Abstract
Proanthocyanidin (PAC) consumption has been linked to better colonic health, but PACs are poorly absorbed, making them a target for colonic metabolism. The resulting metabolites are low molecular weight and could potentially be absorbed. To understand the effects of dietary PACs it would be important to resolve the metabolic issue and link these changes to microbial population changes in a suitable model for human digestion. Here, six crossbred female pigs were fed a diet containing 1% (w/w) of MegaNatural® Gold grape seed extract (GSE) daily for 6 days. Fecal samples were analyzed by normal phase LC coupled to fluorescence detection and LC-MS/ToF. DNA was extracted from pig fecal samples and the V3/V4 region of the 16S rRNA gene was sequenced using an Illumina MiSeq. Intact parent PACs (dimer–pentamer) were observed in the feces on days 3 and 6 at similar high levels (~400 mg kg−1 total) during ingestion of GSE but were absent 48 h post-feeding. The major phenolic metabolites were 4-hydroxyphenylvaleric acid and 3-hydroxybenzoic acid which increased by ~30 and 3 mg kg−1 respectively. The GSE diet also caused an ecological shift in the microbiome, dramatically increasing Lachnospiraceae, Clostridales, Lactobacillus and Ruminococcacceae. The relationship between dietary PACs and colon health may be attributable to the altered bacterial populations or phenolic compounds in the colon.
1 Introduction
Grape seed extract (GSE) contains proanthocyanidins (PACs) which consists of oligomers and polymers of monomeric unit flavan-3-ols (−)-epicatechin or (+)-catechins, (including the gallated forms). PACs are one of the most abundant polyphenols in human diets and are one of the least-absorbed polyphenols mostly due to their size and structure complexity.1 This characteristic is the major limitation for their absorption through the gut barrier, but they may exert a local beneficial effect on the colon and can regulate cell signaling pathways by interacting with cell membrane proteins.1,2 Moreover, animal and in vitro studies suggest that diets rich in PACs may explain the protective effects of fruits and vegetables on colon cancer. There is no consensus on the absorption and metabolism of PACs thus far, although colon is regarded as an important biotransformation site by the gut microbiota.
It has been estimated that more than 90% of ingested polyphenols are not absorbed in the small intestine and thus, remain in the colon at high concentration.3 In the colon, the unabsorbed oligomeric and polymeric PACs are extensively metabolized by gut microbiota to produce smaller phenolic acids including hydroxybenzoic acid, hydroxyphenylacetic acid, hydroxyphenylpropinoic acid, hyrdroxyphenylvaleric acid or hydroxycinnamic acids, with hydroxylation mostly occurring at meta position.1,4 These metabolites can be absorbed and may be conjugated in the liver before being excreted in urine. Due to the low absorption of intact PACs in the human colon, microbial metabolism is likely to play a major role in colonic health and thus the identification of microbial metabolites should be explored further.
In a previous study using a rat model,5 it was found that a total of 11% of parent PAC compounds were still present in the feces, suggesting that PACs undergo extensive but incomplete transformation during the transit through the gastrointestinal tract by the gut microbiota, providing the opportunity to directly and indirectly affect gut physiology. The intestinal tract contains the largest number of immune cells in the body and the gut microbiota operates in a delicate balance with this immune system.6 Therefore, the gut microbiota plays a key role in the health and well-being of the human host. A better understanding of microbial communities in the colon is crucial in developing a greater understanding of the connection between the gut microbiota and the metabolism of PACs by the gastrointestinal microbiota. Only a few studies so far have investigated the influence of PACs on the gut microbiota composition.7–9 To date, no study has characterized the gut bacterial populations after a PACs-rich diet using Illumina sequencing in pigs. This sequencing method provides unprecedented measures of microbial community diversity and massively parallel genus-level measurements that allow for fine-scale characterization of the microbial community. In conjunction with analyses of the gut bacterial communities, the use of a highly sensitive analytical tool is essential for identification and quantification of PACs metabolites. High-accurate mass measurement mass spectrometry techniques, like MS/ToF, have demonstrated to be a reliable tool for the identification of known and unknown compounds in complex matrices.10 Here, the metabolism of PACs and their microbial-derived phenolic metabolites were investigated, as well as changes in the gut microbiome, using a pig model which has a gastrointestinal tract reported to be similar to human.11 This study will provide a detailed description of the gut metabolites of PACs in feces, and an analysis of the resulting changes in microbial populations, leading to a better understanding of how polyphenols could impact colonic health.
2 Experimental
2.1 Chemicals
5-(4-Hydroxyphenyl)pentanoic acid (4-hydroxyphenylvaleric acid) was obtained from Alfa Aesar (Ward Hill, MA), 3-(3-hydroxyphenyl)propionic acid was purchased from Lancaster Synthesis, Inc. (Pelham, NH), 3,4-dihydroxyphenylacetic acid, 3-hydroxyphenylacetic acid, 4-hydroxybenzoic acid were obtained from Acros Organics (Asheville, NC). Ferulic acid was obtained from Calbiochem (Billerica, MA), hydroxyphenylpropionic acid, 3,4-dihydroxyphenylpropionic acid, 3-methoxy-4-hydroxyphenylacetic acid (homovanillic), 4-hydroxyphenylacetic acid, m-coumaric acid, p-coumaric acid, caffeic acid, gallic acid, benzoic acid, hippuric acid, methyl hippuric acid, protocatechuic acid, vanillic acid, 3-hydroxybenzoic acid, 4-hydroxyhippuric acid, 3-hydroxyhippuric acid, 3,6-dimethoxybenzoic acid, (−)-epicatechin (EC), (+)-catechin (CT) and taxifolin were purchased from Sigma Aldrich (St Louis, MO). 5-Phenylvaleric acid, syringic acid, protocatechuic acid-4-glucuronide, protocatechuic acid-3-glucuronide, vanillic acid-4-glucuronide, vanillic acid-3-glucuronide, 4-hydroxybenzoic acid-glucuronide, protocatechuic acid-sulfate, vanillic acid-4-sulfate and 4-hydroxybenzoic acid-sulfate were kindly provided by Dr Colin Kay from University of East Anglia, UK. Water was prepared with a Mili Q system to 18 MΩ resistance (Billerica, MA). All solvents used for sample extraction and chromatography analysis and 0.22 μm syringe filters were obtained from Fisher Scientific (Houston, TX). C18 Sep-Pak cartridges (500 mg) were obtained from Waters (Milford, MA). MegaNatural® Gold Grape seed extract was kindly donated by Polyphenolics Inc. (Madera, CA) and contains 91.9% of total phenolics (w/w) as gallic acids equivalent via the Folin Ciocalteau analysis.
2.2 Animals and treatments
The protocol for pigs and treatments were conducted in accordance with the ILAR Guide for the Care and Use of Laboratory Animals, with approval from the University of California Davis Institutional Animal Care and Use Committee (Protocol #:17257). Six crossbred female pigs weighing (130–150) kg were kept in their standard shared pen throughout the experiment. The animals were fed a standard diet (2 kg per day) composed mostly of corn and soy, neither of which contain PACs, along with nutritional supplements. The treatment diet was the control diet supplemented with 1% w/w Grape Seed Extract (GSE). The pigs were fed with standard diet for 3 days, and then the animals were fed the treatment diet for a period of 6 days, followed by 3 days of post treatment control-feeding. Feces samples were freshly collected daily during their feeding. The feces were put on ice and stored at −80 °C within an hour of collection.
