Abstract
An abundant and diverse collection of bacteria, fungi and viruses inhabit the human skin. These microorganisms have been reported to vary between individuals and between different sites on the skin. The factors responsible for the unique variability of the skin microbiome are only partially understood, but results suggest host genetic and environmental influences play a major role. Today, the steady accumulation of data describing the skin microbiome, combined with experiments designed to test the biological functions of surface microbes, have provided new insights into links between human physiology and skin microbiota. This review describes some of the current information regarding the skin microbiome and their impact on human health. Specifically, this review seeks to summarize the present understanding of the function of microbe–host interactions on the skin and highlight some unique features that distinguish skin commensals from pathogenic microbes.
Keywords: microbiota, microbial diversity, skin diseases, skin health
Skin microbial community
Bacteria, viruses and eukaryotes like fungi and arthropods have been known to inhabit the skin. However, the community of microorganisms on human skin is more complex than once thought. Understanding the composition of the skin microbial community is a significant advance from older classifications of cutaneous microbiota that focused on skin microbes only as pathogens or opportunistic pathogens, and have come from the development of methods based on sequencing technologies that are independent of the need for cultivation of microbes. With this new information the composition of the skin microbiome has been shown to be diverse, loosely organized and variable between different locations of the skin. Most importantly, the more recent descriptions of the composition of the skin microbiome have inspired new work towards understanding the functional significance of the resident microbes on the skin, and with this have brought important new advances in understanding both normal physiology and disease.
First and foremost, the ecological system under normal physiological conditions must maintain homeostasis between the microbiome and host. The mechanisms responsible for this balance remain largely unknown, and have been complicated by observations showing that the exact composition of skin microbes varies from individual to individual, but remains somewhat stable over time [1, 2]. Furthermore, the relevant interactions that define this cutaneous system are not limited to only those between the microbe and the host. The skin is unique among epithelial surfaces for the complex ecological interactions with the environment. Also, competition within and between microbial species are important for the development and maintenance of a healthy microbiome. This review seeks to provide an update on some of the current information describing the skin microbiome and experimental advances towards better understanding the biology and significance of this important system.
The skin is a unique and variable ecosystem
The skin provides many niches in which large populations of microbes are subjected to variable ecological pressures including humidity, temperature, pH and the composition of antimicrobial peptides and lipids. In addition, skin structures such as hair follicles, sebaceous, eccrine, and apocrine glands constitute discrete niches that harbor unique microbiota [3]. Analyzing the topographical diversity of microbes that harbor these niches of the human skin with the use of 16S rRNA gene phylotyping revealed the large effects these habitats have on the composition of microbes. At least nineteen phyla are known to be part of the bacterial skin microbiome. Major examples are Actinobacteria (51.8%), Firmicutes (24.4%), Proteobacteria (16.5%), and Bacteroidetes (6.3%). The majority of the identified genera are Corynebacterium, Propionibacterium and Staphylococus [4]. The abundance of each group is strongly dependent on the characteristics of the appropriate niche. For example, sebaceous sites on the face are predominated by Propionibacterium species and Staphylococcus species. In moist sites such as the axilla, Corynebacterium species predominate, although Staphylococcus species are also present. In contrast, in dry sites mixed populations of bacterial species of β-Proteobacteria and Flavobacteriales are part of the resident microbiota [4, 5]. Findings by classical cultivation methods largely coincide with the findings of new sequencing technologies. For example, older reports of cultures from the skin describe the carriage rate of Staphylococcus aureus among different individuals at about 4 %, whereas its close phylogenetic relative Staphylococcus epidermidis is much more frequently found. This is similar to the results from DNA sequencing [1, 6].
But sequencing and culture methods are not identical. DNA sequencing techniques can detect organisms that are not able to be cultured. This limitation may exist in part by the fact that many of the organisms detected on the skin surface have already been killed by the antimicrobial actions of the skin. In fact, species detected by both techniques tend to be those most resistant to killing by the epidermal environment. In addition, a genomic approach in which samplings of different body sites from 129 males and 113 females were tested uncovered that diversity within single samples (alpha-diversity) differs from comparisons between samples from the same habitat among subjects (beta diversity). In this context, this study indicated that skin is intermediate in alpha diversity and highest in beta diversity compared to the other epithelial surfaces [1]. Thus, because of the variability between the microenvironments of different sites on the skin, and the great variability in external treatment of the skin by different individuals, it has been more difficult to establish clear relationships between the presence of specific organisms and skin functions. Importantly, the uniqueness of each individual’s microbial composition seems to be stable over time, suggesting consistency for the individual subject and study.
Most microbiome studies concentrate on understanding bacterial composition, but the microbes present in human skin habitats is not limited to bacteria. Viruses, fungi and arthropods are also important parts of the skin microbiota. In a small study, the predominant fungus detected by using phylogenetic markers like18S rRNA belongs to the species Malassezia including the most frequent isolates M. globosa, M. restricta, and M. sympodialis [7–9]. Malassezia species are lipophilic microbes that are frequently associated with sebum-rich areas of the skin [10]. Similar to the bacterial distribution on the skin, the distribution of Malassezia is dependent on the characteristics of the respective habitat. For example M. globosa predominates on the back, occiput and inguinal crease, whereas M. restricta is found on the scalp [11], in the external auditory canal, retroauricular crease and glabella [8]. Differences between the species may reflect different lipid requirements [12]. Other areas of the skin like foot sites are colonized with greater diversity (e.g. Aspergillus, Rhodotorula, Cryptococcus and Epicoccum) [8].
Other eukaryotes that colonize the human skin belong to the phylum Arthropoda. Like Malassezia species, Demodex mites favor lipids of the sebum [13]. To date, two of the 0.2–0.4 mm long mite species are known to inhabit human skin. Demodex folliculorum is found in hair follicles in clusters with other mites of the same species. The smaller mite Demodex brevis resides alone in sebaceous glands or in meibomial glands which are located at the rim of the eyelids [14].
