Abstract
Proteolipid protein (PLP1) and its alternatively spliced isoform, DM20, are the major myelin proteins in the CNS, but are also expressed in the PNS. The proteins have an identical sequence except for 35 amino acids in PLP1 (the PLP1-specific domain) not present in DM20. Mutations of PLP1/DM20 cause Pelizaeus-Merzbacher Disease (PMD), a leukodystrophy, and in some instances, a peripheral neuropathy. To identify which mutations cause neuropathy, we have evaluated a cohort of patients with PMD and PLP1 mutations for the presence of neuropathy. As shown previously, all patients with PLP1 null mutations had peripheral neuropathy. We also identified 4 new PLP1 point mutations that cause both PMD and peripheral neuropathy, three of which truncate PLP1 expression within the PLP1-specific domain, but do not alter DM20. The fourth, a splicing mutation, alters both PLP1 and DM20, and is probably a null mutation. Six PLP1 point mutations predicted to produce proteins with an intact PLP1-specific domain do not cause peripheral neuropathy. Sixty-one individuals with PLP1 duplications also had normal peripheral nerve function. These data demonstrate that expression of PLP1 but not DMSO is necessary to prevent neuropathy, and suggest that the 35 amino acid PLP1-specific domain plays an important role in normal peripheral nerve function.
The proteolipid protein 1 gene (PLP1) encodes a 276–amino acid protein, PLP1, as well as its alternatively spliced isoform, DM20 (Fig 1), which are the major structural proteins of central nervous system (CNS) myelin. Both PLP1 and DM20 are hydrophobic transmembrane proteins with four putative membrane spanning domains2 and have identical amino acid sequences except for the presence of an additional 35 amino acids within the second intracellular membrane loop of PLP1 (the PLP1-specific domain) which is absent in DM20 (see Fig 1).3 Interestingly, these proteins are also expressed in the peripheral nervous system (PNS),4,5 where their expression has been shown to be necessary for normal PNS function. Although the biological functions of PLP1/DM20 are not known with certainty, several lines of evidence suggest that PLP1 and DM20 have different roles in the PNS from those in the CNS. In the CNS, for example, PLP1 is the major protein isoform, whereas in the PNS, DM20 is the predominant species.5,6 In the CNS, both DM20 and PLP1 are localized to compact myelin. In contrast, in the PNS, PLP1 is found in the perinuclear cytoplasm as well as in compact myelin, whereas DM20 is found only in areas of uncompacted myelin.7,8 Finally, in both jimpy mice and md rats, animals with PLP1 point mutations, there is evidence of severe CNS dysfunction and dysmyelination, whereas the function and morphology of the peripheral nerves are essentially normal.
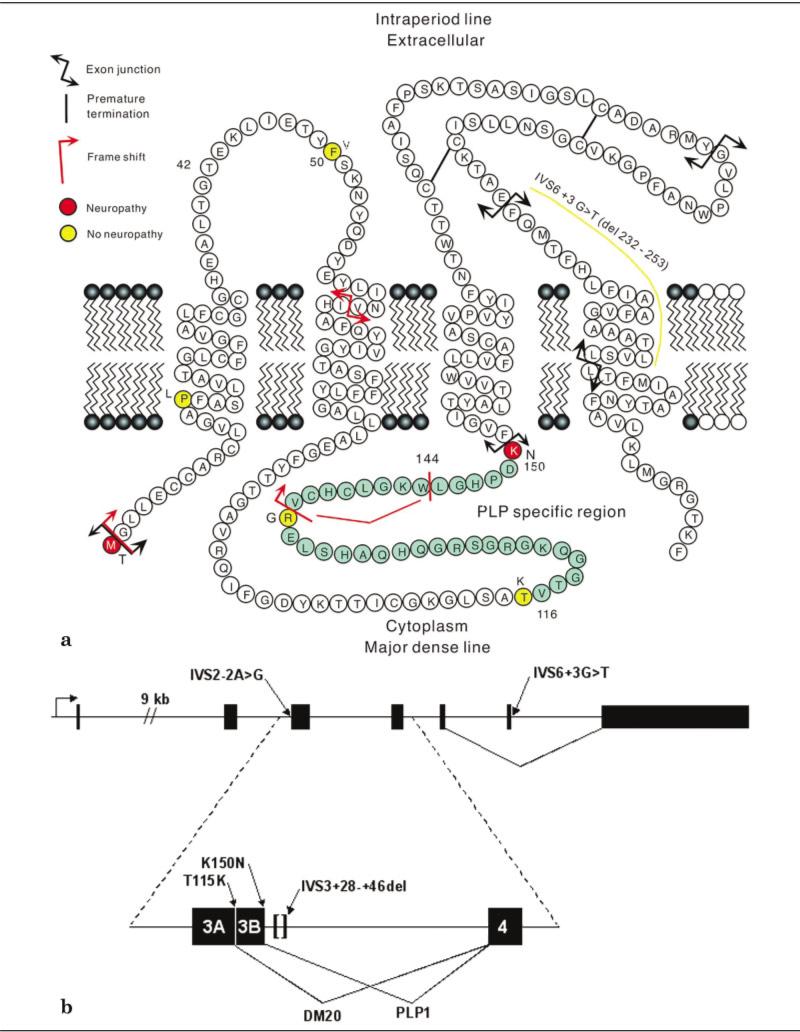
Fig 1.
(A) Schematic representation of PLP1 and DM20. The letters designate the amino acid residues of the proteins. Mutations known to cause neuropathy are shown in red. Mutations associated with normal peripheral nerve function are shown in yellow. The 35 amino acids of the PLP-specific domain are shown in green. (B) Structure of the human PLP1 gene. Use of one of two alternative splice donor sites within exon 3, designated 3A and 3B in the figure, produce either DM20 or PLP1, identical except for the presence of the 35–amino acid PLP1-specific domain in PLP1, which is absent in DM20. The location of mutations that effect PLP1 splicing, discussed further in the text, are shown by arrows.
PLP1 mutations in humans cause Pelizaeus–Merzbacher disease (PMD), an X-linked leukodystrophy.9 We previously have identified a family with the absence of PLP1 and DM20 expression due to a frame-shift mutation in the PLP1 gene (delG1) in which affected male subjects develop both PMD and a demyelinating peripheral neuropathy, demonstrating that these proteins are necessary for normal peripheral nerve myelination.1 To further delineate the function of PLP1/DM20 in the PNS, we evaluated peripheral nerve function in a cohort of families with known PLP1 mutations. We first confirmed the presence of peripheral neuropathy in five additional affected male members of the family with delG1 mutation. In addition, we identified a similar neuropathy in the affected male subjects from three other families with PLP1 null mutations, one with a complete PLP1 gene deletion and two with mutations within the PLP1 initiation codon. We also identified four new PLP1 mutations causing PMD and a demyelinating peripheral neuropathy. Three of these mutations (K150N, 144stop, and 136fs/144stop) truncate PLP1 within the PLP1-specific domain but do not alter DM20 expression. The fourth mutation (IVS2-2A→G) prevents the expression of both PLP1 and DM20 and is probably a null mutation. Six other PLP1 mutations (P14L, F50V, R136Q, T115K, IVS6+3G→T, IVS3+28-+46del), however, in which the PLP1-specific domain is unaffected do not alter peripheral nerve function. In addition, 61 patients with PLP1 duplications also have normal nerve conduction studies. Taken together, these data demonstrate that Schwann cell expression of PLP1, but not DM20, is necessary to prevent demyelinating peripheral neuropathy and suggest that the 35–amino acid PLP1-specific domain plays an important role in this process.