2.3 Sample preparation and extraction
Feces samples were thawed, weighed and extracted using SPE as described previously with slight modification.5 Briefly, for determination of intact parent PACs and microbial phenolic acid metabolites in feces, 3 mL of 50% aqueous MeOH with 1% (v/v) formic acid and 100 μL of 1 mM ascorbic acid was added to 0.1 g feces. The mixture was homogenized with a glass rod, sonicated for 5 min, vortexed and centrifuged for 10 min at 4200 × g, 4 °C in a refrigerated Eppendorf centrifuge 5804 R. The supernatant was collected and the extraction procedure was repeated. The combined supernatant was evaporated to dryness under gentle stream of N2 at 35 °C and reconstituted with 1 mL of 1% (v/v) formic acid in water. Sep-Pak C18 cartridges were conditioned with 5 mL of 100% MeOH followed by 5 mL of 1% (v/v) formic acid in water. Taxifolin and 3,5-dimethylbenzoic acid at 5 μg mL−1 each as internal standards (IS) was spiked in 1 mL of sample of the extract and was loaded into the cartridges. The cartridges were then washed with 5 mL of 2% aqueous MeOH with 1% (v/v) formic acid and eluted with 5 mL of MeOH. The eluate was evaporated to dryness under N2 at 35 °C. The dried extracts were reconstituted with 500 μL 0.1% (v/v) formic acid in water.
2.4 Analysis of intact parent PACs
Feces samples were thawed, weighed and extracted using SPE as described previously with slight modification.5 Briefly, separations were conducted on an Agilent HPLC 1100 series (Agilent Technologies, CA) equipped with an autosampler, binary pump, column heater, and diode array (DAD) and fluorescence (FLD) detectors. The column used was Develosil diol 100 (250 mm × 4.6 mm i.d., 5 μm) from Phenomenex (Torrance, CA). The binary mobile phase consisted of (A) acetonitrile–acetic acid (98 : 2, v/v) and (B) methanol–water–acetic acid (95 : 3 : 2, v/v/v). The elution was performed as follows: 0 min, 7% B; 3 min, 7% B; 50 min, 37.6% B; 53 min, 100% B and hold for 7 min, 66 min, 7% B followed by 10 min of re-equilibration of the column. The UV detection was set to monitor from 200 nm to 400 nm. Fluorescence detection (FLD) was conducted with an excitation and emission wavelengths at 231 nm and 320 nm, respectively. The flow rate and injection volume were 1 mL min−1 and 15 μL respectively, with a total analysis time of 76 min. The concentration of intact parent PACs in feces is expressed as (−)-epicatechin equivalents.
2.5 Analysis of microbial phenolic metabolites
The analysis of microbial derived phenolic acid metabolites was carried out on a Perkin Elmer Flexar HPLC. The analytical column used was a Phenomenex Kinetex PFP Column (4.6 × 100 mm, 2.6 μm particle size) (Torrance, CA), operating at 40 °C at a flow rate of 0.3 mL min−1 and the mobile phases consisted of (A) water–formic acid (99.9 : 0.1, v/v) and (B) acetonitrile–formic acid (99.9 : 0.1, v/v). The separation was performed using the gradient elution program: 0 min, 2% B; 12 min, 20% B; 17 min, 50% B; hold for 3 min at 100% B, followed by 5 min of reequilibration of the column before the next run. The injection volume was 10 μL. The HPLC system was coupled to a AxION® 2 ToF Mass Spectrometer equipped with an ESI interface operating in negative ion trap pulse mode. Mass spectrometer parameters were: flight tube: 10 kV; trap pulse mode: 100–450 m/z; IG exit low (D7): 18 μs, trap pulse delay (D8): 28 μs; capillary exit voltage: −80 V; drying gas heater: 350 °C; drying gas flow: 15 L min−1; and nebulizing gas pressure: 80 PSI. N2 was used as drying gas. Standard curves were prepared at six different concentration levels (0.04–50 mg mL−1) for quantitation of phenolic metabolites in feces of the compounds listed in Table 1. identification of conjugated microbial phenolic metabolites, including protocatechuic acid-4-glucuronide, protocatechuic acid-3-glucuronide, vanillic acid-4-glucuronide, vanillic acid-3-glucuronide, 4-hydroxybenzoic acid-glucuronide, protocatechuic acid-sulfate, vanillic acid-4-sulfate and 4-hydroxybenzoic acid-sulfate was also performed by comparing the retention time with authentic standard using LC-MS/ToF.
Table 1.
List of flavan-3-ols monomer and phenolic acid standards with their theoretical masses, recovery, LOD and LOQ by LC-MS/ToF
| Theoretical [M – H]– | Recovery (%) | LOQ (μg mL–1) | LOD (μg mL–1) | |
|---|---|---|---|---|
| Flavanols monomer | ||||
| (+)-Catechin | 289.0717 | 84 ± 6 | 1 | 0.64 |
| (–)-Epicatechin | 289.0717 | 74 ± 7 | 4 | 1.40 |
| Phenolic acids | ||||
| Hydroxyphenylacetic acids | ||||
| 4-Hydroxyphenylacetic acid | 151.0400 | 83 ± 2 | 1 | 0.72 |
| 3-Hydroxyphenylacetic acid | 151.0400 | 110 ± 4 | 0.2 | 0.01 |
| 3,4-Dihydroxyphenylacetic acid | 167.0349 | 79 ± 2 | 6 | 6.00 |
| Hydroxyphenylpropionic acids | ||||
| 3,4-Dihydroxyphenylpropionic acid | 181.0506 | 110 ± 11 | 0.8 | 0.27 |
| 3-Hydroxyphenylpropionic acid | 165.0557 | 93 ± 3 | 6 | 6.00 |
| Hydroxyphenylvaleric acids | ||||
| 4-Hydroxyphenylvaleric acid | 193.0870 | 91 ± 3 | 0.8 | 0.32 |
| 5-Phenylvaleric acid | 177.0920 | 64 ± 16 | 4 | 1.63 |
| Hydroxycinnamic acids | ||||
| m-Coumaric acid | 163.0400 | 73 ± 5 | 0.1 | 0.03 |
| p-Coumaric acid | 163.0400 | 90 ± 2 | 1 | 0.45 |
| Caffeic acid | 179.0349 | 90 ± 4 | 0.8 | 0.17 |
| Ferulic acid | 193.0506 | 108 ± 1 | 0.2 | 0.09 |
| Hydroxybenzoic acids | ||||
| 4-Hydroxybenzoic acid | 137.0244 | 84 ± 3 | 0.1 | 0.05 |
| 3-Hydroxybenzoic acid | 137.0244 | 95 ± 7 | 0.2 | 0.02 |
| Hippuric acid | 178.0509 | 87 ± 4 | 0.8 | 0.16 |
| 4-Hydroxyhippuric acid | 194.0458 | 88 ± 3 | 0.01 | 0.02 |
| Methyl hippuric | 192.0666 | 85 ± 3 | 0.01 | 0.01 |
| Protocatechuic acid | 153.0193 | 109 ± 7 | 0.8 | 0.18 |
| Homovainillic acid | 181.0506 | 87 ± 2 | 4 | 4.00 |
| Vanillic acid | 167.0349 | 77 ± 6 | 6 | 6.00 |
| Syringic acid | 197.0455 | 66 ± 18 | 4 | 3.47 |
2.6 DNA fecal extraction
Pig fecal microbiota was assessed via culture-independent barcoded next generation sequencing (NGS). DNA from pig fecal samples was extracted according to established protocols via Zymo Research Fecal DNA Miniprep™ (Irvine, CA). Mixed-template amplicons were amplified from these fecal DNA extracts using the F515-R806 (V4) segment of the 16S rRNA gene modified to contain an adapter region for sequencing on the Illumina MiSeq platform, and a unique barcode sequence to enable multiplexing of all samples into a single sequencing run.12 Sequencing was performed on the Illumina MiSeq platform with a sequencing depth of ~10 000 raw reads per sample (top 99.99% of the microbiota).