Less is known about the human virome. This is due to difficulties of amplifying viruses in cell culture, limited antigenic/serological cross-reactivity, or the lack of nucleic acid hybridization to known viral sequences. Detecting methods have also largely focused on metagenomic sequencing of total DNA, making it unlikely to detect RNA viruses. It is also their minuscule genomic size that argues against an easy detection in metagenomic approaches. One approach to facilitate viral discovery is enrichment for small viral particles and removal of contaminating bacterial and human nucleic acids, leaving viral nucleic acids protected within their virion shells [15, 16]. Despite methodological difficulty, a recent high throughput metagenomic sequencing study of skin from five healthy and one Merkel cell carcinoma affected patient revealed a high diversity of DNA viruses on the human skin [17]. However, as this was a small study it is not yet clear if these viruses are actually part of the human skin microbiota or if they serve some mutualistic benefit to the host. Interestingly, it has been hypothesized that even recognized pathogenic viruses such as the human papillomavirus are a normal part of the skin microbiome [17–21].
Host interactions with skin microbiota
It is still too early to say with confidence what the exact assembly of the complex network of human microbiota will look like. From sequencing information we know that microbial communities are more complex than previously expected from culture based studies. These findings raise many new and important questions about the interactions between host and microbes and its relevance to health and disease. As individuals can carry similar compositions of microbes it is highly likely that microbes and their host have co-evolved. Microbes profit from their host as they are provided nutrients and a stable ecological niche. Advantages for the host may include the capacity of the microbe to evolve quickly and therefore help individuals to respond to changes in their environment.
As mentioned, although there are variations of the microbial composition between individuals, the microbial composition of each individual appears to remain relatively stable. Conversely, shifting of microbial communities can alter host–microbiome interactions has been associated with disease [22–24]. To better understand those changes in microbial compositions, the dynamics of microbial interactions at the skin surface will be discussed briefly.
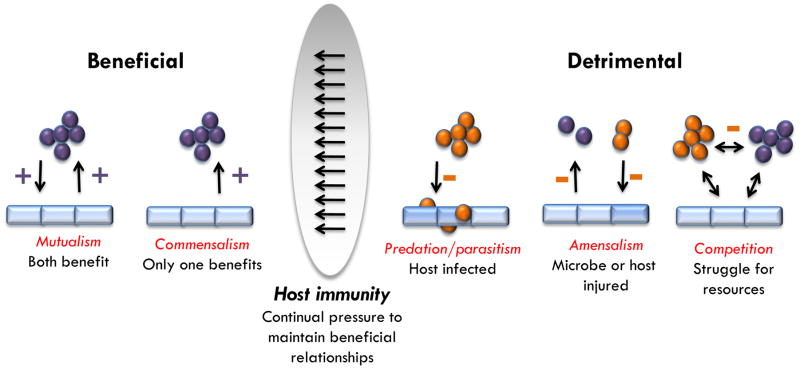
Basically, interactions of microbes within or on the host can be divided in three categories of relationships. The relationship can either have a negative impact, a positive one or no impact on one of the species involved. The possible combinations of these categories allow the classification into various types of interaction [25, 26]. For instance, commensalism describes the situation in which only one species benefits from the relationship while the other one is unaffected. The term mutualism implies that both partners are in a win-win situation. These kinds of interactions are an important part of the skin immune shield. In contrast, in detrimental relationships only one organism benefits while the other is harmed (Figure 1). Cooperative interactions between microbes and their hosts involve microbial participation in host functions.
Figure 1. Dynamics of microbial interactions at the skin surface.
Microorganisms form complex interaction networks. Beneficial situations include mutualistic and commensal relationships. In contrast, detrimental relationships describe correlations in which at least one interaction partner is harmed. The mammalian immune system acts to manage microbial communities and orchestrate beneficial microbe–host relationships.
An example of a beneficial relationship between bacteria and the skin can be seen through the innate capacity of the epithelium to detect microorganisms with Toll-like receptors (TLRs). Stimulation of TLRs induces distinct patterns of gene expression that leads to the activation of a variety of immune responses. Classically, these immune responses were considered to be exclusively pro-inflammatory and designed to defend against the microbe causing infection. It has been shown that the commensal bacterium S. epidermidis modulates TLR3-dependent inflammation by initiating a TLR2-mediated cross-talk mechanism to suppress inflammation [27]. This bacterium also induces keratinocytes to express endogenous antimicrobial peptides (AMP) through a TLR2-dependent mechanism [28]. Furthermore, S. epidermidis has been described to have an autonomous function in controlling and tuning the functions of resident T-lymphocytes [29]. By comparing the production of inflammatory molecules by T cells from germ-free (GF) mice and specific pathogen free (SPF) mice, T cells from GF mice were found to produce lower levels of inflammatory molecules such as interferon-γ (IFN-γ) and interleukin- 17A (IL-17A). Monoassociation of the skin of GF mice with just one bacterium, that is to say S. epidermidis, was enough to reinstate the production of IL-17A by T cells in the skin, but not in the gut. Interestingly, this skin bacterium also allows a protective Th1-response after cutaneous infection with the protozoan parasite Leishmania major, as only GF mice colonized with S. epidermidis are able to mount a proper immune response against the parasite [29].
Various factors can drive disruption of the normal composition of microbes, a state referred to as dysbiosis. Functional dysbiosis can affect host-microbe crosstalk and may result in disease. Host factors such as age, sex, use of medication, lifestyle and hygiene play an essential role [30]. Gender-linked differences in the microbial composition of the skin could be based on physiological and anatomical differences that influence skin properties such as hormone production, sweat rate, sebum production, surface pH, skin thickness, hair growth or cosmetic use [31]. In a recent study of the human epidermis following skin barrier disruption, women showed a significant greater microbial diversity on their hands than men, and this was linked to their less acidic skin surface and use of make-up [31].
In general, more diversity is seen to be more advantageous since it is thought that a diverse ecosystem is more resilient. Studies of the gut microbiome exemplify the enormous influence of westernized lifestyle on the microbial diversity and pathophysiology of many diseases, respectively. Lifestyle choices like the mode of birth delivery and choice of feeding modality [32, 33], improved sanitation, introduction of antibiotics and vaccines [34], Western diets [35], and the consumption of artificial nutrients [36] greatly impact the gut microbiota. A Western lifestyle may also have a similar impact on the skin microbiota. By comparing cutaneous bacterial communities of Amerindians in the Venezuelan Amazon with samplings from volunteers of the United States, one study uncovered significant differences in forearm skin bacterial communities [37]. Using multiplexed V2-targeted 16S rRNA gene pyrosequencing from 112 samples revealed a division of the Amerindian skin microbiota into two major clusters that were not represented in the US samplings. In this context, the alpha diversity of the US samples showed relative equivalencies in species richness as bacteria of only one taxon were presented. In contrast, Cluster B of the Amerindian samples exhibited much higher richness in comparison to the US samples with no recognizable dominant bacterial taxon [37]. Although these observations are interesting, to better understand the impact of modern life on human cutaneous microbiota it will be necessary to sample more groups of individuals of native and Westernized cultures, and more carefully control for the effect of skin care products.