Subjects and Methods
Patient Ascertainment and Evaluation
Ascertainment of PMD patients was obtained through the cooperative efforts of the Pelizaeus–Merzbacher disease Program at Wayne State University and the European Network on Brain Dysmyelinating Diseases. Evaluations consisted of a neurological history and examination, MRI, and nerve conduction study. Testing for PLP1 duplication in each patient was done by quantitative polymerase chain reaction (PCR)10,11 or fluorescence in situ hybridization.12 Identification of PLP1 point mutations was done by DNA sequencing.9 Splicing mutations were named according to recommendations as described.13 Informed consent was obtained from study subjects or their parents/guardians.
Transfection Constructs
The splicing construct was made as described in Hobson and colleagues.14 The patient's mutations were introduced by site-directed mutagenesis. All constructs were verified by automated fluorescent sequence analysis. They all have a base change of A to G at position +549 in intron 2 that destroys a SphI site. The source of PLP1 cDNA was a 1.4kb pair EcoRI fragment comprising the human PLP1 cDNA.8 The DM20 cDNA was constructed by replacing the 330bp BglII/NcoI fragment from the PLP1 cDNA with a 225bp BglII/NcoI fragment from murine DM20 cDNA.15 This fragment lacks the PLP1-specific region but otherwise encodes the identical amino acid sequence as human PLP1.
Constructs for cDNA expression studies were constructed with HA epitope tags on their amino termini. The full-length PLP and DM20 open reading frames (kindly provided by Dr D. Colman, Mt. Sinai School of Medicine, New York), or mutant PLPs were cloned into the pEGFP-N1vector (ClonTech, Palo Alto, CA). To produce an in-frame fusion protein, we blunted the SacI site containing the PLP/DM20 termination codon with T4 DNA Polymerase and ligated it to a Klenow-blunted NcoI site in the EGFP vector. The resulting junction introduces two new amino acids into the fusion protein. DNA sequencing and Western blot analysis were used to confirm the integrity of the fusion protein.
To tag the amino terminus of constructs with HA, we amplified a fragment of the constructs using PCR with a 3′ HA primer including the sequence for the HA tag and a stop codon (encoding YPYDVPDYA stop). This fragment was cloned into PCR-Script (Stratagene). Sequencing of this plasmid was performed to ensure no errors have occurred during PCR. Plasmid DNAs were prepared for transfection by Qiagen (Valencia, CA) maxi-prep kits (splicing constructs) or by double banding in CsCl (cDNA expression constructs).
Cell Culture and Transient Transfections
Schwann cells were isolated from 3-day-old rat pups by the method of Brockes, as modified by Porter and colleagues16 using AraC and complement-mediated lysis with a thy 1.1 antibody to kill fibroblasts. The Schwann cells were expanded in Dulbecco's minimal essential medium supplemented with 10% fetal calf serum, glial growth factor (Cambridge Bioscience, Cambridge, MA), and 2μM forskolin. The cells were passaged no more than three times, grown to confluence in the presence of forskolin to maintain the differentiated phenotype, and then transfected using Lipofectamine (Life Technologies, Bethesda, MD) with 4μg of DNA per 35mm culture dish according to the manufacturer's instructions. At least two independent transfections were performed for each construct. Three days after transfection, the cells were fixed and analyzed by immunocytochemistry studies or harvested for total RNA or protein extraction. PLP1/DM20 expression at the cell surface was identified by the localization of immunofluorescent staining in membrane ruffles that lacked costaining with BiP. At least 20 transfected Schwann cells were analyzed for each PLP construct.
RNA Preparation, Reverse Transcription Polymerase Chain Reaction Analysis, and Sequence Analysis
Three-millimeter sections of frozen nerve biopsy material was used to generate total RNA by standard techniques17 for reverse transcription (RT)–PCR studies. Total RNA was prepared from Schwann cells transfected with the splicing constructs using an RNeasy Kit with QiaShredders (Qiagen) using protocols provided by the manufacturer. RT-PCRs were performed as described by Hobson and colleagues.11,14 RT-PCR products were separated on 4% NuSieve 3 to 1 agarose gels (FMC BioProducts, Rockland, ME), stained with ethidium bromide, and captured using an Eagle Eye II still video system (Stratagene, La Jolla, CA). Alternatively, when Cy5-labeled primers were used, band intensities were captured and quantitated on a Storm Phosphorimaging System (Molecular Dynamics, Sunnyvale, CA). Bands were excised from agarose gels and the DNA was eluted using a QiaQuick Kit (Qiagen) according to the manufacturer's instructions. After further PCR amplification of the eluted DNA as described above, automated fluorescent sequence analysis was performed on a Prism 377 DNA Sequencer (Applied Biosystems, Foster City, CA). Primers sequences used for sequence analysis are available at http://cmmg.biosci.wayne.edu/jgarbern/primers.html.
Immunocytochemistry
Cover slips bearing the transfected cells were placed on slides in phosphate-buffered saline (PBS) and allowed to dry before fixing for 30 minutes with 4% paraformaldehyde. After washing, the slides were postfixed in ice-cold acetone for 10 minutes and washed in PBS before immunolabeling. Slides were treated with blocking solution (5% normal goat serum, 0.1% Triton X-100 [Sigma, St. Louis, MO] in PBS) and incubated with primary antibodies for 18 hours at 4°C. Cells were treated with optimized dilutions of primary antibody (1:50), washed in PBS, and then incubated with specific fluorochrome conjugated secondary antibodies (1:100) for 1 hour at room temperature. Control slides were stained with secondary antibodies alone. Antibodies included rat monoclonal antibodies to the carboxy terminus of PLP (gift from K. Ikenaka), a polyclonal rat antibody to BiP18 to permit localization of the endoplasmic reticulum (ER), and a mouse monoclonal antibody to HA (Santa Cruz Biotechnologies, Santa Cruz, CA). Confocal images resulting from double immunostained cells were collected from two channels. To examine the distribution of two antigens, we generated data from both channels that produced a single image in which regions of colocalization were indicated in yellow. All image processing was done using Scanware on a Leica TCS 4D (Deerfield, IL) confocal scanning microscope and Adobe Photoshop software (Mountainview, CA).
Western Analysis
Western blots of proteins isolated from transfected Schwann cells were performed by standard conditions. Proteins from transfected cells were homogenized in 4% sodium dodecyl sulfate at a 1 to 20 wt/vol dilution, and 50 to 100μg of protein was loaded per lane and separated by electrophoresis in 10% acrylamide gels19 and transferred to a nitrocellulose membrane.20 Membranes were blocked with 5% nonfat dry milk in PBS and 0.1% Tween 20 (PBS-Tween), washed, incubated in primary antibody (diluted 1:2,000 in PBS-Tween) overnight at 4°C, washed, incubated for 1 hour in secondary antibody (diluted 1:5,000 dilution in PBS-Tween), washed, and processed for chemiluminescence using the Amersham ECL kit according to the manufacturer's instructions.
