31 (4.3%) | 17 (2.4%) | 1.633 (0.881-3.029) | 0.119 |
| Ins/del | TT | 19 (2.6%) | 16 (2.2%) | 1.064 (0.535-2.117) | 0.860 |
| Del/del | TT | 4 (0.6%) | 1 (0.1%) | 3.583 (0.398-32.288) | 0.255 |
significant.
Association between combinations of NFKB1 -94 ins/del ATTG polymorphism and NFKBIA -881A>G polymorphism and lung cancer risk is shown in Table-III. Similar to the above, three combinations showed significantly decreased risk association with lung cancer, namely ins/del-AA(P=0.004), del/del-AA (P<0.001)and del/del-AG (P=0.001)genotypes.
Table-III.
Association between combinations of NFKB1 -94 ins/del ATTG polymorphism and NFKBIA -881A>G polymorphism and lung cancer risk.
| Genotype | Case | Control | OR (95% CI) | P | |
|---|---|---|---|---|---|
| -94 ins/del ATTG | -881A>G | ||||
| Ins/ins | AA | 254 (35.4%) | 228 (31.8%) | 1.000 | -- |
| Ins/del | AA | 128 (17.8%) | 176 (24.5%) | 0.653 (0.489-0.872) | 0.004* |
| Del/del | AA | 20 (2.8%) | 50 (7.0%) | 0.359 (0.208-0.621) | <0.001* |
| Ins/ins | AG | 148 (20.6%) | 105 (14.6%) | 1.265 (0.930-1.721) | 0.134 |
| Ins/del | AG | 103 (14.3%) | 98 (13.6%) | 0.943 (0.679-1.311) | 0.729 |
| Del/del | AG | 6 (0.8%) | 24 (3.3%) | 0.224(0.090-0.559) | 0.001* |
| Ins/ins | GG | 32(4.5%) | 19 (2.6%) | 1.512(0.834-2.741) | 0.174 |
| Ins/del | GG | 21 (2.9%) | 16 (2.2%) | 1.178 (0.600-2.313) | 0.634 |
| Del/del | GG | 6 (0.8%) | 2 (0.3%) | 2.693(0.538-13.476) | 0.228 |
significant.
The combinations of NFKBIA -826C>T polymorphism and NFKBIA -881A>G polymorphism and their association with lung cancer risk is shown in Table-IV. Of the nine possible combinations, two combinations showed significantly increased risk association, namely CT-AG (P=0.043)and TT-GG (P=0.017)genotypes.
Table-IV.
Association between combinations of NFKBIA -826C>T polymorphism and NFKBIA -881A>G polymorphism and lung cancer risk.
| Genotype | Case | Control | OR (95% CI) | P | |
|---|---|---|---|---|---|
| -826C>T | -881A>G | ||||
| CC | AA | 400 (55.7%) | 452 (63.0%) | 1.000 | -- |
| CT | AA | 2 (0.3%) | 2 (0.3%) | 1.130(0.158-8.059) | 0.903 |
| TT | AA | 0 (0.0%) | 0 (0.0%) | --- | -- |
| CC | AG | 8 (1.1%) | 6 (0.8%) | 1.507(0.518-4.380) | 0.452 |
| CT | AG | 246 (34.3%) | 220 (30.6%) | 1.264(1.008-1.584) | 0.043* |
| TT | AG | 3 (0.4%) | 1 (0.1%) | 3.390 (0.351-32.722) | 0.291 |
| CC | GG | 5 (0.7%) | 3 (0.4%) | 1.883(0.447-7.931) | 0.388 |
| CT | GG | 3 (0.4%) | 1 (0.1%) | 3.390 (0.351-32.722) | 0.291 |
| TT | GG | 51 (7.1%) | 33 (4.6%) | 1.746(1.105-2.761) | 0.017* |
significant.
Analysis of NFKB1 risk association by gender and smoking status
The stratified association between the NFKB1 polymorphism and lung cancer risk according to gender and smoking status is shown in Table-V. The heterozygote genotype appeared to decrease the risk of lung cancer among females(P=0.005), but not males. However, the variant genotype decreased the lung cancer risk both in males(P=0.004) and females(P<0.001). The heterozygous genotype was also associated witha decreased risk of lung cancer among smokers(P=0.003), but not non-smokers. The variant genotype was associated with decreased lung cancer risk in both smokers (P<0.001)and non-smokers(P=0.001).
Table-V.