The skin microbiome in human disease
Disturbance of the homeostasis between microbiome and host has been associated with disease. For example, the antibiotic vancomycin, when used in early life, can increase the incidence and severity of allergic asthma [38]. However, only the neonatal, but not adult, exposure to certain antibiotics appears to promote susceptibility to experimental allergic asthma, a time point when key immunological thresholds are established. The following section will review current information regarding the relevance of microbe-host interactions from the perspective of specific human skin disorders.
Acne
Although this chronic inflammatory disease of the pilosebaceous unit is not yet completely understood, altered bacterial colonization is considered to be one of the main elements contributing to the development of acne [39]. The basic disease mechanism is thought to involve androgen-induced increase sebum production, altered keratinization, inflammation, and dysbiosis of the facial skin. The primary disease-associated bacterium is Propionibacterium acnes. It colonizes sebaceous follicles that contain microcomedones providing the bacterium an anaerobic and lipid-rich environment [40, 41]. Secretion of several enzymes such as hyaluronidases, lipases and proteases, causes local injury and inflammation [42].
In a comparison at the strain and genome level of P. acnes based on samplings of nose pilosebaceous units from 49 acne patients and 52 healthy individuals showed no significant statistical difference in the relative abundance of P. acnes. However, only certain P. acnes strains, based on their phylotype, were highly associated with acne. In contrast, those strains were less abundant on healthy skin, and other strains were enriched [5]. In another study different P. acnes phylotypes were visualized in sebaceous follicles of facial skin biopsies. Samples from patients with acne presented higher amounts of P. acnes and follicles containing P. acnes compared to control samples [43].
Interestingly, acne is absent in non-Westernized people in Papua New Guinea and Paraguay. Here, it is speculated that high glycemic loads in Western diet could lead to increased androgens, increased insulin-like growth-factor 1, and altered retinoid signaling [44, 45]. In this context, such variables may also influence the skin microbiome.
Atopic dermatitis
Atopic dermatits (AD) is a chronic, inflammatory skin disease whose prevalence is increasing in industrialized countries [46, 47]. A hallmark of AD that has been well known for decades is that patients have increased colonization with bacteria, and are particularly susceptible to infections with S. aureus and viruses such as herpes and vaccinia [48, 49]. It has been hypothesized that the alteration in surface microbial composition is due to dysfunction of the skin barrier. These disruptions include mutations in the gene that encodes for filaggrin, a protein involved in cornification [50]. A greater susceptibility during the course of this disease may also be attributable to decreased expression of antimicrobial proteins (AMPs) in the skin [51]. Once the skin barrier is impaired and the expression of AMPs is reduced it is likely that the homeostasis between host and microbe is shifted.
Several recent findings in patients with atopic dermatitis (AD) reveal a dramatic change in microbial community structures compared to healthy volunteers [23, 52]. AD patients can occasionally obtain benefit from the use of certain antibiotics in combination with corticosteroids and diluted bleach baths [23, 53], but the excessive use of antibiotics has also come under criticism for negatively affecting the microbiome and potentially disrupting their beneficial function. Worsening disease and lower skin bacterial diversity is strongly associated with each other and microbial shifts are known to be localized to sites of disease predilection. The latter may suggest that not only specific ecological niches such as the anticubital and popliteal crease are important for the initiation of disease, but also influenced by the microbial communities that live in it. Interestingly, effective treatment against AD is associated with a higher bacterial diversity, indicating that current treatments that promote bacterial diversity promote improvement of the disease. Increases of specific bacterial genera, such as Corynebacterium, Streptococcus and Propionibacterium, are observed during therapy indicating more complex species relationships during AD than known from culture-based methods [23]. Some studies have also suggested that Malassezia are more often associated with AD [7, 12].
Psoriasis
In comparison to AD, less is known about the role of the skin microbiome in psoriasis. Psoriasis is an idiopathic inflammatory skin disorder that affects 2 % of the world’s population. Hallmarks of this disease include hyperkeratosis, hyperproliferation of keratinocytes, infiltration of skin by immune cells, and angiogenesis [54]. Similar to AD, Psoriasis results from a combination of genetic and environmental factors. Overall ecological diversity of the microbial population overlaying the lesions is greater than for healthy skin. Firmicutes are predominant whereas Actinobacteria are underrepresented in psoriatic lesions in comparison to healthy skin [55, 56]. It is not known if these alterations in the microbiome in psoriasis are a consequence of the disease, or contributes to its pathogenesis.
Rosacea
3 % of the world’s population suffers from rosacea [57]. Most of the patients are fair-skinned and have their origins from Northern Europe. The clinical presentations of the disease include flushing, nontransient erythema, papules, pustules, telangiectasia, and inflammatory nodules, mainly found in the facial skin. In rosacea, microbes other than bacteria may take advantage of impaired homeostasis between host and skin microbiota. The Demodex mite found on healthy skin is significantly increased on the skin of rosacea patients [58, 59].
Dysregulation of the immune system has been described in rosacea. These include altered TLR2 expression, high levels of the serine protease kallikrein 5 (KLK5) and abnormal expression of the cathelicidin antimicrobial peptide (AMP) LL-37 [60, 61]. As these elements respond to, and influence the composition of the skin microbiome, it is logical to speculate that skin microbes play an important part in this disease. Skin inflammation also correlates with mite density on the skin of rosacea-affected patients [62]. Thus, the mite may participate in the exacerbation of disease either by disrupting the skin barrier [14, 63], or by triggering TLR2 activation through chitin in the insect’s cuticle [64, 65]. Furthermore, it has been reported that bacteria that live in the digestive tract of Demodex are released into the surrounding skin tissues, thereby triggering further tissue degradation and inflammation [66, 67]. All of these mechanisms may participate in various degrees. However, it appears most likely that a genetic predisposition in the host changes certain ecological characteristics of the skin that result in a shift of the microbiome. Demodex may take advantage of those changes and evoke a response that makes the skin susceptible to other inflammatory triggers such as UV exposure, alcohol, hormone fluctuations and bacteria that overgrow the developing papules (e.g. S. epidermidis). As rosacea affects patients at a certain age it is also possible that age-specific modulations in TLR expression might play an essential role in the course of this disease [68].