Results
PLP/DM20 Expression Is Necessary for Normal Peripheral Nervous System Function
We previously identified a family with PMD in which the affected male proband also had a demyelinating pe ripheral neuropathy.1 The PLP1 mutation identified in this patient, a deletion of the first nucleotide of the coding sequence of the PLP1 gene (delG1), causes a shift in the reading frame of the PLP/DM20 messenger RNA and creates a new stop codon two amino acids downstream from the start site of protein translation. For this reason, neither PLP nor DM20 protein can be produced in this subject, and the mutation is functionally a ‘null.’
To confirm that expression of PLP1 and DM20 is necessary for normal peripheral nerve function, we evaluated nerve conduction velocities and amplitudes in five additional male subjects from the delG1 family as well as in three other patients with PLP1 null mutations: two subjects from two kindreds with a Met to Thr mutation within the initiation codon and another subject with a complete PLP1 gene deletion. As shown in Table 1, all affected male subjects with a PLP1 null mutation, regardless of age, had a mild to moderate multifocal motor and sensory demyelinating neuropathy similar to that described previously in the proband.1 There was no conduction block or temporal dispersion noted. PLP1 gene expression, including both PLP1 and DM20, thus is necessary for normal PNS function.
Table 1.
Null Kindreds with Affected Male Subjects
| Motor Conduction Velocities |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ulnar Nerve |
Median Nerve |
Peroneal Nerve |
||||||||||||
| Subject | Age (yr) | Side | DML (ms) | Vel (m/s) | Amp (mV) | F (ms) | DML (ms) | Vel (m/s) | Amp (mV) | F (ms) | DML (ms) | Vel (m/s) | Amp (mV) | F (ms) |
| DelG1 family | ||||||||||||||
| VI-7 | 17 | R | ||||||||||||
| L | 4.0 | 36.0 | 7.0 | 42.8 | 5.0 | 5.1 | 9.5 | 29.0 | 0.9 | |||||
| VI-17 | 18 | R | 3.9 | 30.0 | 5.5 | 48.4 | 6.1 | 38.0 | 3.0 | 10.0 | 27.0 | 3.2 | ||
| L | 6.2 | 31.0 | 9.1 | 52.0 | 8.1 | 25.0 | 0.7 | 10.3 | 20.0 | 0.3 | ||||
| VI-6 | 20 | R | 3.3 | 47.0 | 8.8 | 34.6 | 5.4 | 44.0 | 6.9 | 36.6 | 7.0 | 24.0 | 2.7 | 66.2 |
| V-5 | 39 | 7.0 | 42.0 | 6.9 | 28.0 | 0.6 | ||||||||
| V-29 | 36 | 5.2 | 38.0 | |||||||||||
| VI-13 | 16 | L | 4.5 | 38.0 | 6.5 | 38.6 | 5.7 | 37.0 | 5.1 | 35.4 | 5.4 | 27.0 | 6.3 | |
| M1T | ||||||||||||||
| Family 1 | 4 | R | 4.6 | 45.0 | 2.5 | 34.2 | 4.4 | 57.0 | 29.6 | 6.9 | 23.0 | 0.7 | ||
| Family 2 | 14 | R | 5.8 | 36 | 4.8 | 33 | ||||||||
| Same patient, age 20 yr | 20 | R | 4.2 | 42 | 9.8 | 33 | ||||||||
| PLP1 Del family | 6.5 | 41 | 11.1 | 34 | ||||||||||
| 1-1 | 40 | R | 3.9 | 40 | 1.7 | 30.5 | 6.4 | 30 | 10.1 | |||||
| Normal values | <3.5 | >50 | >6.0 | <33 | <4.5 | >48 | >4.0 | <32 | <6.0 | >41 | >2.0 | <56 | ||
| Sensory Conduction Velocities |
||||||||
|---|---|---|---|---|---|---|---|---|
| Ulnar Nerve |
Median Nerve |
Sural Nerve |
||||||
| Subject | Age (yr) | Side | Vel (m/s) | Amp (μV) | Vel (m/s) | Amp (μV) | Vel (m/s) | Amp (μV) |
| DelG1 family | ||||||||
| VI-7 | 17 | R | 43.0 | 11.0 | 42.0 | 15.9 | ||
| L | 38.0 | 11.6 | 38.0 | 4.3 | 33.0 | 6.1 | ||
| VI-17 | 18 | R | 38.0 | 5.1 | ||||
| L | 27.0 | 8.3 | ||||||
| VI-5 | 20 | R | 48.0 | 30.0 | ||||
| V-29 | 36 | L | 40.0 | 4.9 | ||||
| VI-13 | 16 | L | 47.0 | 14.4 | 60.0 | 31.1 | 42.0 | 9.0 |
| Normal values | >54 | >20 | >50 | >20 | >35 | >6.0 | ||
Abnormal values are in boldface, italic type.
DML = distal motor latency; Vel = nerve conduction velocity; Amp = compound action motor potential; F = F-wave latency.
Mutations That Alter PLP1, but Not Its Alternatively Spliced Isoform, DM20, Cause Pelizaeus–Merzbacher Disease and Peripheral Neuropathy
To identify additional patients with PMD and peripheral neuropathy, we analyzed a cohort of patients with various PLP1 mutations, including duplications and point mutations, for the presence of peripheral neuropathy by measuring their nerve conduction velocities and amplitudes. As shown in Table 2, we found four new PLP1 mutations associated with peripheral neuropathy. The neuropathy had electrophysiological features of both demyelination and axonal loss, but there was no evidence of conduction block or temporal dispersion. Because few subjects were studied in this group, however, at this time we cannot determine whether demyelination or axonal loss is the primary pathological process. The clinical features of neuropathy in these subjects were subtle, and masked by neurological signs of CNS disease. These mutations include a point mutation in intron 2 (IVS2-2A→G) and three different point mutations within exon 3 (K150N,21,22 144stop,23,24 and 136fs/144stop). The mutation at the –2 position of intron 2 (IVS2-2A→G) is located within the splice acceptor site of this intron and probably alters mRNA splicing, producing PLP1 and DM20 proteins with altered carboxy termini, suggesting this mutation is functionally a null. The three point mutations within exon 3, however, fall within the PLP1-specific domain not expressed in DM20 and are predicted to produce altered or truncated PLP1, but normal DM20. The location of these three mutations within the PLP1-specific domain suggests that PLP1 expression is necessary for normal peripheral nerve function, but that DM20 expression is not. Further analysis of each of these mutations, as shown below, supports this conclusion.
Table 2.