2.7 Sequence analysis
Raw Illumina fastq files were demultiplexed, quality-filtered, and analyzed using QIIME 1.8.0,13 as described previously.14 Alpha diversity rarefactions were calculated by observed species diversity estimates and compared using a two sample t-test. Individual samples were rarified based on alpha-diversity estimates, to ensure even sequencing depth for diversity and relative abundance measurements. Beta diversity metrics were calculated using weighted discrete UNIFRAC estimates and collapsed to 3D principal coordinate analysis (PCoA) plots for visualization of bacterial communities. Significant associations between phenolic end products and specific operational taxonomic unit (OTUs) were calculated by linear regression analysis in Graph Pad Prism 6. Significant changes in OTU relative abundance were calculated using g-test in the QIIME package, and treatment means are shown. Network analyses were visualized in Cytoscape 3.0.2.15
2.8 Statistical methods
Statistical analysis was performed using IBM SPSS Statistics version 17.0 Mann–Whitney test for unrelated samples was performed to compare changes in feces concentration between treatment days. Statistical tests were two-tailed and the significance level was considered as (p < 0.05).
3 Results
3.1 Extraction recovery of phenolic acids
To assess method accuracy, feces were spiked at different concentrations with the following compounds: 4-hydroxyphenylacetic acid (4-HPA), 3-hydroxyphenylacetic acid (3-HPA), 3,4-dihydroxyphenylacetic acid (3,4-diHPA), 3,4-dihydroxyphenylpropionic acid (3,4-diHPPA), 3-hydroxyphenylpropionic acid (3-HPPA), 4-hydroxyphenylvaleric acid (4-HPVA), 5-phenylvaleric acid (5-PVA), m-coumaric acid (m-CA), p-coumaric acid (p-CA), caffeic acid (CA), ferulic acid (FA), 4-hydroxybenzoic acid (4-HBA), 3-hydroxybenzoic acid (3-HBA), hippuric acid (HA), 4-hydroxyhippuric acid (4-HHip), methyl-hippuric acid (mehip), protocatechui acid (PCA), homovanillic acid (HVA), vanillic acid (VA), syringic acid (SA), (−)-epicatechin (EC) and (+)-catechin (CT). Analyses were performed in triplicates in two concentrations, low and high. Briefly, the above compounds were pooled into a solution and spiked into baseline feces. Ascorbic acid (1 mM) was added to the samples to prevent oxidation during extraction. The feces were extracted as described in methods. Table 1 showed the recovery expressed as the mean value ± SD (%), LOD and LOQ for each compound. The recovery for the phenolic metabolites ranged from 64 to 110%.
3.2 Limits of detection, quantification of phenolic acids
The instrumental limits of detection (LOD) and limit of quantification (LOQ) for the ToF analysis were calculated based on signal to noise ratio of 3 (S/N ≥ 3) and 10 × S/N respectively. The LODs varied between 0.001 (me-HA) to 0.681 mg kg−1 (HVA) and generally <0.120 mg kg−1 for most phenolic acid metabolites. Similarly, LOQs varied between 0.006 to 4 mg kg−1.
3.3 Analysis of intact parent PACs
Our results show that PACs oligomers (dimer–pentamer) were absent at baseline, but then present in feces after 3 days and remain constant after 6 days of GSE intake. The maximum increase of total fecal concentration of PACs oligomers (441 ± 15 mg kg−1) were observed after 3 days of GSE administration, in which the highest contributors were trimers (117.2 ± 22.3 mg kg−1) and the lowest were dimers (100.6 ± 7.6 mg kg−1). The concentration of PACs oligomers in feces (dimer–pentamer) showed significant differences between baseline and following 3 days and 6 days of GSE consumption, showing that these oligomeric forms are not absorbed completely and could be targets for metabolism by the gut microbiota. In the 72 h post feeding samples, PACs (dimers–pentamers) were not observed in feces, indicating that these polar compounds are eliminated rapidly from the GI tract.
3.4 Analysis of parent PAC monomers and microbial phenolic metabolites
The selected LC parameters using a PFP column allowed separation of 21 phenolic metabolites and further peak confirmation by MS/ToF. The list of flavan-3-ols monomer and phenolic acid standards with their recoveries, spike levels and theoretical masses are presented in Table 1. From the LC-MS/ToF analysis, intact monomers at day 3 of GSE intake were detected and quantified, substances absent in baseline samples. The fecal concentration of monomer (−)-epicatechin and (+)-catechin increased 8.17 ± 2.89 mg kg−1 and 5.46 ± 1.71 mg kg−1 (p < 0.05) after 3 days consuming GSE and the excretion remained stable during the GSE intervention period. A total of 11 phenolic metabolites were identified in the feces following 24 h consumption of GSE and most of the phenolics were already present at baseline feces. The most abundant microbial phenolic metabolites were 4-hydroxyphenylvaleric acid (4-HPVA) and 3-hydroxybenzoic acid (3-HBA). Additionally, other phenolic metabolites were also detected in pig feces: 3-hydroxyphenyl acetic acid (3-HPA), 4-hydroxyphenylacetic acid (4-HPA), 3,4-dihydroxyphenylpropionic acid (3,4-diHPP), 4-hydroxybenzoic acid (4-HBA), syringic acid (SA), protocatechuic acid (PCA), caffeic acid (CA) and hippuric acid (HA) (Table 2). However, some phenolic metabolites including 3,4-dihydroxyphenylacetic acid (3,4-diHPA), 3-hydroxyphenylpropionic acid (3-HPPA), 5-phenylvaleric acid (5-PVA), p-coumaric acid (p-CA), ferulic acid (FA), gallic acid (GA), 4-hydroxyhippuric acid (4-HHip), homovanillic acid (HVA) and methyl hippuric acid (me-Hip) were not detected in any of the feces.
Table 2.