Seborrheic dermatitis
The predominant fungus of the skin microbiota, Malassezia, is postulated to take part in seborrhoeic dermatitis (SD). This chronic inflammatory skin disorder is often first diagnosed around puberty due to an increase in cutaneous lipids resulting from an androgen-driven sebaceous gland development and sebum secretion. The disease also often occurs in patients older than 50. The prevalence of SD in the overall population is estimated at a range from 1 to 5 % and can affect any ethnic group, and is more often diagnosed in men than in women. High-risk population includes patients with acquired immunity-deficient syndrome (AIDS). The clinical presentation is characterized by erythematous patches associated with greasy scaling. The commonly affected sites include the anterior hair line, eyebrows, ears, glabella, chest and scalp [69]. It is reported that the disease may also occur in combination with other skin diseases such as AD and psoriasis, leading to complications in diagnosis.
Dandruff is a commonly applied term for seborrhea of the scalp. It is mainly associated with M. restricta and M. globosa [10, 11], and has a very high prevalence of nearly 50 % of the population. Improvement of the disease is achieved by therapeutically applying antifungal but not antibacterial agents. The underlying mechanisms of pathogenicity are incompletely understood. Impaired skin barrier function facilitates the course of disease [70]. The fungus is known to secrete a lipase that splits triglycerides into irritant fatty acids that may induce hyperproliferation and scaling or releases arachidonic acid, which is also involved in inflammation [71]. Given the current literature it can be speculated that the fungus, which is part of the normal skin microbiota, switches to a pathogenic state when its growth is not controlled. What these control factors are and how they are dampened is not yet understood.
Microbe–microbe interactions on the skin
Altered ecological interactions between pathogenic microbes and their hosts can lead to disease, but the consequence of microbes interacting with each other has been underestimated and not well-studied. This section provides some examples of specific microbe–microbe interactions and their impact on the skin.
Bacterium–bacterium interactions
S. epidermidis has been proposed to protect the skin from pathogens by producing AMPs such as phenol-soluble modulins (PSMs). PSMδ and PSMγ interact with lipid membranes similar to mammalian AMPs. By secreting these peptides, S. epidermidis exerts selective antimicrobial action against skin pathogens such as S. aureus and Group A streptococci [72]. Moreover, S. aureus biofilm formation and nasal colonization is inhibited by the serine protease Esp produced by S. epidermidis [73]. But the interaction between different Staphylococcus species seems to be more complex. In a recent study, Kong et al. [23] discovered that the proportion of S. epidermidis consistently increased during the condition of AD when no treatment was applied, opening new hypotheses about the relationship between staphylococci. One possibility is that S. epidermidis not only has a mutualistic relationship with its eukaryotic host but also shares a mutualistic or commensal relationship with other Staphylococcus species such as S. aureus [23]. Thus, bacteria may take advantage of the specific situation. For example, the relative decrease of other bacteria such as Streptococcus, Corynebacterium and Propionibacterium in AD could be due to the combined action of Staphylococcus species.
Interaction between species of one common genus is also observed in propionibacteria. As mentioned above, the distribution of different Propionibacterium strains is significantly different on the skin of healthy volunteers in comparison to acne-affected skin indicating a possible communication between bacterial strains [5].
Virus–bacterium interactions
In addition to bacterium–bacterium interactions, bacterial communities also interact with the bacteriophages that infect them. These viruses influence the bacterial community structure and function by several mechanisms, including killing their host and mediating genetic exchanges. Several phages are known in Staphylococcus, Pseudomonas and Propionibacterium species [74–76]. So far it is not clear to what extent bacteriophages have an impact on the skin microbiome. Phages have been shown to reduce microbial colonization and pathology in a host-independent way. In the gut, they attach to specific glycoproteins in the luminal mucus via a specific capsid protein thereby creating an antimicrobial layer that reduces bacterial attachment to and colonization of the mucus, which in turn lessens epithelial cell death [77].
Bacterium fungi–interactions
Complexes containing bacteria and fungi are varied and wide-ranging. Industry uses those interactions for the production of food such as cheese and beer. These interactions are also found in many parts of the human body such as the oral cavity [78] and in the gastrointestinal tract [79]. However, so far most of the clinical studies to date have focused on bacterial interaction with Candida albicans, a yeast that is common in the human microbiome but which can also cause several infections. In this context, mixed communities have virulence and resistance properties significantly different from those of single species communities. In example, biofilms containing S. epidermidis and C. albicans in medical device-associated infections are significant more resistant to antimicrobials as single-organism biofilms [80]. In contrast, Pseudomonas aeruginosa biofilm formation leads to the death of the fungal cell. One explanation of communication between bacteria and fungi is based on quorum sensing systems [80, 81]. Furthermore, P. aeruginosa is able to grow on skin at the expense of dermatophyte fungi associated with skin and nail infections [82].
These examples show that interactions between microbes have a strong influence in health and disease. However, although it is known that fungi are part of the healthy human skin, less is known about their ecological interactions in the state of health. A recent study revealed that bacterial and fungal richness are not linearly correlated, but both kingdoms live in clusters in close relation in the same location [8]. In accordance to their observations the authors provided a preliminary evaluation of major fungal–bacterial associations in the skin. For example, co-occurrence analysis of foot sites enlightened the anti-correlation of Actinobacteria with resident Ascomycota and Basidiomycota. In contrast, Firmicutes and Proteobacteria are positively correlated with these fungal taxa [8].
From commensal to pathogen and unique propensities of skin microbes
Commensal microorganisms can become pathogens, thus no microbe can really be considered to be exclusively beneficial. The most prominent and best studied examples are S. epidermidis and C. albicans. Others are less understood such as M. restricta and M. globosa. But how does a commensal become a pathogen? And even more importantly, what might be the unique factors that allow a commensal to be tolerated by the host? This can be illustrated with the example of S. epidermidis.