Additional Mutations Causing Neuropathy
| Motor Conduction Velocities |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ulnar Nerve |
Median Nerve |
Peroneal Nerve |
||||||||||||
| Subject | Age (yr) | Side | DML (ms) | Vel (m/s) | Amp (mV) | F (ms) | DML (ms) | Vel (m/s) | Amp (mV) | F (ms) | DML (ms) | Vel (m/s) | Amp (mV) | F (ms) |
| IVS2-2A→G: disrupts exon 3 splice acceptor | 9 | R | 4.1 | 41.0 | 11.7 | 6.0 | 43.0 | 16.9 | 30.8 | 6.6 | 33 | 2.4 | 49.9 | |
| 136fs/144 stop | 11 | R | 3.0 | 49.0 | 0.2 | 5.1 | 35 | 2.0 | ||||||
| 136fs/144 stop | 14 | R | 3.0 | 37.0 | 0.1 | 5.0 | 48 | 7.5 | ||||||
| K150N | 9 | L | 2.1 | 43 | 3.7 | 2.3 | 38.0 | 4.1 | ||||||
| 144stop | 19 | L | 3.2 | 68 | 11.8 | 25.4 | 5.0 | 58.0 | 9.8 | 26.9 | 4.8 | 47 | 7.4 | 45.1 |
| Normal values | <3.5 | >50 | >6.0 | <33 | <4.5 | >48 | >4.0 | <32 | <6.0 | >41 | >2.0 | <56 | ||
| Sensory Conduction Velocities |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ulnar Nerve |
Median Nerve |
Sural Nerve |
||||||||
| Subject | Age (yr) | Side | Vel (m/s) | Lat (ms) | Amp (μV) | Vel (m/s) | Lat (ms) | Amp (μV) | Vel (m/s) | Amp (μV) |
| IVS2-2A→G: disrupts exon 3 splice acceptor | 9 | R | 4.1 | 34.4 | 4.1 | 49.2 | ||||
| 144stop | 19 | L | 58.0 | 25.9 | 50.0 | 40.5 | 45 | 21 | ||
| Normal values | >54 | <2.9 | >20 | >50 | <3.2 | >20 | >35 | >6.0 | ||
Abnormal values are in boldface, italic type.
DML = distal motor latency; Vel = nerve conduction velocity; Amp = compound action motor potential; F = F-wave latency.
Analysis of the IVS2-2A→G Mutation
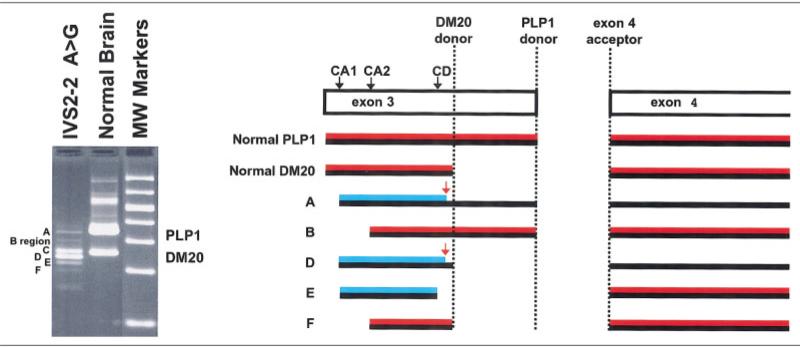
To evaluate the effect of the point mutation within intron 2 on PLP1/DM20 expression, we analyzed the PLP1 and DM20 cDNAs obtained from the sural nerve of an affected patient. Six different ‘PLP1’ bands were identified after RT-PCR; five were PLP/DM20-specific cDNAs, whereas the sixth (band C) was a heteroduplex of bands D and E (Fig 2). The significance of the amount of each of these five bands, however, is not known. The DNA sequence of the five PLP1/DM20-specific cDNAs then were determined, as shown in Figure 2. Each of these five cDNAs encodes an abnormal PLP1 or DM20 protein, probably because of the use of alternative splice acceptor sites within exon 3 (see Fig 2). Two of the abnormal cDNAs encode proteins that are missing the initial 19 amino acids of exon 3A, which encodes most of the second transmembrane domain of both PLP1 and DM20. Three others encode proteins that are missing the first five amino acids of exon 3A and have an altered sequence downstream of this point because of a frame-shift. Of these three, two have altered carboxy termini caused by an aberrant stop codon in exon 3A. In the third, activation of a cryptic donor site in exon 3A before the stop codon in the altered reading frame results in restoration of the reading frame at the beginning of exon 4. Thus, this cDNA has normal exons 1 and 2, as well as exons 4 through 7, but does not contain exon 3, which is responsible for the PLP1-specific domain of the mature protein. No cDNAs encoding normal PLP1 or DM20 were found. The IVS2-2A→G mutation thus alters PLP1 RNA splicing, producing an array of abnormal PLP and DM20 proteins.
Fig 2.
Reverse transcription polymerase chain reaction (RT-PCR) analysis of sural nerve RNA from a patient with the IVS2-2A→G mutation. The agarose gel electrophoresis of the RT-PCR products is shown in the first lane in the left panel; the structure of the cDNAs, determined by DNA sequence analysis, is diagrammed in the right panel. Product C is a heteroduplex of products D and E (not shown). The cDNA products in the diagram are shown by solid black lines. Conceptual translation of the cDNA products is indicated in red. The PLP1/DM20 amino acid sequence is shown by a solid red line. Frameshift products are shown by diagonal striped lines. Premature stop codons are indicated by red arrows. The IVS2-2A→G mutation alters the splice acceptor site within intron 2 and results in utilization of two cryptic splice acceptor sites (CA1 and CA2) and one cryptic splice donor site (CD) located within exon 3. RT-PCR analysis of human brain RNA is shown in the second lane in the left panel; molecular weight markers are displayed in the third lane in the left panel.
To determine whether any of these mutant proteins could be expressed, we cloned each of the altered PLP1 and DM20 cDNAs with an HA tag sequence at the 5′ end into an expression vector with a cytomegalovirus promoter and transfected them into COS-7 cells. Analysis of the transfected cells by immunohistochemistry demonstrated that none of the mutant cDNAs derived from the CA1 splice site which contain both a deletion and frameshift express either the amino terminal epitope, recognized by an anti–HA tag antibody, or the carboxy terminal epitope of PLP1 recognized by the AA3 antibody (not shown). The cDNAs derived from the CA2 splice site, however, have an in-frame deletion and are detected at low levels using the anti–HA tag antibody. These proteins lack the initial 19 amino acids of the second transmembrane domain and thus probably are not inserted into the cell membrane. These data suggest that the IVS2-2A→G point mutation in the PLP1 gene is a null mutation, leading to the absence of both PLP1 and DM20 expression and are consistent with our previous finding that the absence of PLP and DM20 expression causes peripheral neuropathy.