Fecal concentration (mg kg–1) of flavanols and phenolic metabolites detected in pigs following intake of 1% (w/w) GSEa
| Feces excretion (μg mL–1) |
|||||
|---|---|---|---|---|---|
| Baseline |
Treatment |
Post-feeding |
|||
| d 0 |
d 1 |
d 3 |
d 6 |
d 9 |
|
| Compounds | Mean ± SEM | Mean ± SEM | Mean ± SEM | Mean ± SEM | Mean ± SEM |
| Proanthocyanidins | |||||
| Dimer | 0 | 0 | 101 ± 7.61 | 97.4 ± 6.34 | 0 |
| Trimer | 0 | 0 | 117 ± 22.3 | 117 ± 25.7 | 0 |
| Tetramer | 0 | 0 | 113 ± 7.95 | 103 ± 11.2 | 0 |
| Pentamer | 0 | 0 | 110 ± 16.8 | 106 ± 20.8 | 0 |
| Total | 0 | 0 | 441 ± 15 | 423.4 ± 16 | 0 |
| Flavanol monomers | |||||
| (+)-Catechin | 0 | 0 | 5.46 ± 1.71 | 5.72 ± 1.55 | 0 |
| (–)-Epicatechin | 0 | 0 | 8.17 ± 2.89 | 7.56 ± 2.54 | 0 |
| Phenolic acids | |||||
| Hydroxyphenylacetic acids | |||||
| 4-Hydroxyphenylacetic acid | 3.55 ± 0.97 | 3.62 ± 0.58 | 2.96 ± 1.77 | 2.54 ± 0.44 | 3.45 ± 1.2* |
| 3-Hydroxyphenylacetic acid | 5.39 ± 3 | 5.99 ± 2.82 | 5.86 ± 2.95 | 5.02 ± 1.07 | 3.87 ± 1.07 |
| 3,4-Dihydroxyphenylacetic acid | ND | ND | ND | ND | ND |
| Hydroxyphenylpropionic acids | |||||
| 3,4-Dihydroxyphenylpropionic acid | 0.401 ± 0.22 | 0.354 ± 0.25 | 0.149 ± 0.22 | 0.287 ± 0.14 | 0.415 ± 0.27 |
| 3-Hydroxyphenylpropionic acid | ND | ND | ND | ND | ND |
| Hydroxyphenylvaleric acids | |||||
| 4-Hydroxyphenylvaleric acid | 0.619 ± 1.06 | 5.26 ± 2.93* | 34.8 ± 15.1* | 42.2 ± 21.5* | 1.36 ± 0.66 |
| 5-Phenylvaleric acid | ND | ND | ND | ND | ND |
| Hydroxycinnamic acids | |||||
| m-Coumaric acid | 0.013 ± 0.004 | 0.035 ± 0.029 | 0.015 ± 0.004 | 0.006 ± 0.009 | 0.007 ± 0.015 |
| p-Coumaric acid | ND | ND | ND | ND | ND |
| Caffeic acid | 0.112 ± 0.14 | 0 | 0 | 0.132 ± 0.265 | 0.143 ± 0.199 |
| Ferulic acid | ND | ND | ND | ND | ND |
| Hydroxybenzoic acids | |||||
| 4-Hydroxybenzoic acid | 0.165 ± 0.186 | 0.077 ± 0.096 | 0.3 ± 0.426 | 0.477 ± 0.494 | 0.687 ± 1.54 |
| 3-Hydroxybenzoic acid | 5.09 ± 3.78 | 5.87 ± 3.52 | 8.69 ± 3.53* | 8.32 ± 2.47* | 5.31 ± 3.67 |
| Hippuric acid | 0.118 ± 0.278 | 0.293 ± 0.416 | 0.171 ± 0.293 | 4.84 ± 9.39 | 0.004 ± 0.008 |
| 4-Hydroxyhippuric acid | ND | ND | ND | ND | ND |
| Methyl hippuric | ND | ND | ND | ND | ND |
| Protocatechuic acid | 0.06 ± 0.071 | 0.599 ± 0.982 | 0.115 ± 0.066 | 0.148 ± 0.061 | 0.066 ± 0.062 |
| Homovainillic acid | ND | ND | ND | ND | ND |
| Vanillic acid | 0 | 0.202 ± 0.233 | 0.124 ± 0.173 | 0.084 ± 0.167 | 0.168 ± 0.232 |
| Syringic acid | 7.88 ± 5.31 | 11.2 ± 3.31 | 6.63 ± 5 | 4.31 ± 1.99 | 4.43 ± 1.35 |
| Gallic acid | ND | ND | ND | ND | ND |
| Total | 23.4 ± 3.15 | 33.5 ± 3.54 | 73.4 ± 9.92 | 81.6 ± 11.95 | 19.9 ± 2.18 |
Data represent the mean (n = 3–6) ± SD.
Values with * are significantly different compared with control diet group (p < 0.05) according to Mann–Whitney test. ND, not detected.
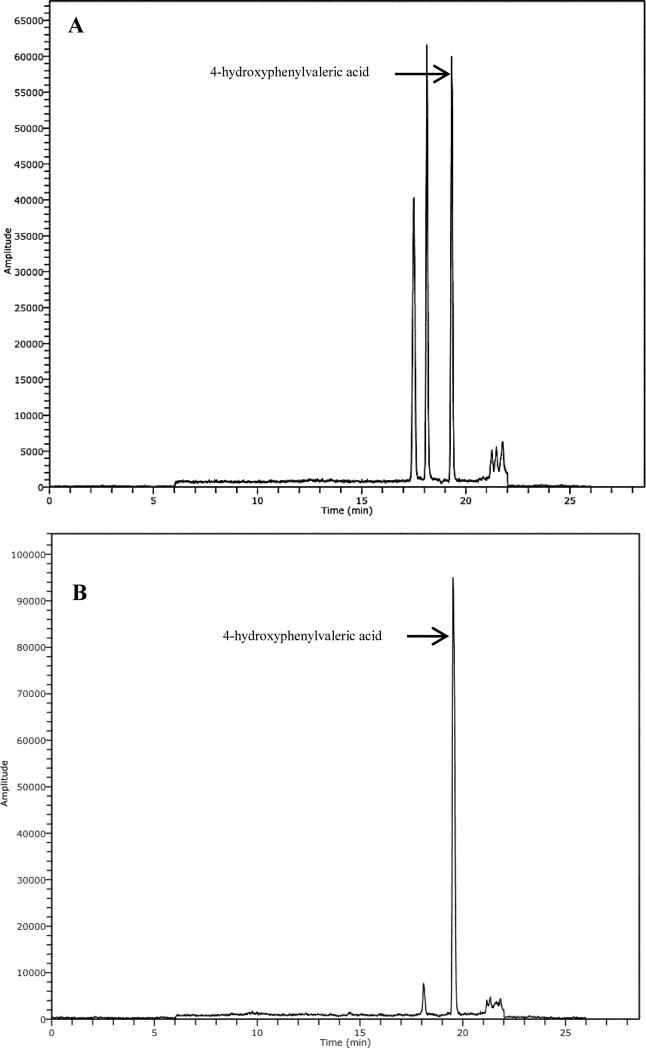
In general, of those 11 phenolic acids metabolites detected during the GSE intervention period, only 4-HPVA and 3-HBA showed a statistical increment compare to baseline (p < 0.05). Fig. 1 shows the extracted ion chromatogram (EIC) of 4-hydroxyphenylvaleric acid in blank feces and in pig feces collected at day 1 of GSE intake.