Among coagulase-negative staphylococci (CoNS), S. epidermidis causes the greatest number of infections. It is commonly regarded as the most frequent causative agent of infections of indwelling medical devices such as peripheral or central venous catheters [83]. And indeed, staphylococci are transferred into the patient’s body from the skin of the patient or that of health care personnel during device insertion. Once the bacteria have entered the body they use various virulence factors to facilitate interactions with host tissues and subvert the host’s immune system. The most essential staphylococcal virulence factor is the characteristic biofilm formation on medical devices. Biofilms are multicellular, surface-attached agglomerations of microorganisms [84]. Their regulation is liable to quorum sensing systems and is not yet completely understood. But do all S. epidermidis species on the skin have the potential of becoming a pathogen? Previous studies based on epidemiological analysis and genetic studies suggest that S. epidermidis isolates in the hospital environment differ from those obtained outside of medical facilities in biofilm formation, antibiotic resistance, and the presence of mobile DNA elements. For example, most disease-associated S. epidermidis biofilms are dependent on the expression of polysaccharide intercellular adhesion (PIA). However, the intercellular adhesion operon (ica) necessary for PIA production is rarely found in isolates obtained outside of hospital settings [85, 86]. To date, icaA, mecA and IS256 are used as markers for invasive nosocomial strains. Acquisition of these markers is associated with intra- and interspecifically horizontal gene transfer. In this context, S. epidermidis can be regarded as highly flexible organism on an evolutionary level.
Indeed, a high diversity of S. epidermidis skin commensal and hospital infection-associated isolates was recently uncovered [87]. By comparing specially created draft genomes from several commensal and nosocomial isolates, commensals were found to have an open pan-genome with 80 % core genes and 20 % variable genes, comprising mobile DNA elements, transcription factors and transporters. The formate dehydrogenase gene (fdh) was present in 23 % of the commensal strains, but only in 4 % in pathogens. These 4 % of fdh-positive nosocomial strains were characterized by exhibiting less virulence influenced by less abundance of the markers icaA, mecA and IS256. Thus the authors suggest that these strains could represent skin commensal contaminants from venipuncture.
Although there is less known about how skin commensals and mutualistic bacteria can be distinguished from pathogens, some possibilities have been put forward. The virulence factors of the pathogen group A streptococcus (GAS) mediate the in vivo changes from non-invasive GAS serotype M1T1 to the invasive phenotype. This process is potentiated by spontaneous mutations within a specific two-component system resulting in an upregulation of numerous virulence-associated genes. One of those genes includes the operon for the synthesis of the hyaluronic acid capsule. The glucuronic-β-1,3-N-acetylglucosamine chain has the identical structure to the human hyaluronan molecule and provides the bacterium a form of mimicry that impairs immune recognition and phagocytic clearance [88]. Indeed, hyaluronan is the most abundant glycosaminoglycan in the skin and it is plausible that commensal bacteria use a similar mechanism to mask themselves and not be recognized by the immune system.
Epigenetic mechanisms regulate expression of the genome to generate multiple cell types during development or orchestrate cellular responses to external stimuli. To date, it has been widely accepted that pathogenic bacteria and viruses can induce epigenetic changes in host cells by influencing various epigenetic factors to their own benefit. In this context, it is likely that commensal microbes can interfere with histone modification or DNA methylation mechanisms. Indeed, epigenetic control of host genes by commensal bacteria in the gut has been reported to contribute to the maintenance of intestinal symbiosis [89, 90]. Therefore, it would be highly interesting to test if skin commensals can similarly regulate epigenetic mechanisms.
Concluding remarks and future perspectives
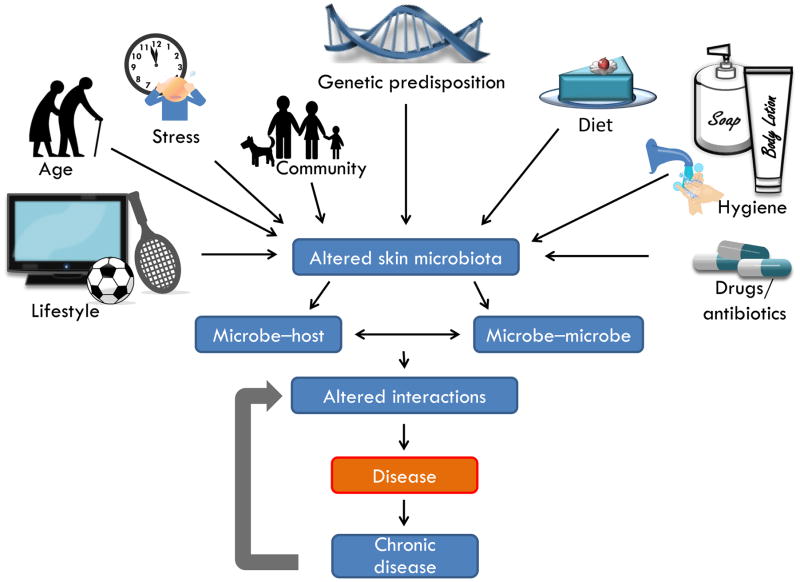
Modern technology has taught us to rethink our sense of self, and consider our normal physiology to be a complex dynamic interaction between many organisms. Initial descriptions of the skin microbiota have revealed an important role for genetics and environmental variation in shaping patterns of diversity. The resulting specific composition of microbes consequently determines the interaction between host, microbe and other microbes which in turn are strongly associated with the state of health and disease (Figure 2). Once a change in genetics or in the environment occurs, the microbiota can change rapidly. A better understanding of interactions between host, microbes and disease-causing organisms may lead to better strategies against skin diseases. To this end, many details remain for further work to fill in. Future research will include overcoming current methodological limitations in describing the composition of microbes (Box 1).
Figure 2. Environment and interactions predict functions of the skin microbiome.
The ecology of the skin surface is influenced by many variables including lifestyle, age, emotion, community, genetics, diet, hygiene products and drugs. Under unfavorable conditions, an altered community of microbes can arise on the skin, thus leading to conditions of dysbiosis. Both interactions between microbes and between microbe and host can promote disease. Dysfunction of the capacity to reestablish a normal microbial community may perpetuate chronic disease.
Box 1. Outstanding questions.
What is the composition of the complex network of organisms that comprise the human skin microbiome?
How do factors such as diet, stress and lifestyle interfere with skin microbial composition?