Analysis of the 136fs/144stop and 144stop Mutations
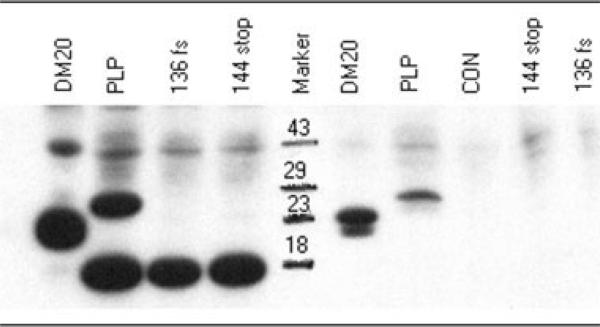
To evaluate the biochemical consequence of the 136fs/144stop and 144stop mutations within exon 3B, encoding the PLP1-specific domain, we constructed cDNAs encoding these mutant proteins, and transfected them into COS7 cells. Neither of these mutant proteins was detected in transfected cells by either Western blotting (Fig 3) or immunohistochemistry utilizing an antibody to the carboxy terminus of PLP1 (not shown). Using an antibody to the HA tag located at the amino terminus of the protein, however, we identified a protein of approximately 14kDa, smaller than normal PLP1, on the Western blot (see Fig 3), although no normal-sized protein was detected. This smaller protein is the predicted size of PLP1 truncated at amino acid 144, the site of the stop codon in both mutants. Interestingly, a similar-sized protein fragment is also seen in both control lanes along with the normal-sized protein of 30kDa. A proteolipid of approximately 14kDa has been identified previously in purified preparations of PLP1 and shown to be the result of proteolytic cleavage, so that this protein is also likely an amino terminal fragment of PLP1. The approximately 43kDa band observed in all the lanes is probably because of nonspecific binding. These results demonstrate that both the 136fs/144stop and the 144stop mutations produce a truncated PLP1, consistent with the notion that PLP1, but not DM20, is necessary for normal PNS function.
Fig 3.
Western blot analysis of protein extracts from COS7 cells transiently transfected with PLP1/DM20 and mutant 136fs/144stop and 144stop cDNA constructs. The lanes to the left of the markers were probed with an antibody to the HA tag, located on the amino terminus of the protein; the lanes to the right of the markers were probed with an antibody to the carboxy terminus shared by both PLP1 and DM20. Protein extract on the control (CON) lane was prepared from nontransfected COS7 cells.
Analysis of the K150N Mutation
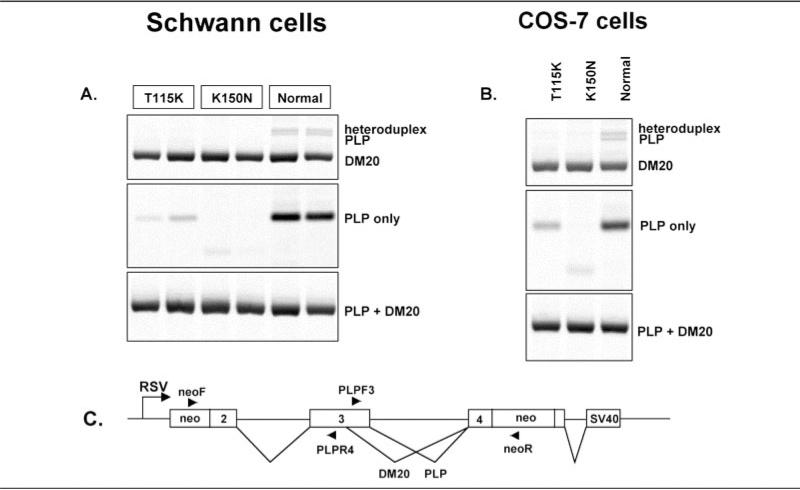
The third mutation in exon 3B causing neuropathy, K150N, is an amino acid substitution. Amino acid 150, however, is the last amino acid in exon 3B and includes the splice donor site. Splice donor site mutations are known to cause skipping of the preceding exon or activation of a cryptic splice site, so that this coding mutation could lead to an alteration of PLP1 splicing. Because we could not obtain a nerve biopsy from this patient, we introduced the K150N mutation into an expression construct containing a portion of the PLP1 gene encoding part of exon 2, all of exon 3, part of exon 4, and intervening sequences 2 and 3 under control of the cytomegalovirus promoter. This construct then was transfected into both COS-7 cells and Schwann cells, and the resulting spliced RNA was assayed by RT-PCR. As can be seen in Figure 4, mRNA encoding DM20 was produced in both cell types, but PLP1 mRNA was not detected in either, suggesting that the K150N mutation does alter mRNA splicing. A unique, smaller band was seen in two of the lanes that were isolated and shown to encode an altered PLP1 lacking amino acids 136 to 150, produced by utilization of an alternative splice donor site within exon 3B. These data suggest that this patient expresses normal DM20 but an altered PLP1 and support the hypothesis that PLP1, but not DM20, is necessary for normal peripheral nerve function.
Fig 4.
Analysis of PLP1 splicing in Schwann cells and COS7 cells. Reverse transcription polymerase chain reaction (RT-PCR) analysis of RNA from (A) Schwann cells and (B) COS7 cells transfected with constructs encoding either a normal or mutant PLP1 (T115K; K150N) minigene constructs containing exons 2, 3, and 4 of the PLP1 gene. The structure of the PLP1 minigene and the primer pairs used in the RT-PCR analysis are shown in panel C.
Analysis of the T115K Mutation
We also assayed the T115K mutation25 in our in vitro splicing assay even though the patient carrying this mutation does not have neuropathy. This mutation is located near the alternative splice donor site at the end of exon 3A, and, like the K150N mutation, might also affect RNA splicing. Analysis of this mutation thus is an important control for our hypothesis that PLP1 is required for normal peripheral nerve function. The results of this experiment, shown in Figure 4, demonstrate that the T115K mutation does not alter the pattern of PLP1 splicing, because both PLP1 and DM20 are expressed from this construct, although the amount of PLP1 cDNA is reduced. These data further support the hypothesis that PLP1 expression is necessary for normal peripheral nerve function. In addition, they suggest that small amounts of PLP1 are sufficient to maintain normal peripheral nerve function.
PLP1 Gene Duplications Do Not Cause Demyelinating Peripheral Neuropathy
The most common genetic mechanism causing PMD is PLP1 gene duplication, present in 60 to 70% of PMD patients, which probably produces disease caused by PLP1/DM20 overexpression.10 To determine whether PLP1 duplications also can cause peripheral neuropathy, we screened 61 PLP1 duplication patients, aged 3 to 45 years, for the presence of nerve conduction abnormalities. No evidence of neuropathy was found in any of these subjects with PLP1 duplications, suggesting that PLP1/DM20 overexpression is not sufficient to produce peripheral nerve dysfunction.
Some PLP1 Point Mutations Cause PMD but Do Not Affect the Peripheral Nervous System
We have identified six PLP1 mutations that cause signs and symptoms of PMD but do not cause peripheral neuropathy. These include the point mutations P14L,26,27 F50V, R136Q, T115K,25 a point mutation in the splice donor site of intron 6 (IVS6+3G→T),11 and a 19bp deletion in intron 3 (IVS3+28-+46del).11 Interestingly, two of these mutations, P14L and IVS6+3G→T, produce particularly severe CNS disease, demonstrating that some PLP1 mutations can cause severe CNS disease without affecting PNS function.