Association between NFKB1 -94 ins/del ATTG polymorphism and lung cancer risk according to sex and smoking status.
| Genotype | Case | Control | OR (95% CI) | P |
|---|---|---|---|---|
| Male | ||||
| Ins/ins | 220 (58.8%) | 188 (50.3%) | 1.000 | -- |
| Ins/del | 137 (36.6%) | 150 (40.1%) | 0.781 (0.577-1.056) | 0.108 |
| Del/del | 17 (4.5%) | 36 (9.6%) | 0.404 (0.220-0.742) | 0.004* |
| Female | ||||
| Ins/ins | 214 (62.2%) | 164 (47.7%) | 1.000 | -- |
| Ins/del | 115 (33.4%) | 140 (40.7%) | 0.630 (0.457-0.867) | 0.005* |
| Del/del | 15 (4.4%) | 40 (11.6%) | 0.287 (0.154-0.538) | <0.001* |
| Smoker | ||||
| Ins/ins | 234 (60.2%) | 181 (46.5%) | 1.000 | -- |
| Ins/del | 135 (34.7%) | 164 (42.2%) | 0.637 (0.472-0.859) | 0.003* |
| Del/del | 20 (5.1%) | 44 (11.3%) | 0.352 (0.200-0.617) | <0.001* |
| Non-smoker | ||||
| Ins/ins | 200 (60.8%) | 171 (52.0%) | 1.000 | --- |
| Ins/del | 117 (35.6%) | 126 (38.3%) | 0.794 (0.574-1.098) | 0.163 |
| Del/del | 12 (3.6%) | 32 (9.7%) | 0.321 (0.160-0.642) | 0.001* |
significant.
Analysis of NFKBIA -826C>T risk association by gender and smoking status
The association between NFKBIA -826C>T polymorphism and lung cancer risk according to sex and smoking status is shown in Table-VI.. Interestingly, the risk association of the heterozygous and variant genotypes appeared to be lost after stratification by gender and smoking status.
Table-VI.
Association between NFKBIA -826C>T polymorphism and lung cancer risk according to sex and smoking status.
| Genotype | Case | Control | OR (95% CI) | P |
|---|---|---|---|---|
| Male | ||||
| CC | 215 (57.5%) | 241 (64.4%) | 1.000 | --- |
| CT | 133 (35.6%) | 116 (31.0%) | 1.285 (0.943-1.752) | 0.112 |
| TT | 26 (7.0%) | 17 (4.5%) | 1.714 (0.905-3.246) | 0.098 |
| Female | ||||
| CC | 198 (57.6%) | 220 (64.0%) | 1.000 | --- |
| CT | 118 (34.3%) | 107 (31.1%) | 1.225 (0.886-1.695) | 0.220 |
| TT | 28 (8.1%) | 17 (4.9%) | 1.830 (0.972-3.445) | 0.061 |
| Smoker | ||||
| CC | 219 (56.3%) | 245 (63.0%) | 1.000 | --- |
| CT | 138 (35.5%) | 124 (31.9%) | 1.245 (0.919-1.686) | 0.157 |
| TT | 32 (8.2%) | 20 (5.1%) | 1.790 (0.995-3.222) | 0.052 |
| Non-smoker | ||||
| CC | 194 (59.0%) | 216 (65.7%) | 1.000 | --- |
| CT | 113 (34.3%) | 99 (30.1%) | 1.271 (0.912-1.772) | 0.157 |
| TT | 22 (6.7%) | 14 (4.3%) | 1.750 (0.871-3.515) | 0.116 |
significant.
Analysis of NFKBIA -881A>G risk association by gender and smoking status
The association between NFKBIA -881A>G polymorphism and the risk of lung cancer according to sex and smoking status is shown in Table-VII. Similar to the NFKBIA -826C>T polymorphism, no association was observed for the heterozygous and variant genotypes and lung cancer risk in both males and females after stratification by sex. However, after stratification by smoking status, significant association was observed only for the variant genotype with lung cancer risk among smokers(P=0.032).
Table-VII.
Association between NFKBIA -881A>G polymorphism and lung cancer risk according to sex and smoking status.
| Genotype | Case | Control | OR (95% CI) | P |
|---|---|---|---|---|
| Male | ||||
| AA | 208 (55.6%) | 235 (62.8%) | 1.000 | --- |
| AG | 136 (36.4%) | 120 (32.1%) | 1.280 (0.941-1.743) | 0.116 |
| GG | 30 (8.0%) | 19 (5.1%) | 1.784 (0.975-3.264) | 0.060 |
| Female | ||||
| AA | 194 (56.4%) | 219 (63.7%) | 1.000 | --- |
| AG | 121 (35.2%) | 107 (31.1%) | 1.277 (0.923-1.765) | 0.140 |
| GG | 29 (8.4%) | 18 (5.2%) | 1.819 (0.979-3.378) | 0.058 |
| Smoker | ||||
| AA | 214 (55.0%) | 240 (61.7%) | 1.000 | --- |
| AG | 140 (36.0%) | 128 (32.9%) | 1.227 (0.907-1.660) | 0.185 |
| GG | 35 (9.0%) | 21 (5.4%) | 1.869 (1.055-3.310) | 0.032* |
| Non-smoker | ||||
| AA | 188 (57.1%) | 214 (65.0%) | 1.000 | --- |
| AG | 117 (35.6%) | 99 (30.1%) | 1.345 (0.966-1.874) | 0.080 |
| GG | 24 (7.3%) | 16 (4.9%) | 1.707 (0.881-3.311) | 0.113 |
significant.
