SUMMARY
Designing and producing novel proteins that fold into stable structures and provide essential biological functions are key goals in synthetic biology. In initial steps toward achieving these goals, we constructed a combinatorial library of de novo proteins designed to fold into 4-helix bundles. As described previously, screening this library for sequences that function in vivo to rescue conditionally lethal mutants of E. coli (auxotrophs) yielded several de novo sequences, termed SynRescue proteins, which rescued four different E. coli auxotrophs. In an effort to understand the structural requirements necessary for auxotroph rescue, we investigated the biophysical properties of the SynRescue proteins, using both computational and experimental approaches. Results from circular dichroism, size exclusion chromatography, and NMR demonstrate that the SynRescue proteins are α-helical and relatively stable. Surprisingly, however, they do not form well-ordered structures. Instead, they form dynamic structures that fluctuate between monomeric and dimeric states. These findings show that a well-ordered structure is not a prerequisite for life-sustaining functions, and suggest that dynamic structures may have been important in the early evolution of protein function.
Keywords: de novo protein design, helix bundle, synthetic biology, artificial proteomes
Graphical Abstract
INTRODUCTION
The two central challenges of protein design are (i) to devise novel amino acid sequences that fold into stable 3-dimensional structures and (ii) to devise sequences that perform chemically and/or biologically significant functions. Early work in protein design began approximately 25 years ago, with attempts to design 4-helix bundles 1; 2. Those pioneering studies focused exclusively on folding and stability, and paid little attention to protein function. This seemed reasonable at the time, because it was assumed that achieving a well-ordered structure was an essential prerequisite for protein function. Because of this assumption, it was only in recent years, as the design of stably folded structures achieved some level of success 3; 4; 5; 6; 7; 8, that protein designers began to consider the possibility of devising novel proteins that bind targets and/or catalyze reactions 9; 10; 11; 12.
The presumption that uniquely folded structures are essential for function arose from the pioneering achievements of structural biology. The first crystal structures, solved more than half a century ago, revealed ordered structures with well-defined active sites that accounted for their biochemical functions 13. After observing such structures, it is not surprising that researchers assumed that a well-ordered structure was a prerequisite for a well-defined function. Indeed, these early findings led to a central paradigm of structural biology: Amino acid sequence determines 3-dimensional structure, and structure – typically denoting a well-ordered structure – determines function.
In recent years, however, numerous studies have demonstrated that many natural proteins responsible for essential cellular functions are, in fact, intrinsically disordered and/or dynamic 14; 15. In light of these findings, it may be time to reconsider assumptions about the relationship between well-ordered structures and biological function – both for naturally evolved proteins and for proteins designed de novo.
In the current study, we question these assumptions by probing the structural and biophysical properties of several α-helical proteins, which were designed de novo in our laboratory, and shown previously to function in vivo by providing life-sustaining activities in E. coli 16. Using a range of experimental techniques, we probe whether these functional de novo proteins fold into well-ordered, kinetically stable structures, or alternatively, fluctuate between dynamic states.
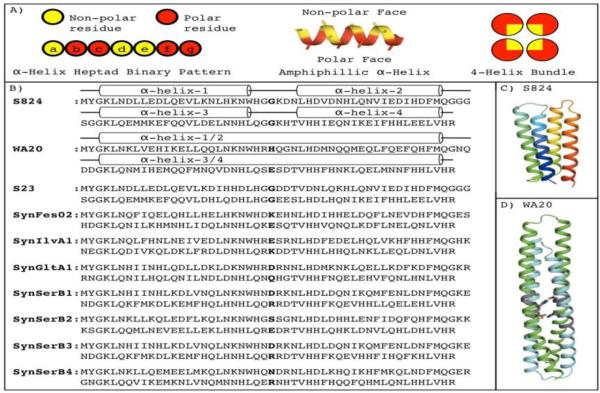
The de novo α-helical proteins that are the subject of the current study were drawn from a large combinatorial library of binary patterned sequences that we described previously 16; 17; 18. Briefly, binary patterning is a strategy for protein design, which is built on the premise that the overall structure of a protein can be specified by designing the sequence periodicity of polar and nonpolar amino acids to match the structural periodicity of the desired secondary structure. Thus, a pattern that places a nonpolar amino acid every 3 or 4 residues along a sequence would match the structural repeat of 3.6 residues/turn of a canonical α-helix, and thereby generate an amphiphilic α-helical segment. When four such helices are linked together, the hydrophobic effect drives them to pack against one another, thereby forming a 4-helix bundle with nonpolar residues pointing towards the protein core and polar residues exposed to solvent (Figure 1A). Since only the type of residue – polar vs. nonpolar – is designed explicitly, the strategy is inherently binary. Yet, because the identities of the polar and nonpolar side chains are not specified, the strategy in inherently combinatorial, and facilitates the construction of vast libraries of novel sequences.