Gow and Lazzarini have suggested that the severity of PMD is a consequence of the misfolding of PLP1 and DM20. They demonstrated that both mutant PLP1 and mutant DM20 from patients with severe PMD are misfolded and retained within the ER in COS7 cells, whereas only PLP1 is misfolded and retained28 from mildly affected patients. Consistent with these data, mutant Plp1 from myelin-deficient (md) rats, a rodent model of severe PMD, is retained within the ER in oligodendrocytes (reviewed in Griffiths and colleagues29), whereas significant amounts of mutant Plp1 from the rumpshaker mouse (rsh) with a milder disease can be transported to the cell surface. Because the abnormal trafficking of PLP1 and DM20 is a consequence of mutation and protein misfolding, one also would expect mutant PLP1 and DM20 to be retained within the Schwann cell ER. As stated above, we have identified five patients with PLP1 missense mutations and severe PMD who do not have neuropathy. These data suggest either that misfolded PLP1 and DM20 are processed differently in Schwann cells from in other cell types or that the mutant proteins are retained within the Schwann cell ER but can still subserve a portion of their normal function at that location.
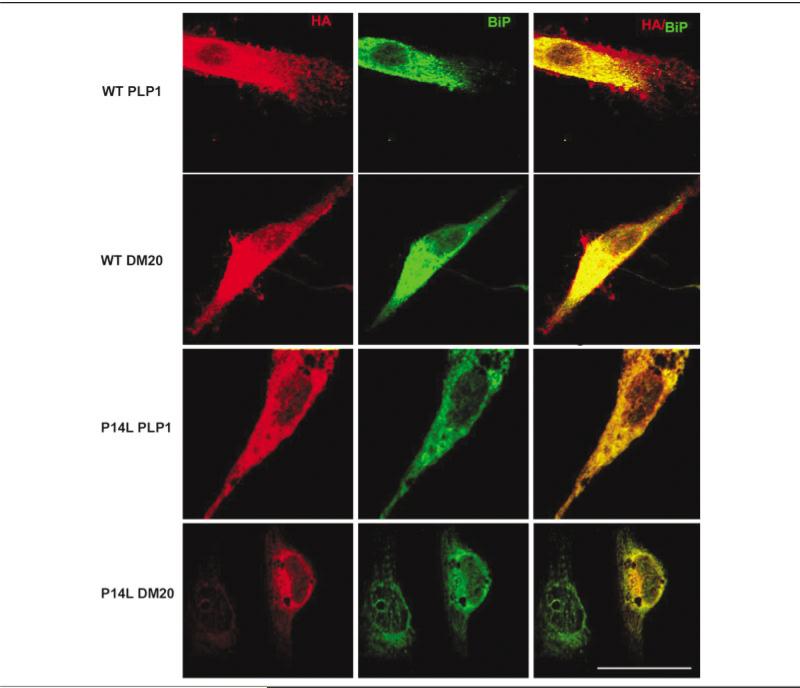
To test whether misfolded PLP1 and DM20 are processed differently in Schwann cells from in COS7 cells, we expressed both the normal PLP1/ DM20 and PLP1/DM20 carrying the P14L mutation in differentiated Schwann cells and COS7 cells and evaluated their cellular localization by immunohistochemistry as described in Subjects and Methods. The P14L mutation causes severe CNS disease and is known to undergo protein misfolding as a consequence of this nonconservative amino acid substitution in the first transmembrane domain.30 As expected, normal PLP1 and DM20 were transported to the cell surface in Schwann cells (Fig 5) as they are in COS7 cells (data not shown). In contrast, consistent with the findings of Gow and Lazzarini, the PLP1 and DM20 proteins carrying the P14L mutation colocalized with BiP, a marker of the ER30 in both COS7 cells (data not shown) and Schwann cells (see Fig 5) and were not found on the cell surface. These data demonstrate that these mutant proteins are similarly misfolded and arrested in their intracellular transport in Schwann cells as they are in COS7 cells, arguing against Schwann cell–specific protein processing. Because the P14L mutation does not cause peripheral neuropathy, these mutant proteins may provide at least a portion of their normal function in Schwann cells within the ER.
Fig 5.
Analysis of cultured Schwann cells transiently transfected with normal and mutant P14L PLP1 and DM20. The cells were immunostained with antibodies to the carboxy terminus of PLP (rhodamine) and BiP (FITC). As shown in the top two panels, normal PLP1 and DM20 are detected within the ER, throughout the cytoplasm, and on the cell surface. In the bottom two panels, both P14L PLP1 and P14L DM20, however, are retained within the ER, colocalize with BiP, and are not found at the cell surface. Scale bar = 10μm.
Discussion
In this article, we have confirmed the presence of demyelinating peripheral neuropathy in five affected male members of a family with a PLP1 null mutation caused by a nucleotide deletion causing a frameshift in the PLP1 coding region. In addition, we have identified a similar neuropathy in the affected male members of three additional PMD families, one with a PLP1 deletion and two with a frameshift mutation within the PLP1 initiation codon, both of which also abolish PLP1 and DM20 expression. Expression of PLP1 thus is necessary for normal peripheral nerve myelination.
We have also identified four additional subjects with PLP1 mutations who have both PMD and electrophysiological evidence for peripheral neuropathy. In three of these subjects, the PLP1 mutation is predicted to truncate PLP1 within the PLP1-specific domain downstream of amino acid 144 but does not alter DM20 expression. In the fourth patient, the mutation alters the coding sequence of both PLP1 and DM20 because of the use of one of two cryptic splice acceptor sites within exon 3, abolishing expression of both proteins. Six additional subjects with PLP1 mutations are predicted to produce proteins with an intact PLP1-specific domain, however, have normal peripheral nerve function, as do 61 patients with PLP1 duplications. Transfection studies of mutant and normal PLP1 and DM20 do not demonstrate Schwann cell–specific processing of these proteins, some of which, as previously shown in COS7 cells, also are sequestered within the ER of Schwann cells. Taken together, these data demonstrate that Schwann cell expression of PLP1 is necessary for normal peripheral nerve myelination. DM20 expression alone is not sufficient to maintain normal nerve function. Mutations that specifically alter the PLP1 expression thus are predicted to cause both PMD and peripheral neuropathy.
The above data from patients with PLP1 mutations suggest that PLP1, but not DM20 expression in Schwann cells, plays a role in the process of myelination. Consistent with this notion we have demonstrated previously that PLP1, but not DM20, is coordinately expressed with the set of myelin-specific proteins P0, MAG, P2, and Cx32 in the PNS.31 In addition, expression of both the myelin-specific genes and PLP1 depends on continuous Schwann cell–axonal contact, whereas DM20 is expressed in Schwann cells in the absence of axons.31 Finally, we also have shown that PLP1 is located within compact myelin.1 The function of PLP1 in the PNS myelination, however, still remains to be elucidated.