Can the surface microbiome be used to diagnose or predict disease?
Is dysbiosis of the skin microbiome a causative element in the pathophysiology of some diseases?
Can skin microbes cause differential epigenetic changes in the host?
Can manipulation of skin microbial communities be a beneficial therapeutic approach to disease?
As this advances, the composition of microbes during several skin diseases will be further revealed, thus generating hypothesis related to their role in the pathophysiology of each disorder.
Differences in the genome of pathogenic and non-pathogenic bacteria at the strain level may have an important role. Subtle variations could have great consequences during interactions between host and microbe or between microbe and microbe. One should not underestimate the opportunity for microbes such as bacteria and viruses to cause epigenetic changes may result in important functional changes in the host.
Current available information already supports the conclusion that some treatments in dermatologic medicine should be rethought. The indiscriminate use of antibiotics has a cost to the long-term health of the whole microbiota. It can take up to years until the normal microbiota has recovered [91]. In this context, it is plausible that the application of several medications intended for acute treatments could lead to dysbiosis and therefore promote long-term susceptibility to certain diseases. Moreover, exposure to pharmacological doses of antimicrobial agents over years can alter bacterial species in the population and possibly lead to a shift in the human microbiome and higher susceptibility to pathogenic microbes [56]. Helicobacter pylori is such a bacterium, which is becoming rarer in the Western population. The bacterium has long been regarded as exclusively pathogenic but is now proposed to also have several protective functions [92]. The skin has many such microbial inhabitants whose biology and relationship to the host is incompletely understood. Some of these are associated with disease, but as long it is not clear if they belong to the normal human microbiota or not, elimination of these microbes should not be an option. Furthermore, modern lifestyle also leads to changes in the skin microbiome and therefore might present a parameter in skin microbiome research.
Manipulation of microbial communities to enhance the abundance of beneficial species is a hot topic in human therapy. Such manipulations may aid us by directly promoting health or by reducing of the presence of undesirable pathogens. Although not yet described in human dermatology, adding an antifungal bacterial species to the skins of the frog Rana muscosa successfully prevents morbidity and mortality caused by the pathogen Batrachochytrium dendrobatidis [93]. This fungus is the cause of chytridiomycosis which has been shown to lead to dramatic declines of many amphibian populations. More cause-effect experiments are needed that will allow understanding of whether changes in the human skin microbiome are indeed responsible for disease initiation or progression. Only the future will reveal to what extent recognition of ourselves as a colony of mutualistic organisms will aid in the treatment of disease.
Highlights.
A new understanding has emerged of links between human physiology and skin microbiota.
Microbial diversity is governed by genetics and diverse skin environments.
Host and microbe interactions shape and influence skin health.
Commensal microorganisms exhibit unique behaviors that permit tolerance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grice EA, et al. A diversity profile of the human skin microbiota. Genome research. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fitz-Gibbon S, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. The Journal of investigative dermatology. 2013;133:2152–2160. doi: 10.1038/jid.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuehnert MJ, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. The Journal of infectious diseases. 2006;193:172–179. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]
- 7.Gioti A, et al. Genomic insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis. mBio. 2013;4:e00572–00512. doi: 10.1128/mBio.00572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Findley K, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulino LC, et al. Analysis of Malassezia microbiota in healthy superficial human skin and in psoriatic lesions by multiplex real-time PCR. FEMS yeast research. 2008;8:460–471. doi: 10.1111/j.1567-1364.2008.00359.x. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, et al. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18730–18735. doi: 10.1073/pnas.0706756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clavaud C, et al. Dandruff is associated with disequilibrium in the proportion of the major bacterial and fungal populations colonizing the scalp. PloS one. 2013;8:e58203. doi: 10.1371/journal.pone.0058203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saunders CW, et al. Malassezia fungi are specialized to live on skin and associated with dandruff, eczema, and other skin diseases. PLoS pathogens. 2012;8:e1002701. doi: 10.1371/journal.ppat.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacey N, et al. Demodex mites--commensals, parasites or mutualistic organisms? Dermatology. 2011;222:128–130. doi: 10.1159/000323009. [DOI] [PubMed] [Google Scholar]
- 14.Lacey N, et al. Under the lash: Demodex mites in human diseases. The biochemist. 2009;31:2–6. [PMC free article] [PubMed] [Google Scholar]
- 15.Allander T, et al. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11609–11614. doi: 10.1073/pnas.211424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delwart E. A roadmap to the human virome. PLoS pathogens. 2013;9:e1003146. doi: 10.1371/journal.ppat.1003146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foulongne V, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PloS one. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh S, et al. The role of human endogenous retroviruses in melanoma. The British journal of dermatology. 2009;161:1225–1231. doi: 10.1111/j.1365-2133.2009.09415.x. [DOI] [PubMed] [Google Scholar]
- 19.Rosenthal M, et al. Skin microbiota: microbial community structure and its potential association with health and disease. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2011;11:839–848. doi: 10.1016/j.meegid.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonsson A, et al. Prevalence and type spectrum of human papillomaviruses in healthy skin samples collected in three continents. The Journal of general virology. 2003;84:1881–1886. doi: 10.1099/vir.0.18836-0. [DOI] [PubMed] [Google Scholar]