Fig. 1.
Extracted ion chromatogram (EIC) of 4-hydroxyphenylvaleric acid in blank feces (A) and in pig feces collected at day 1 after intake of 1% (w/w) GSE for 6 days (B).
The maximum total fecal concentration of microbial phenolic metabolites was 73.4 ± 9.92 mg kg−1 observed after 3 days of GSE administration, in which the highest contributor was 4-HPVA (34.8 ± 15.1 mg kg−1) while the lowest was m-CA (0.015 ± 0.004 mg kg−1).
Moreover, conjugated microbial metabolites such as protocatechuic acid-4-glucuronide, protocatechuic acid-3-glucuronide, vanillic acid-4-glucuronide, vanillic acid-3-glucuronide, 4-hydroxybenzoic glucuronide, protocatechuic acid-sulfate, vanillic acid-4-sulfate and 4-hydroxybenzoic acid-sulfate were not found in feces. It has been described elsewhere that conjugated metabolites of EC and microbial derived phenolic metabolites including 3,4-dihydroxyphenylpropionic acid-sulfate and 3- or 4-hydroxybenzoic acid-sulfate were observed previously in feces of rats 24 h after ingesting a phenolic-rich dietary fiber mix.16 However, differences in the gut microbiota of rats compared to pigs may explain why these metabolites were not detected.
3.5 Microbial community analyses
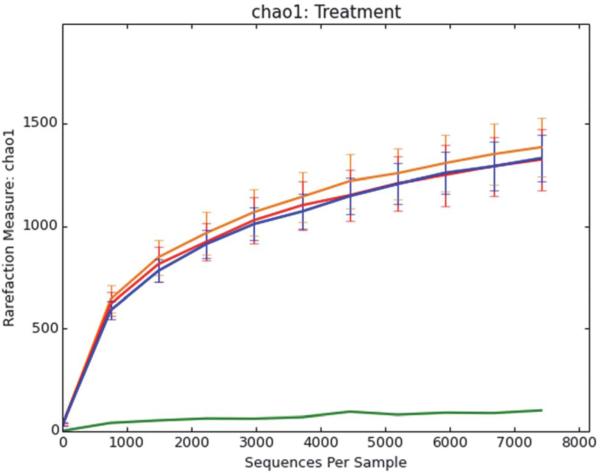
Rarified samples were compared by alpha diversity estimates by observed species (Fig. 2). No difference in overall diversity based on treatment (baseline, GSE intervention and post-feeding period) were observed when evenly sampled (p = 1).
Fig. 2.
Rarefaction curves graphing within-sample (Alpha) Chao1 diversity of pig fecal bacterial populations. Red, orange, and blue curves are shown for samples that are before, during, and after (respectively) GSE treatment. Pig feed was also sampled (green curve).
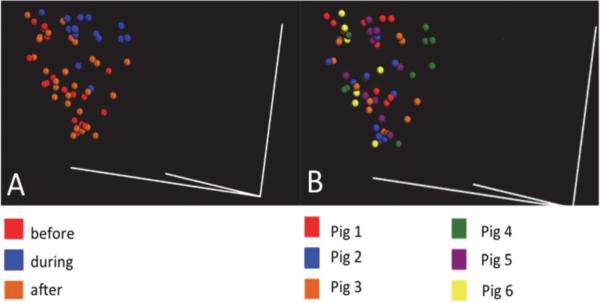
Principal coordinate analysis (PCoA) was used to visualize relationships among samples and in a three-dimensional space. PCoA is an ordination method similar to principal component analysis (PCA) but it uses distance metric instead of covariances. PCoA plots variance of the data long orthogonal axes or principal components. For bacterial population, samples appeared to show the greatest discrimination as a function of time, with the before and after intervention showing the greatest difference (Fig. 3A), while animals consuming GSE, during treatment, shows a transition state between these two (Fig. 3B). Similar comparisons by individual animal were evenly distributed throughout the same space (figure not shown).
Fig. 3.
3-Dimensional PCoA plot showing weighted UniFrac distance of fecal samples. Samples showed a greater difference with (A) before and after GSE treatment compared to differences among pigs (B).
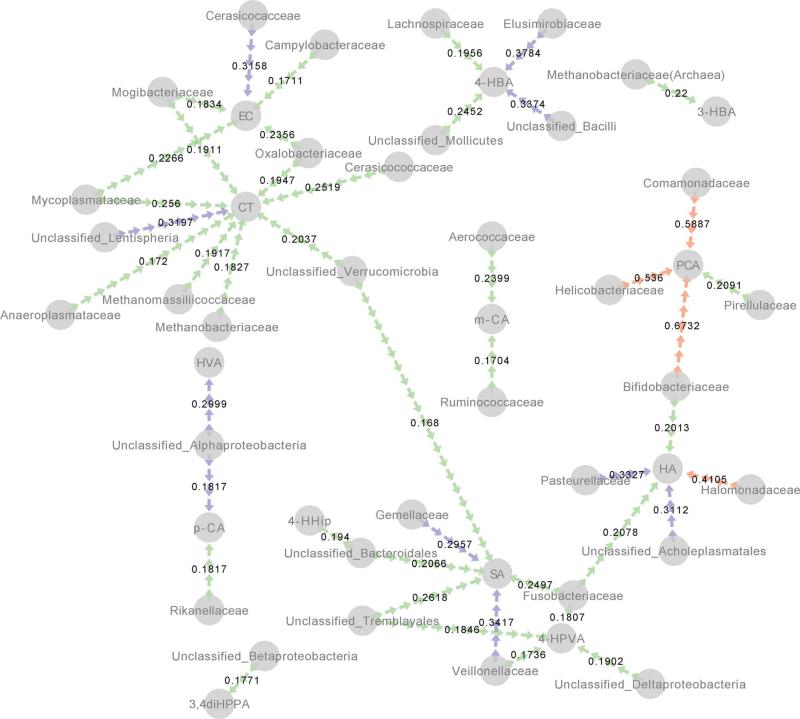
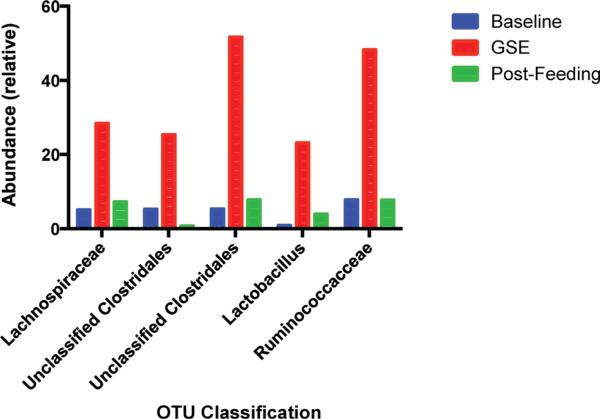
Relationships between phenolic breakdown products measured on day 0, 3, 5, 8, and 10 were correlated with the sequencing results from cognate fecal samples (Fig. 4), showing an association of a core group of taxa which are associated with increased phenolic breakdown products. Among these products and taxa, the strongest correlations were found between Bifidobacteriaceae and Comamonadaceae and PCA (r2 = 0.6732, 0.5887, respectively). Interestingly, these taxa did not show significant increases in relative abundance during feeding (data not shown), instead the Lachnospiraceae, Unclassified Clostridales, Lactobacillus, and Ruminococcus operational taxonomic units (OTUs) were found to significantly increase (p < 0.05) during feeding, relative to before and after the GSE intervention (Fig. 5).