The molecular consequences of the K150N mutation at the end of exon 3B further demonstrate the importance of PLP1 in PNS myelination. Mutation of the final nucleotide of exon 3B from a G to a T (G450T) changes the coding sequence of PLP1 at amino acid 150 from lysine to asparagine (AAG to AAT; K150N), causing both PMD and a demyelinating peripheral neuropathy. Codon 150, however, is also part of a cononical splice donor sequence recognized by the U1 snRNA, 5′ ... (exon 3B) ... AGguragu ... (IVS 3) ... 3′, so that the G to T transversion should also alter RNA splicing, reducing the use of the normal splice donor site and possibly uncovering a cryptic splice site upstream.32 Analysis of the G to T mutation in vitro is consistent with this mechanism: no cDNA encoding PLP1 can be detected, although there is cDNA formed by utilizing an upstream cryptic splice site within exon 3B. Two additional patients with PMD also have been identified with mutations in codon 150. One of these mutations alters the final nucleotide of exon 3B from a G to a C (G450C; AAG to AAC) producing the same amino acid change, K150N; the second mutation alters the final nucleotide of exon 3B from a G to an A (G450A; AAG to AAA) and is a silent mutation (unpublished data). Both of these mutations probably affect PLP1 splicing in a similar manner as in our patient, producing a PLP1 with an internal, in-frame 14–amino acid deletion but without changing DM20. Whether either of these two patients also have a demyelinating peripheral neuropathy is not known currently but is under investigation.
The molecular consequences of T115K, caused by a point mutation at the end of exon 3A also provide evidence for the role of PLP1 in PNS myelination. Mutation of nucleotide 344 from a C to an A changes amino acid 115 at the beginning of exon 3A from a threonine to a lysine (T115K; C344A; ACG to AAG) and causes PMD but not a demyelinating peripheral neuropathy. Codon 115 contributes to the alternative splice donor site sequence recognized by the U1 snRNA, 5′ ... exon 3A ... (A/C)Gguragu ... (exon 4) ... 3′.32 The C344A mutation changes the second base of the splice donor sequence from C to the more favored A, strengthening the splice donor site. Because alternative splice sites are frequently weak sites with a poor match to the consensus (reviewed in Berget33 and Smith and Valcarcel34), a change to a consensus site could result in increased use of this site.35 Analysis of the consequence of this C to A mutation in vitro is consistent with this mechanism: correctly spliced PLP1 and DM20 cDNAs are both present after transfection of the mutated minigene construct, but the amount of PLP1 cDNA is reduced. Although the in vivo effect of the T115K mutation on PLP1 splicing is not known, there must be sufficient amounts of PLP1 to maintain normal peripheral nerve function. Analysis of mRNA from peripheral nerve from this patient will be necessary to investigate this point further.
Because of the change in mRNA splicing, the molecular consequences of the K150N mutation are similar to those of the 144stop and the 136fs/144stop mutations, both of which also are predicted to produce normal DM20, but a PLP1 truncated within the PLP1-specific domain. Interestingly, the subject with the K150N mutation is clinically much more severely affected than either subject with the 144stop or 136fs/144stop mutations. Although the reasons for this disparity are not known, one possibility is that the K150N mutation produces an altered PLP1 with a gain of function effect in oligodendrocytes, whereas the 144stop and 136fs/144stop mutations do not. Clearly, further study will be needed to correlate the phenotype with genotype in these three patients.
Although truncation of PLP1 within the PLP1-specific domain is associated with peripheral neuropathy, several mutations that cause misfolding and inhibit trafficking of both PLP1 and DM20 do not affect peripheral nerve function. Because our transfection data demonstrate that trafficking of both mutant and normal PLP1 and DM20 in Schwann cells is similar to that in other cell types, these mutant proteins probably accumulate within the ER of Schwann cells. These data imply either that PLP1 and DM20 can function in Schwann cells within an intracellular compartment or that sufficient amounts of mutant PLP1 and DM20 to rescue nerve function are expressed on the cell surface. Nave and colleagues recently reported that the gene encoding the tetramembrane spanning protein, M6B, similar in both structure and sequence to PLP1 and DM20, is alternatively spliced, producing at least eight separate M6B protein isoforms that are differentially expressed in neurons and oligodendrocytes.36 Six of these isoforms are tetraspan membrane proteins that differ by highly conserved amino- and carboxy-terminal domains. Interestingly, one of these domains, called the β-domain, stabilizes tetraspan proteolipids at the cell surface. In contrast, non-β isoforms are more abundant in intracellular compartments, including the ER, suggesting that these proteins have an intracellular role. Perhaps PLP1 and/or DM20 have a similar intracellular function in myelinating Schwann cells.
Acknowledgments
This work was supported in part by a grant from the Nemours Foundation (G.H).
We thank the patients and families with PMD for their help in performing these studies.
Appendix
Members of the Clinical European Network on Brain Dysmyelinating Disease (ENBDD): M. Baethmann, E. Bertini, B. Chabrol, J. M. Cuisset, J. Gaertner, F. Hanefeld, A. Kohlschutter, P. Landrieu, J. M. Lopes-Terradas, M. Mayer, S. Peudenier, J. M. Prats-Vinas, D. Rodriguez, D. Rating, R. Surtees, G. Uziel, L. Vallee, and T. Voit, with the collaboration of J. Aicardi, P. Amarenco, I. Bernard, J. Boulloche, M. L. Chauvet, G. Cioni, I. Desguerre, C. DeSouza, D. Fontan, A. Gal, C. Hubner, I. Krageloh-Mann, G. Kurlemann, D. Lacombe, C. LeBerre, B. Lemarec, J. Lopez-Pison, C. Moraine, K. Muller, A. Nivelon-Chevalier, J. P. Nuyts, K. Pohl, E. Scalais, W. Schrank, G. Sebire, M. Spada, H. Steinbock, S. Stocker, M. Tardieu, A. Toutain, and L. Zelante.