DISCUSSION
Given the important role of NF-κB and IκB in diverse biological pathways, genetic variations within key genes of the NF-κB pathway could obstruct the normal functioning of the protein products which in turn influences the risk of cancers. The NFKB1 -94 ins/del ATTG, NFKBIA -826C>T and NFKBIA -881A>G polymorphisms have been described. These functional polymorphisms have been shown to affect the level of the proteins produced, with the deletion allele, T allele and G allele of the three polymorphisms are associated with the reduced production of the respective proteins products.16,17
We have showed that the variant del genotype of the NFKB1 -94 ins/del ATTG polymorphism could significantly reduce lung cancerrisk. Our results are in disagreement with those reported by Huang et al.18 who found no association between the NFKB1 polymorphism and lung cancer risk in a Chinese population. However, our results concur with Huo et al.19 who investigated the association of the polymorphism with ovarian cancer among Chinese.
We have also showed in the current study that the variant genotypes of NFKBIA -826C>T and -881A>G polymorphisms could increase lung cancerrisk. Similar to the NFKB1 -94 ins/del ATTG polymorphism, our results are contrary to the results of Huang et al.18 who also found no association between the -826C>T polymorphism and lung cancer risk. However, when stratified by gender, we found that our results concur with the former, in that there was a lack of association of the polymorphism and cancer risk in both males and females. A similar lack of association was observed when stratified by smoking status, with the exception of NFKBIA -881GG variant genotype among smokers. This was in partial agreement with Umar et al.20 who found that the CT and CT+TT genotype of the -826C>T polymorphism was associated with a decreased risk of esophageal squamous cell carcinoma in an Indian population. On the other hand, Lin et al.21 showed that the heterozygous genotype of both the NFKBIA -826C>T and -881A>G polymorphisms increased the risk of oral cancer among Chinese. The disagreements among the findings reported in the literature could be due to the different backgrounds (age, sex, ethnic group, smoking status and geography) of the study subjects recruited, as well as the properness of subject matching.
The strengths of this study are: (i) it is the first study which establishes an association between the polymorphism and lung cancer risk in China, and (ii) it was conducted in a relatively properly-matched population. However, the limitation is that only three SNPs were analyzed.
CONCLUSION
We have showed that the variant alleles of NFKB1 -94 ins/del ATTG polymorphism, NFKBIA -826C>T polymorphism and NFKBIA -881A>G polymorphism could influence the risk of lung cancer in China.
Footnotes
Declaration of interest: None.
Source of Funding: No fund was received for the present study. The study was supported by the personal funds of the authors.
Authors’ contribution
Xiao-Yan Sun conceived and planned the study.
Jing-Wei Zhang, Qiu-Sheng Chen and Jian-Xia Zhai performed lab work and interpreted the evidence it presents.
Qiu-Sheng Chen and Peng-JuLv wrote the manuscript.
Qiu-Sheng Chen takes the responsibility and is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors have approved the final version of the manuscript.