- 21.Delwart EL. Viral metagenomics. Reviews in medical virology. 2007;17:115–131. doi: 10.1002/rmv.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srinivas G, et al. Genome-wide mapping of gene-microbiota interactions in susceptibility to autoimmune skin blistering. Nature communications. 2013;4:2462. doi: 10.1038/ncomms3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong HH, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome research. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fry L, et al. Is chronic plaque psoriasis triggered by microbiota in the skin? The British journal of dermatology. 2013;169:47–52. doi: 10.1111/bjd.12322. [DOI] [PubMed] [Google Scholar]
- 25.Faust K, Raes J. Microbial interactions: from networks to models. Nature reviews Microbiology. 2012;10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 26.Cogen AL, et al. Skin microbiota: a source of disease or defence? The British journal of dermatology. 2008;158:442–455. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai Y, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nature medicine. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai Y, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. The Journal of investigative dermatology. 2010;130:2211–2221. doi: 10.1038/jid.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naik S, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fredricks DN. Microbial ecology of human skin in health and disease. The journal of investigative dermatology. Symposium proceedings/the Society for Investigative Dermatology, Inc [and] European Society for Dermatological Research. 2001;6:167–169. doi: 10.1046/j.0022-202x.2001.00039.x. [DOI] [PubMed] [Google Scholar]
- 31.Giacomoni PU, et al. Gender-linked differences in human skin. Journal of dermatological science. 2009;55:144–149. doi: 10.1016/j.jdermsci.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 32.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eloe-Fadrosh EA, et al. Impact of oral typhoid vaccination on the human gut microbiota and correlations with S. Typhi-specific immunological responses. PloS one. 2013;8:e62026. doi: 10.1371/journal.pone.0062026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hold GL. Western lifestyle: a ‘master’ manipulator of the intestinal microbiota? Gut. 2013 doi: 10.1136/gutjnl-2013-304969. [DOI] [PubMed] [Google Scholar]
- 36.Payne AN, et al. Gut microbial adaptation to dietary consumption of fructose, artificial sweeteners and sugar alcohols: implications for host-microbe interactions contributing to obesity. Obesity reviews: an official journal of the International Association for the Study of Obesity. 2012;13:799–809. doi: 10.1111/j.1467-789X.2012.01009.x. [DOI] [PubMed] [Google Scholar]
- 37.Blaser MJ, et al. Distinct cutaneous bacterial assemblages in a sampling of South American Amerindians and US residents. The ISME journal. 2013;7:85–95. doi: 10.1038/ismej.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russell SL, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO reports. 2012;13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bojar RA, Holland KT. Acne and Propionibacterium acnes. Clinics in dermatology. 2004;22:375–379. doi: 10.1016/j.clindermatol.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 40.Williams HC, et al. Acne vulgaris. Lancet. 2012;379:361–372. doi: 10.1016/S0140-6736(11)60321-8. [DOI] [PubMed] [Google Scholar]
- 41.Iinuma K, et al. Involvement of Propionibacterium acnes in the augmentation of lipogenesis in hamster sebaceous glands in vivo and in vitro. The Journal of investigative dermatology. 2009;129:2113–2119. doi: 10.1038/jid.2009.46. [DOI] [PubMed] [Google Scholar]
- 42.McDowell A, et al. A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology. 2011;157:1990–2003. doi: 10.1099/mic.0.049676-0. [DOI] [PubMed] [Google Scholar]
- 43.Jahns AC, et al. An increased incidence of Propionibacterium acnes biofilms in acne vulgaris: a case-control study. The British journal of dermatology. 2012;167:50–58. doi: 10.1111/j.1365-2133.2012.10897.x. [DOI] [PubMed] [Google Scholar]
- 44.Abulnaja KO. Changes in the hormone and lipid profile of obese adolescent Saudi females with acne vulgaris. Brazilian journal of medical and biological research = Revista brasileira de pesquisas medicas e biologicas/Sociedade Brasileira de Biofisica … [et al ] 2009;42:501–505. doi: 10.1590/s0100-879x2009000600005. [DOI] [PubMed] [Google Scholar]
- 45.Thiboutot DM, Strauss JS. Diet and acne revisited. Archives of dermatology. 2002;138:1591–1592. doi: 10.1001/archderm.138.12.1591. [DOI] [PubMed] [Google Scholar]
- 46.Shaw TE, et al. Eczema prevalence in the United States: data from the 2003 National Survey of Children’s Health. The Journal of investigative dermatology. 2011;131:67–73. doi: 10.1038/jid.2010.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hanifin JM, et al. A population-based survey of eczema prevalence in the United States. Dermatitis: contact, atopic, occupational, drug: official journal of the American Contact Dermatitis Society, North American Contact Dermatitis Group. 2007;18:82–91. doi: 10.2310/6620.2007.06034. [DOI] [PubMed] [Google Scholar]
- 48.Leung DY. New insights into atopic dermatitis: role of skin barrier and immune dysregulation. Allergology international: official journal of the Japanese Society of Allergology. 2013;62:151–161. doi: 10.2332/allergolint.13-RAI-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim BE, et al. IL-25 enhances HSV-1 replication by inhibiting filaggrin expression, and acts synergistically with Th2 cytokines to enhance HSV-1 replication. The Journal of investigative dermatology. 2013 doi: 10.1038/jid.2013.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Regan GM, et al. Filaggrin in atopic dermatitis. The Journal of allergy and clinical immunology. 2008;122:689–693. doi: 10.1016/j.jaci.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 51.Harder J, et al. Enhanced expression and secretion of antimicrobial peptides in atopic dermatitis and after superficial skin injury. The Journal of investigative dermatology. 2010;130:1355–1364. doi: 10.1038/jid.2009.432. [DOI] [PubMed] [Google Scholar]
- 52.Hata TR, Gallo RL. Antimicrobial peptides, skin infections, and atopic dermatitis. Seminars in cutaneous medicine and surgery. 2008;27:144–150. doi: 10.1016/j.sder.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang JT, et al. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics. 2009;123:e808–814. doi: 10.1542/peds.2008-2217. [DOI] [PubMed] [Google Scholar]
- 54.Schon MP, Boehncke WH. Psoriasis. The New England journal of medicine. 2005;352:1899–1912. doi: 10.1056/NEJMra041320. [DOI] [PubMed] [Google Scholar]
- 55.Gao Z, et al. Substantial alterations of the cutaneous bacterial biota in psoriatic lesions. PloS one. 2008;3:e2719. doi: 10.1371/journal.pone.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature reviews Genetics. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jarmuda S, et al. Potential role of Demodex mites and bacteria in the induction of rosacea. Journal of medical microbiology. 2012;61:1504–1510. doi: 10.1099/jmm.0.048090-0. [DOI] [PubMed] [Google Scholar]
- 58.Forton F, Seys B. Density of Demodex folliculorum in rosacea: a case-control study using standardized skin-surface biopsy. The British journal of dermatology. 1993;128:650–659. doi: 10.1111/j.1365-2133.1993.tb00261.x. [DOI] [PubMed] [Google Scholar]