Fig. 4.
Significant positive correlations between selected taxa and phenolic acids end products visualized by network analysis. Edges (arrows) are colored by significance (red, p < 0.001; blue, p < 0.01; green, p < 0.05) and r2 values are shown for each relationship.
Fig. 5.
Significant relative abundance differences of selected taxa during consumption of GSE.
4 Discussion
To our knowledge, this is the first study that has identified the intact parent PACs and their microbial phenolic metabolites in feces and shows that a PACs-rich diet modulates the gut bacteria population after ingestion of GSE. When investigating the bioavailability of polyphenols, most studies are focused on the analysis of plasma or urine11,17–19 and few in feces.20–22
In this study, the monomer (−)-epicatechin and (+)-catechin and PACs oligomers (dimer–pentamer) were detected in feces on day 3 after ingestion of GSE. It was continued through day 6, and was absent in 48 h post-feeding fecal samples. These findings indicate that the monomers and parent PACs oligomers were not completely absorbed and consequently present in the gastrointestinal tract for at least 72 h during gastric transit before excretion in feces. Similar results were demonstrated by Jimenez-Ramsey et al., 1994,23 albeit in chickens, in which monomeric and oligomeric PACs are partially absorbed in the intestinal tract while the polymeric PACs are detected in the feces. The PACs compounds detected in feces, which have been in contact with the colonic epithelium cells, may exhibit beneficial direct activities in the gut including anti-inflammatory24 and anti-tumor25 properties. Inhibition of TNFα-induced NF-κB activation occurred at about 20 mg L−1 of dimer in model system,26 but that is about 1/5 the amount observed here for each compound. Such data may shed light on recent epidemiological data showing that increased consumption of a PAC-rich diet is linked to decrease risk of colorectal cancer (CRC).27,28
It has been reported elsewhere that direct absorption through the gut barrier may not be required for PACs to protect the gut epithelium from oxidative stress or carcinogens.29 It is also important to note that there are neither PACs oligomers nor polymers observed in baseline feces and their presence is due to intake of GSE PACs. The potential health benefits and bioavailability of these partially absorbed PACs may thus be explained by microbial metabolism, that lead to colonic degradation products, such as phenolic acids and valerolactones. While numerous in vivo studies have showed the presence of microbial derived phenolic metabolites in urine after intake of PACs-rich diets, few have addressed fecal content. Some of the principal microbial metabolites found in urine include mono- and di-hydroxyphenylpropionic acids and phenylacetic acids, as well as hydroxyhippuric acids.30
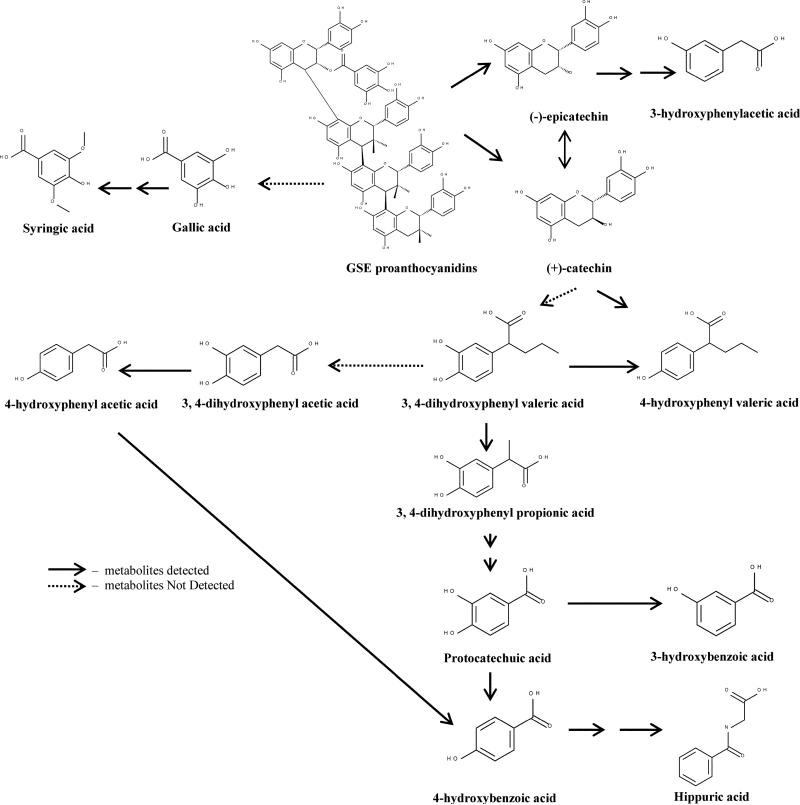
The major microbial phenolic metabolites identified and quantified after ingestion of GSE were 4-HPVA and 3-HBA. Other phenolic metabolites were also detected but at much lower concentrations, including 3-HPA, 4-HPA, 3,4-diHPP, 4-HBA, SA, PCA, CA and HA. Our findings are in accordance with others, who also detected hydroxylated phenolic acids as the main microbial metabolites.29,31 This study is the first to identify 4-HPVA as one of the principal microbial phenolic metabolites following ingestion of a PAC-rich diet. In a previous study, a diet supplemented with procyanidins trimer C2 and polymer, the investigators were able to detect 3-HPVA in the urine of rats but at a low level.29 Conjugated forms of phenylvaleric acid has been detected in urine from rats32 suggesting phenylvaleric acid was absorbed and later conjugated in the liver. Others have reported that CT is degraded mainly to hydroxyphenylvaleric acids when using rat and rabbit microbiota.29,33 4-HPVA can be further oxidized and form 3,4-diHPPA, 4-HPPA and 4-HBA. PCA is formed by decarboxylation of 3,4-diHPPA and further dehydroxylation will produce 3-HBA and 4-HBA29 (Fig. 6). 4-HBA will then eventually convert to HA while it has been proposed that SA was probably derived from a secondary metabolite pathway.34 It also has been reported that SA was produced in vitro by the pig gut microbiota.35 3-HPA is most likely converted from EC and dehydroxylated to phenylacetic acid, as observed in both CT and EC colonic fermentation models using rat colonic microflora.36 Nonetheless, the presence of both parent PACs and microbial-derived metabolites in feces suggest that either of these classes could be providing protective effects on colonic health as these compounds are in contact with the colon epithelial cells. Others have also detected microbial phenolic metabolites in feces, cecal content and colonic tissues, and have suggested these substances are partly responsible for improved gut health following intake of PAC-rich diets.16
Fig. 6.
Proposed metabolic pathway of GSE proanthocyanidins by pig gut microbiota. The detected metabolites in feces is indicated with  , and metabolites not detected is indicated with
, and metabolites not detected is indicated with  .
.