REFERENCES
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. GLOBOCAN 2008 v2.0, Cancer Incidence & Mortality Worldwide: IARC CancerBase No. 10. Lyon, France: International Agency for Research on Cancer; 2010. [Google Scholar]
- 2.Wang YC, Wei LJ, Liu JT, Li SX, Wang QS. Comparison of cancer incidence between China & the USA. Cancer Biol Med. 2012;9(2):128–132. doi: 10.3969/j.issn.2095-3941.2012.02.009. doi: 10.3969/j.issn.2095-3941.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 4.Marshall AL, Christiani DC. Genetic susceptibility to lung cancer--light at the end of the tunnel? Carcinogenesis. 2013;34(3):487–502. doi: 10.1093/carcin/bgt016. doi: 10.1093/carcin/bgt016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz AG, Prysak GM, Bock CH, Cote ML. The molecular epidemiology of lung cancer. Carcinogenesis. 2007;28(3):507–518. doi: 10.1093/carcin/bgl253. [DOI] [PubMed] [Google Scholar]
- 6.Kundu JK, Surh YJ. Emerging avenues linking inflammation & cancer. Free RadicBiol Med. 2012;52(9):2013–2037. doi: 10.1016/j.freeradbiomed.2012.02.035. doi: 10.1016/j.freeradbiomed.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 7.DiDonato JA, Mercurio F, Karin M. NF-κB & The link between inflammation & cancer. Immunol Rev. 2012;246(1):379–400. doi: 10.1111/j.1600-065X.2012.01099.x. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 8.Eiró N, Vizoso FJ. Inflammation & cancer. World J Gastrointest Surg. 2012;27;4(3):62–72. doi: 10.4240/wjgs.v4.i3.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tak PP, Firestein GS. NF-kappaB: A key role in inflammatory diseases. J Clin Invest. 2001;107(1):7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors & its regulation. Cold Spring Harb Perspect Biol. 2009;1(4):a000034. doi: 10.1101/cshperspect.a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lind DS, Hochwald SN, Malaty J, Rekkas S, Hebig P, Mishra G, et al. Nuclear factor-kappa B is upregulated in colorectal cancer. Surgery. 2001;130(2):363–369. doi: 10.1067/msy.2001.116672. [DOI] [PubMed] [Google Scholar]
- 12.Chen CD, Sawyers CL. NF-kappa B activates prostate-specific antigen expression & is upregulated in androgen-independent prostate cancer. Mol Cell Biol. 2002;22(8):2862–2870. doi: 10.1128/MCB.22.8.2862-2870.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biswas DK, Dai SC, Cruz A, Weiser B, Graner E, Pardee AB. The nuclear factor kappa B (NF-kappa B): a potential therapeutic target for estrogen receptor negative breast cancers. Proc Natl Acad Sci USA. 2001;98(18):10386–10391. doi: 10.1073/pnas.151257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzairi MSM, Tan SC, Aizat AAA, Aminudin MM, Nurfatimah MSS, Andee ZD, et al. The functional -94 insertion/deletion ATTG polymorphism in the promoter region of NFKB1 gene increases the risk of sporadic colorectal cancer. Cancer Epidemiol. 2013;37(5):634–638. doi: 10.1016/j.canep.2013.05.007. doi: 10.1016/j.canep.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Tan SC, Suzairi MSM, Aizat AAA, Aminudin MM, Nurfatimah MSS, Bhavaraju VMK, et al. Gender-specific association of NFKBIA promoter polymorphisms with the risk of sporadic colorectal cancer. Med Oncol. 2013;30(4):693. doi: 10.1007/s12032-013-0693-6. doi: 10.1007/s12032-013-0693-6. [DOI] [PubMed] [Google Scholar]
- 16.Karban AS, Okazaki T, Panhuysen CI, Gallegos T, Potter JJ, Bailey-Wilson JE, et al. Functional annotation of a novel NFKB1 promoter polymorphism that increases risk for ulcerative colitis. Hum Mol Genet. 2004;13(1):35–45. doi: 10.1093/hmg/ddh008. [DOI] [PubMed] [Google Scholar]
- 17.Abdallah A, Sato H, Grutters JC, Veeraraghavan S, Lympany PA, Ruven HJ, et al. Inhibitor kappa B-alpha (IkappaB-alpha) promoter polymorphisms in UK & Dutch sarcoidosis. Genes Immun. 2003;4(6):450–454. doi: 10.1038/sj.gene.6364001. [DOI] [PubMed] [Google Scholar]
- 18.Huang D, Yang L, Liu Y, Zhou Y, Guo Y, Pan M, et al. Functional polymorphisms in NFκB1/IκBαpredict risks of chronic obstructive pulmonary disease & lung cancer in Chinese. Hum Genet. 2013;132(4):451–460. doi: 10.1007/s00439-013-1264-9. doi: 10.1007/s00439-013-1264-9. [DOI] [PubMed] [Google Scholar]
- 19.Huo ZH, Zhong HJ, Zhu YS, Xing B, Tang H. Roles of functional NFKB1&β-TrCP insertion/deletion polymorphisms in mRNA expression & epithelial ovarian cancer susceptibility. Genet Mol Res. 2013;12(3):3435–3443. doi: 10.4238/2013.March.11.6. doi: 10.4238/2013.March.11.6. [DOI] [PubMed] [Google Scholar]
- 20.Umar M, Upadhyay R, Kumar S, Ghoshal UC, Mittal B. Association of common polymorphisms in TNFA, NFkB1&NFKBIA with risk & prognosis of esophageal squamous cell carcinoma. PLoS One. 2013;8(12):e81999. doi: 10.1371/journal.pone.0081999. doi: 10.1371/journal.pone.0081999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CW, Hsieh YS, Hsin CH, Su CW, Lin CH, Wei LH, et al. Effects of NFKB1&NFKBIA gene polymorphisms on susceptibility to environmental factors & the clinicopathologic development of oral cancer. PLoS One. 2012;7(4):e35078. doi: 10.1371/journal.pone.0035078. doi: 10.1371/journal.pone.0035078. [DOI] [PMC free article] [PubMed] [Google Scholar]