- 59.Casas C, et al. Quantification of Demodex folliculorum by PCR in rosacea and its relationship to skin innate immune activation. Experimental dermatology. 2012;21:906–910. doi: 10.1111/exd.12030. [DOI] [PubMed] [Google Scholar]
- 60.Yamasaki K, et al. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nature medicine. 2007;13:975–980. doi: 10.1038/nm1616. [DOI] [PubMed] [Google Scholar]
- 61.Yamasaki K, et al. TLR2 expression is increased in rosacea and stimulates enhanced serine protease production by keratinocytes. The Journal of investigative dermatology. 2011;131:688–697. doi: 10.1038/jid.2010.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Georgala S, et al. Increased density of Demodex folliculorum and evidence of delayed hypersensitivity reaction in subjects with papulopustular rosacea. Journal of the European Academy of Dermatology and Venereology: JEADV. 2001;15:441–444. doi: 10.1046/j.1468-3083.2001.00331.x. [DOI] [PubMed] [Google Scholar]
- 63.Forton FM. Papulopustular rosacea, skin immunity and Demodex: pityriasis folliculorum as a missing link. Journal of the European Academy of Dermatology and Venereology: JEADV. 2012;26:19–28. doi: 10.1111/j.1468-3083.2011.04310.x. [DOI] [PubMed] [Google Scholar]
- 64.Da Silva CA, et al. TLR-2 and IL-17A in chitin-induced macrophage activation and acute inflammation. Journal of immunology. 2008;181:4279–4286. doi: 10.4049/jimmunol.181.6.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koller B, et al. Chitin modulates innate immune responses of keratinocytes. PloS one. 2011;6:e16594. doi: 10.1371/journal.pone.0016594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Reilly N, et al. Demodex-associated Bacillus proteins induce an aberrant wound healing response in a corneal epithelial cell line: possible implications for corneal ulcer formation in ocular rosacea. Investigative ophthalmology & visual science. 2012;53:3250–3259. doi: 10.1167/iovs.11-9295. [DOI] [PubMed] [Google Scholar]
- 67.Lacey N, et al. Mite-related bacterial antigens stimulate inflammatory cells in rosacea. The British journal of dermatology. 2007;157:474–481. doi: 10.1111/j.1365-2133.2007.08028.x. [DOI] [PubMed] [Google Scholar]
- 68.Iram N, et al. Age-related changes in expression and function of Toll-like receptors in human skin. Development. 2012;139:4210–4219. doi: 10.1242/dev.083477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gaitanis G, et al. The Malassezia genus in skin and systemic diseases. Clinical microbiology reviews. 2012;25:106–141. doi: 10.1128/CMR.00021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harding CR, et al. Dandruff: a condition characterized by decreased levels of intercellular lipids in scalp stratum corneum and impaired barrier function. Archives of dermatological research. 2002;294:221–230. doi: 10.1007/s00403-002-0323-1. [DOI] [PubMed] [Google Scholar]
- 71.Gupta AK, et al. Skin diseases associated with Malassezia species. Journal of the American Academy of Dermatology. 2004;51:785–798. doi: 10.1016/j.jaad.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 72.Cogen AL, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. The Journal of investigative dermatology. 2010;130:192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iwase T, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. [DOI] [PubMed] [Google Scholar]
- 74.Goerke C, et al. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. Journal of bacteriology. 2009;191:3462–3468. doi: 10.1128/JB.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ceyssens PJ, Lavigne R. Bacteriophages of Pseudomonas. Future microbiology. 2010;5:1041–1055. doi: 10.2217/fmb.10.66. [DOI] [PubMed] [Google Scholar]
- 76.Marinelli LJ, et al. Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates. mBio. 2012;3 doi: 10.1128/mBio.00279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barr JJ, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10771–10776. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Avila M, et al. The oral microbiota: living with a permanent guest. DNA and cell biology. 2009;28:405–411. doi: 10.1089/dna.2009.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iliev ID, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336:1314–1317. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peleg AY, et al. Medically important bacterial-fungal interactions. Nature reviews Microbiology. 2010;8:340–349. doi: 10.1038/nrmicro2313. [DOI] [PubMed] [Google Scholar]
- 81.Bandara HM, et al. Pseudomonas aeruginosa inhibits in-vitro Candida biofilm development. BMC microbiology. 2010;10:125. doi: 10.1186/1471-2180-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Foster KW, et al. A bipartite interaction between Pseudomonas aeruginosa and fungi in onychomycosis. Archives of dermatology. 2005;141:1467–1468. doi: 10.1001/archderm.141.11.1467. [DOI] [PubMed] [Google Scholar]
- 83.Otto M. Molecular basis of Staphylococcus epidermidis infections. Seminars in immunopathology. 2012;34:201–214. doi: 10.1007/s00281-011-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Otto M. Staphylococcus epidermidis--the ‘accidental’ pathogen. Nature reviews Microbiology. 2009;7:555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ziebuhr W, et al. Nosocomial infections by Staphylococcus epidermidis: how a commensal bacterium turns into a pathogen. International journal of antimicrobial agents. 2006;28(Suppl 1):S14–20. doi: 10.1016/j.ijantimicag.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 86.Rohde H, et al. Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. Journal of clinical microbiology. 2004;42:5614–5619. doi: 10.1128/JCM.42.12.5614-5619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Conlan S, et al. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome biology. 2012;13:R64. doi: 10.1186/gb-2012-13-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cole JN, et al. Molecular insight into invasive group A streptococcal disease. Nature reviews Microbiology. 2011;9:724–736. doi: 10.1038/nrmicro2648. [DOI] [PubMed] [Google Scholar]
- 89.Haller D, et al. Transforming growth factor-beta 1 inhibits non-pathogenic Gram negative bacteria-induced NF-kappa B recruitment to the interleukin-6 gene promoter in intestinal epithelial cells through modulation of histone acetylation. The Journal of biological chemistry. 2003;278:23851–23860. doi: 10.1074/jbc.M300075200. [DOI] [PubMed] [Google Scholar]
- 90.Takahashi K, et al. Epigenetic control of the host gene by commensal bacteria in large intestinal epithelial cells. The Journal of biological chemistry. 2011;286:35755–35762. doi: 10.1074/jbc.M111.271007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ahmed N, et al. Helicobacter pylori--a seasoned pathogen by any other name. Gut pathogens. 2009;1:24. doi: 10.1186/1757-4749-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Harris RN, et al. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. The ISME journal. 2009;3:818–824. doi: 10.1038/ismej.2009.27. [DOI] [PubMed] [Google Scholar]