To date, the knowledge of which microbial species are responsible for the bioconversion of phenolic compounds in the human colon is scarce, however a few studies have confirmed that polyphenol rich diets cause a shift in the bacterial populations in the colon.7,8,37 Emerging molecular technologies based on the 16S ribosomal RNA (rRNA), have remodeled the field of gut microbial ecology. Novel high-throughput diversity approaches, such as barcoded “next generation” sequencing38 can be a powerful tool to monitor gut community shifts induced by polyphenols with greater sensitivity than former fingerprinting methods, such as PCR-DGGE.39,40 Moreover, next-generation DNA sequencing allows culture-independent microbiomics methods to determine the gut microbiome.41,42
Here it was observed that Lachnospiraceae, Unclassified Clostridales, Lactobacillus, and Ruminococcus OTUs increased during GSE intervention period. To our knowledge, this is the first time that Lachnospiraceae and Ruminococcus are shown to be modulated by polyphenols. Lachnospiraceae and Ruminococcus represent the major butyrate and propionate producers from human fecal samples.43 Short chain fatty acids are known to play an important role in the colonocytes health since they are absorbed by the host and used as an energy source. In particular, butyrate has been proposed to revert cancer and inflammation processes in colonocytes.44,45 Therefore, grape seed polyphenols may be able to increase butyrate-producers species and thus modulate colonic health. In previous studies, it was reported that polyphenol rich foods, such as wine and cocoa, could increase Lactibacillus spp. populations. Dolara et al. 2005 demonstrated that rats treated with red wine polyphenols showed higher levels of Lactibacillus spp.46 Tzounis et al. 2011 found significant increase in Lactobacillus spp. after the consumption of high cocoa flavanol drink via in vivo and in vitro studies.37
Moreover, our finding indicates that some taxa were significantly associated with the appearance of phenolic compounds during feeding (Fig. 4). Protocatechuic acid was highly associated with Bifidobacteriaceae and Comamondaceae (r2 = 0.6732, 0.5887, respectively, p < 0.001). However, other positive correlations between taxa and phenolic acids were observed with weaker associations. Hippuric acid was associated with Halomonadaceae (r2 = 0.4105, p < 0.001), catechin was correlated with Lentispheria (r2 = 0.3197, p < 0.01), and syringic acid was associated with Veillonellaceae (r2 = 0.3417, p < 0.01). 4-Hydroxyphenylvaleric acid, the major PACs metabolite, was positively associated with Deltaproteobacteria, Tremblayales, Fusobacteriaceae and Veillonellaceae (r2 = 0.1902, 0.1846, 0.1807, 0.1736, respectively, p < 0.05). Future studies examining how these significantly associated species could break down PACs under gut conditions are thus of interest.
5 Conclusions
In conclusion, our study shows that in a porcine model, parent PACs oligomers (dimer–pentamer) were only partially metabolized by the gut microbiota, producing phenolic metabolites that are known to be more readily absorbed. Thus the entire gut is exposed to the native PACs as well as the simple metabolites at high levels (>100 mg kg−1 for some individual compounds in feces). The GSE diet also caused bacterial population shifts in the GI tract and this change may exert beneficial effects on the colon. So, it appears likely that the relationship between dietary PACs and colon health may be attributable to the phenolic compounds or altered bacterial populations. Future studies should attempt to disentangle this interactive system.
Acknowledgements
We would like to thank Kent Parker from the Swine Center, University of California Davis for his assistance, Karen Kalanetra for preparing samples for microbial community sequencing, and Polyphenolics Inc. for their generous supply of Grape seed extract. Paola Quifer thanks the predoctoral fellowships awarded by the Generalitat de Catalunya (FI-DRG). Ying Yng Choy and Paola Quifer-Rada declare equal contribution to the work. D.A.M. acknowledges National Institutes of Health award R01AT007079.
Abbreviations
- PACs
Proanthocyanidins
- GSE
Grape seed extract
- 4-HPA
4-Hydroxyphenylacetic acid
- 3-HPA
3-Hydroxyphenylacetic acid
- 3,4-diHPA
3,4-Dihydroxyphenylacetic acid
- 3,4-diHPPA
3,4-Dihydroxyphenylpropionic acid
- 3-HPPA
3-Hydroxyphenylpropionic acid
- 4-HPVA
4-Hydroxyphenylvaleric acid
- 5-PVA
5-Phenylvaleric acid
- m-CA
m-Coumaric acid
- p-CA
p-Coumaric acid
- CA
Caffeic acid
- FA
Ferulic acid
- 4-HBA
4-Hydroxybenzoic acid
- 3-HBA
3-Hydroxybenzoic acid
- HA
Hippuric acid
- 4-HHip
4-Hydroxyhippuric acid
- me-hip
Methyl-hippuric acid
- PCA
Protocatechuic acid
- HVA
Homovanillic acid
- VA
Vanillic acid
- SA
Syringic acid
- EC
(−)-Epicatechin
- CT
(+)-Catechin
Footnotes
Conflict of interest
The authors have declared no conflict of interest.
References
- 1.Manach C, Williamson G, Morand C, Scalbert A, Rémésy C. Am. J. Clin. Nutr. 2005;81:230S–242S. doi: 10.1093/ajcn/81.1.230S. [DOI] [PubMed] [Google Scholar]
- 2.Verstraeten SV, Jaggers GK, Fraga CG, Oteiza PI. Biochim. Biophys. Acta, Biomembr. 2013;1828:2646–2653. doi: 10.1016/j.bbamem.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 3.Clifford MN. Planta Med. 2004;70(1103):1114. doi: 10.1055/s-2004-835835. [DOI] [PubMed] [Google Scholar]
- 4.Appeldoorn MM, Vincken J-P, Aura A-M, Hollman PCH, Gruppen H. J. Agric. Food Chem. 2009;57:1084–1092. doi: 10.1021/jf803059z. [DOI] [PubMed] [Google Scholar]
- 5.Choy YY, Jaggers GK, Oteiza PI, Waterhouse AL. J. Agric. Food Chem. 2012;61:121–127. doi: 10.1021/jf301939e. [DOI] [PubMed] [Google Scholar]
- 6.O'Hara AM, Shanahan F. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee HC, Jenner AM, Low CS, Lee YK. Res. Microbiol. 2006;157:876–884. doi: 10.1016/j.resmic.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Smith AH, Mackie RI. Appl. Environ. Microbiol. 2004;70:1104–1115. doi: 10.1128/AEM.70.2.1104-1115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pozuelo MJ, Agis-Torres A, Hervert-Hernández D, Elvira López-Oliva M, Muñoz-Martínez E, Rotger R, Goni I. J. Food Sci. 2012;77:H59–H62. doi: 10.1111/j.1750-3841.2011.02520.x. [DOI] [PubMed] [Google Scholar]