References
- 1.Garbern JY, Cambi F, Tang XM, et al. Proteolipid protein is necessary in peripheral as well as central myelin. Neuron. 1997;19:205–218. doi: 10.1016/s0896-6273(00)80360-8. [DOI] [PubMed] [Google Scholar]
- 2.Greer JM, Dyer CA, Pakaski M, et al. Orientation of myelin proteolipid protein in the oligodendrocyte cell membrane. Neurochem Res. 1996;21:431–440. doi: 10.1007/BF02527707. [DOI] [PubMed] [Google Scholar]
- 3.Nave KA, Lai C, Bloom FE, Milner RJ. Splice site selection in the proteolipid protein (PLP) gene transcript and primary structure of the DM-20 protein of central nervous system myelin. Proc Natl Acad Sci USA. 1987;84:5665–5669. doi: 10.1073/pnas.84.16.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamholz J, Sessa M, Scherer S, et al. Structure and expression of proteolipid protein in the peripheral nervous system. J Neurosci Res. 1992;31:231–244. doi: 10.1002/jnr.490310204. [DOI] [PubMed] [Google Scholar]
- 5.Pham-Dinh D, Birling MC, Roussel G, et al. Proteolipid DM-20 predominates over PLP in peripheral nervous system. Neuroreport. 1991;2:89–92. doi: 10.1097/00001756-199102000-00006. [DOI] [PubMed] [Google Scholar]
- 6.Ikenaka K, Kagawa T, Mikoshiba K. Selective expression of DM-20, an alternatively spliced myelin proteolipid protein gene product, in developing nervous system and in nonglial cells. J Neurochem. 1992;58:2248–2253. doi: 10.1111/j.1471-4159.1992.tb10970.x. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths IR, Montague P, Dickinson P. The proteolipid protein gene. Neuropathol Appl Neurobiol. 1995;21:85–96. doi: 10.1111/j.1365-2990.1995.tb01034.x. [DOI] [PubMed] [Google Scholar]
- 8.Puckett C, Hudson L, Ono K, et al. Myelin-specific proteolipid protein is expressed in myelinating Schwann cells but is not incorporated into myelin sheaths. J Neurosci Res. 1987;18:511–518. doi: 10.1002/jnr.490180402. [DOI] [PubMed] [Google Scholar]
- 9.Cailloux F, Gauthier-Barichard F, Mimault C, et al. Genotypephenotype correlation in inherited brain myelination defects due to proteolipid protein gene mutations. Clinical European Network on Brain Dysmyelinating Disease. Eur J Hum Genet. 2000;8:837–845. doi: 10.1038/sj.ejhg.5200537. [DOI] [PubMed] [Google Scholar]
- 10.Mimault C, Giraud G, Courtois V, et al. Proteolipoprotein gene analysis in 82 patients with sporadic Pelizaeus-Merzbacher disease: duplications, the major cause of the disease, originate more frequently in male germ cells, but point mutations do not. The Clinical European Network on Brain Dysmyelinating Disease. Am J Hum Genet. 1999;65:360–369. doi: 10.1086/302483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobson GM, Davis AP, Stowell NC, et al. Mutations in non-coding regions of the proteolipid protein gene in Pelizaeus-Merzbacher disease. Neurology. 2000;55:1089–1096. doi: 10.1212/wnl.55.8.1089. [DOI] [PubMed] [Google Scholar]
- 12.Hodes ME, Woodward K, Spinner NB, et al. Additional copies of the proteolipid protein gene causing Pelizaeus-Merzbacher disease arise by separate integration into the X chromosome. Am J Hum Genet. 2000;67:14–22. doi: 10.1086/302965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonarakis SE. Recommendations for a nomenclature system for human gene mutations. Nomenclature Working Group. Hum Mutat. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 14.Hobson GM, Huang Z, Sperle K, et al. A PLP splicing abnormality is associated with an unusual presentation of PMD. Ann Neurol. 2002;52:477–488. doi: 10.1002/ana.10320. [DOI] [PubMed] [Google Scholar]
- 15.Timsit SG, Bally-Cuif L, Colman DR, Zalc B. DM-20 mRNA is expressed during the embryonic development of the nervous system of the mouse. J Neurochem. 1992;58:1172–1175. doi: 10.1111/j.1471-4159.1992.tb09378.x. [DOI] [PubMed] [Google Scholar]
- 16.Porter S, Clark MB, Glaser L, Bunge RP. Schwann cells stimulated to proliferate in the absence of neurons retain full functional capability. J Neurosci. 1986;6:3070–3078. doi: 10.1523/JNEUROSCI.06-10-03070.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 18.Bole DG, Dowin R, Doriaux M, Jamieson JD. Immunocyto-chemical localization of BiP to the rough endoplasmic reticulum: evidence for protein sorting by selective retention. J Histochem Cytochem. 1989;37:1817–1823. doi: 10.1177/37.12.2685110. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pratt VM, Naidu S, Dlouhy SR, et al. A novel mutation in exon 3 of the proteolipid protein gene in Pelizaeus-Merzbacher disease. Neurology. 1995;45:394–395. doi: 10.1212/wnl.45.2.394. [DOI] [PubMed] [Google Scholar]
- 22.Carango P, Funanage VL, Quiros RE, et al. Overexpression of DM20 messenger RNA in two brothers with Pelizaeus-Merzbacher disease. Ann Neurol. 1995;38:610–617. doi: 10.1002/ana.410380409. [DOI] [PubMed] [Google Scholar]
- 23.Hodes ME, Blank CA, Pratt VM, et al. Nonsense mutation in exon 3 of the proteolipid protein gene (PLP) in a family with an unusual form of Pelizaeus-Merzbacher disease. Am J Med Genet. 1997;69:121–125. [PubMed] [Google Scholar]
- 24.Osaka H, Kawanishi C, Inoue K, et al. Novel nonsense proteolipid protein gene mutation as a cause of X-linked spastic paraplegia in twin males. Biochem Biophys Res Commun. 1995;215:835–841. doi: 10.1006/bbrc.1995.2539. [DOI] [PubMed] [Google Scholar]
- 25.Nance MA, Boyadjiev S, Pratt VM, et al. Adult-onset neuro-degenerative disorder due to proteolipid protein gene mutation in the mother of a man with Pelizaeus-Merzbacher disease. Neurology. 1996;47:1333–1335. doi: 10.1212/wnl.47.5.1333. [DOI] [PubMed] [Google Scholar]
- 26.Trofatter JA, Dlouhy SR, DeMyer W, et al. Pelizaeus-Merzbacher disease: tight linkage to proteolipid protein gene exon variant. Proc Natl Acad Sci USA. 1989;86:9427–9430. doi: 10.1073/pnas.86.23.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeman W, DeMyer W, Falls HF. Pelizaeus-Merzbacher disease: a study in nosology. J Neuropathy Exp Neurol. 1964:334–354. doi: 10.1097/00005072-196404000-00008. [DOI] [PubMed] [Google Scholar]
- 28.Gow A, Lazzarini R. A cellular mechanism governing the severity of Pelizaeus-Merzbacher disease. Nat Genet. 1996;13:422–427. doi: 10.1038/ng0896-422. [DOI] [PubMed] [Google Scholar]
- 29.Griffiths I, Klugmann M, Anderson T, et al. Axonal swellings and degeneration in mice lacking the major proteolipid of myelin. Science. 1998;280:1610–1613. doi: 10.1126/science.280.5369.1610. [DOI] [PubMed] [Google Scholar]
- 30.Southwood C, Gow A. Molecular pathways of oligodendrocyte apoptosis revealed by mutations in the proteolipid protein gene. Microsc Res Tech. 2001;52:700–708. doi: 10.1002/jemt.1054. [DOI] [PubMed] [Google Scholar]
- 31.Jiang H, Duchala CS, Awatramani R, et al. Proteolipid protein mRNA stability is regulated by axonal contact in the rodent peripheral nervous system. J Neurobiol. 2000;44:7–19. doi: 10.1002/1097-4695(200007)44:1<7::aid-neu2>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 32.Philips AV, Cooper TA. RNA processing and human disease. Cell Mol Life Sci. 2000;57:235–249. doi: 10.1007/PL00000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 34.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 35.Hastings ML, Ingle HA, Lazar MA, Munroe SH. Post-transcriptional regulation of thyroid hormone receptor expression by cis-acting sequences and a naturally occurring antisense RNA. J Biol Chem. 2000;275:11507–11513. doi: 10.1074/jbc.275.15.11507. [DOI] [PubMed] [Google Scholar]
- 36.Werner H, Dimou L, Klugmann M, et al. Multiple splice iso-forms of proteolipid M6B in neurons and oligodendrocytes. Mol Cell Neurosci. 2001;18:593–605. doi: 10.1006/mcne.2001.1044. [DOI] [PubMed] [Google Scholar]