- 10.Ferrer I, Thurman EM, Fernández-Alba AR. Anal. Chem. 2005;77:2818–2825. doi: 10.1021/ac048458x. [DOI] [PubMed] [Google Scholar]
- 11.Rzeppa S, Bittner K, Döll S, Dänicke S, Humpf H-U. Mol. Nutr. Food Res. 2012;56:653–665. doi: 10.1002/mnfr.201100471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Proc. Natl. Acad. Sci. U. S. A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bokulich NA, Bamforth CW, Mills DA. PLoS One. 2012;7:e35507. doi: 10.1371/journal.pone.0035507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Touriño S, Pérez-Jiménez J, Mateos-Martiín M. a. L., Fuguet E, Vinardell M. a. P., Cascante M, Torres J. L. s. J. Agric. Food Chem. 2011;59:5955–5963. doi: 10.1021/jf200159f. [DOI] [PubMed] [Google Scholar]
- 17.Rios LY, Gonthier M-P, Rémésy C, Mila I, Lapierre C, Lazarus SA, Williamson G, Scalbert A. Am. J. Clin. Nutr. 2003;77:912–918. doi: 10.1093/ajcn/77.4.912. [DOI] [PubMed] [Google Scholar]
- 18.Urpi-Sarda M, Monagas M, Khan N, Llorach R, Lamuela-Raventós RM, Jáuregui O, Estruch R, Izquierdo-Pulido M, Andrés-Lacueva C. J. Chromatogr. A. 2009;1216:7258–7267. doi: 10.1016/j.chroma.2009.07.058. [DOI] [PubMed] [Google Scholar]
- 19.Borges G, Mullen W, Mullan A, Lean MEJ, Roberts SA, Crozier A. Mol. Nutr. Food Res. 2010;54:S268–S277. doi: 10.1002/mnfr.200900611. [DOI] [PubMed] [Google Scholar]
- 20.Tsang C, Auger C, Mullen W, Bornet A, Rouanet J-M, Crozier A, Teissedre P-L. Br. J. Nutr. 2005;94:170–181. doi: 10.1079/bjn20051480. [DOI] [PubMed] [Google Scholar]
- 21.Mateos-Martín ML, Pérez-Jiménez J, Fuguet E, Torres JL. Mol. Nutr. Food Res. 2012;56:671–675. doi: 10.1002/mnfr.201100672. [DOI] [PubMed] [Google Scholar]
- 22.Saínchez-Pataín F, Monagas M. a., Moreno-Arribas MV, Bartolomé B. a. J. Agric. Food Chem. 2011;59:2241–2247. doi: 10.1021/jf104574z. [DOI] [PubMed] [Google Scholar]
- 23.Jimenez-Ramsey LM, Rogler JC, Housley TL, Butler LG, Elkin RG. J. Agric. Food Chem. 1994;42:963–967. [Google Scholar]
- 24.Larrosa M, Luceri C, Vivoli E, Pagliuca C, Lodovici M, Moneti G, Dolara P. Mol. Nutr. Food Res. 2009;53:1044–1054. doi: 10.1002/mnfr.200800446. [DOI] [PubMed] [Google Scholar]
- 25.Zhao J, Wang J, Chen Y, Agarwal R. Carcinogenesis. 1999;20:1737–1745. doi: 10.1093/carcin/20.9.1737. [DOI] [PubMed] [Google Scholar]
- 26.MacKenzie G, Delfino J, Keen CL, Fraga CG, Oteiza PI. Biochem. Pharmacol. 2009;78:1252–1262. doi: 10.1016/j.bcp.2009.06.111. [DOI] [PubMed] [Google Scholar]
- 27.Wang ZJ, Ohnaka K, Morita M, Toyomura K, Kono S, Ueki T, Tanaka M, Kakeji Y, Maehara Y, Okamura T, Ikejiri K, Futami K, Maekawa T, Yasunami Y, Takenaka K, Ichimiya H, Terasaka R. World J. Gastroenterol. 2013;7(19):2683–2690. doi: 10.3748/wjg.v19.i17.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theodoratou E, Kyle J, Cetnarskyj R, Farrington SM, Tenesa A, Barnetson R, Porteous M, Dunlop M, Campbell H. Cancer Epidemiol., Biomarkers Prev. 2007;16:684–693. doi: 10.1158/1055-9965.EPI-06-0785. [DOI] [PubMed] [Google Scholar]
- 29.Gonthier M-P, Donovan JL, Texier O, Felgines C, Remesy C, Scalbert A. Free Radical Biol. Med. 2003;35:837–844. doi: 10.1016/s0891-5849(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 30.Monagas M, Urpi-Sarda M, Sanchez-Patan F, Llorach R, Garrido I, Gomez-Cordoves C, Andres-Lacueva C, Bartolome B. Food Funct. 2010;1:233–253. doi: 10.1039/c0fo00132e. [DOI] [PubMed] [Google Scholar]
- 31.Urpi-Sarda M, Monagas M, Khan N, Lamuela-Raventos R, Santos-Buelga C, Sacanella E, Castell M, Permanyer J, Andres-Lacueva C. Anal. Bioanal. Chem. 2009;394:1545–1556. doi: 10.1007/s00216-009-2676-1. [DOI] [PubMed] [Google Scholar]
- 32.Mateos-Martín ML, Pérez-Jiménez J, Fuguet E, Torres JL. Br. J. Nutr. 2012;108:290–297. doi: 10.1017/S0007114511005678. [DOI] [PubMed] [Google Scholar]
- 33.Scheline RR. Biochim. Biophys. Acta, Gen. Subj. 1970;222:228–230. doi: 10.1016/0304-4165(70)90373-9. [DOI] [PubMed] [Google Scholar]
- 34.Farah A, Monteiro M, Donangelo CM, Lafay S. J. Nutr. 2008;138:2309–2315. doi: 10.3945/jn.108.095554. [DOI] [PubMed] [Google Scholar]
- 35.Forester SC, Waterhouse AL. J. Agric. Food Chem. 2008;56:9299–9304. doi: 10.1021/jf801309n. [DOI] [PubMed] [Google Scholar]
- 36.Serra A, Macià A, Romero M-P, Anglés N, Morelló J-R, Motilva M-J. Food Chem. 2011;126:1127–1137. [Google Scholar]
- 37.Tzounis X, Rodriguez-Mateos A, Vulevic J, Gibson GR, Kwik-Uribe C, Spencer JP. Am. J. Clin. Nutr. 2011;93:62–72. doi: 10.3945/ajcn.110.000075. [DOI] [PubMed] [Google Scholar]
- 38.Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P, Engstrand L. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Metzker ML. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 40.van Duynhoven J, Vaughan EE, Jacobs DM, Kemperman RA, van Velzen EJJ, Gross G, Roger LC, Possemiers S, Smilde AK, Doré J, Westerhuis JA, Van de Wiele T. Proc. Natl. Acad. Sci. U. S. A. 2011;108(Suppl 1):4531–4538. doi: 10.1073/pnas.1000098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turnbaugh PJ, Gordon JI. J. Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, Taylor TD, Noguchi H, Mori H, Ogura Y, Ehrlich DS, Itoh K, Takagi T, Sakaki Y, Hayashi T, Hattori M. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP, Flint HJ, Louis P. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Aliment. Pharmacol. Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 45.Berni Canani R, Di Costanzo M, Leone L. Clin. Epigenet. 2012;4:4. doi: 10.1186/1868-7083-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dolara P, Luceri C, Filippo CD, Femia AP, Giovannelli L, Caderni G, Cecchini C, Silvi S, Orpianesi C, Cresci A. Mutat. Res., Fundam. Mol. Mech. Mutagen. 2005;591:237–246. doi: 10.1016/j.mrfmmm.2005.04.022. [DOI] [PubMed] [Google Scholar]