Figure 1. The binary code strategy for protein design, and the sequences of the characterized proteins.
(A) The binary code strategy designs amino acid sequences by placing polar (red) and nonpolar (yellow) residues to match the structural periodicity of an α-helix. Thus, helix heptad positions a, d, & e are designed to be nonpolar, while positions b, c, f, & g are polar. This binary patterning can direct four amphiphilic α-helices to assemble into a 4-helix bundle. (B) The sequences of the control proteins of S824 and WA20 are shown with their α-helices shown as cylinders. (C) Structure of S824 4. (D) Structure of WA20 22. Protein S824 forms a monomer and WA20 forms an extended domain swapped dimer. In WA20, the buried polar amino acids H26 and E78, which form a set of buried hydrogen bonds, are shown as sticks in WA20 and the positions 26 and 78 are bolded for all sequences.
The combinatorial diversity of the protein library is encoded at the DNA level by using degenerate codons, such as NTN (N = A, T, C, or G) to encode five nonpolar amino acids (Phe, Leu, Ile, Met, and Val), and VAN (V = A, C, or G) to encode six polar amino acids (His, Glu, Gln, Asp, Asn, and Lys). These degenerate codons can be assembled in a pattern compatible with the desired structure to produce a collection of synthetic genes, which can be translated in E. coli to produce a large library of de novo proteins.
Previously, we reported the construction of three binary patterned libraries of sequences designed to fold into 4-helix bundles 17; 19; 20. The sequences in these libraries do not share homology with naturally occurring proteins. They were not selected by eons of evolution, and may share features with primordial sequences that existed in the early history of life on earth.
Previous studies of proteins from these binary patterned libraries showed that many of the sequences fold into stable structures 20. Three structures were determined by NMR or crystallography to reveal 4-helix bundles with hydrophobic interiors and polar surfaces, as envisioned by the binary patterned design. Two proteins from our 2nd generation library formed monomeric 4-helix bundles 4; 21, while an X-ray structure solved from a sequence from the 3rd generation library revealed a domain swapped dimer 22. We have also identified de novo proteins from these libraries that bind small molecules, including drugs and cofactors 18; 23. Furthermore, we identified sequences that possess weak catalytic activity for simple reactions and substrates, such as the hydrolysis of p-nitrophenyl esters 18.
The results summarized in the previous paragraph demonstrated that proteins from binary patterned libraries possess structural and functional properties in vitro resembling those of natural proteins. More recently, we have become interested in the possibility of designing collections of novel sequences as an initial step toward constructing artificial “proteomes.” This interest led to experiments probing the ability of our novel sequences to provide essential functions in vivo. Since the proteins in our libraries were designed for structure, but not explicitly designed for any particular function, we used unbiased high throughput genetic selections to search for novel sequences that functioned in vivo. These selections relied on a series of E. coli auxotrophs; strains that are deleted for individual genes that encode enzymes necessary for survival on minimal medium. In a typical auxotroph rescue experiment, an E. coli auxotrophic strain was transformed with a binary patterned library encoding 106 de novo proteins. In most cases, the auxotroph was not rescued by sequences from our library, however, four auxotrophic strains of E. coli were rescued by sequences from our 3rd generation binary patterned library 16. The four rescued auxotrophic strains are deleted for a range of functions: Δfes is missing enterobactin esterase, ΔilvA is missing threonine deaminase, ΔserB is missing phosphoserine phosphatase, and ΔgltA is missing citrate synthase. In all, more than 20 de novo sequences were found to rescue one of these four deletion strains. We denote these novel sequences the SynRescue proteins because they are synthetic (not derived from nature) and they rescue the given deletion strain. Individual proteins are named SynΔstrain#, such that SynFes2 is the second de novo protein identified that rescued Δfes.
It is tempting to assume the SynRescue proteins rescue the deletion strains in a direct manner by performing the same biochemical activity as the deleted protein. However, this need not be the case. It is also possible for a SynRescue protein to compensate for a deleted protein by increasing the expression, enhancing the activity, or altering the specificity of an endogenous E. coli protein. Irrespective of the mechanism of rescue, structural and biophysical characterization of the SynRescue proteins may help elucidate their rescue mechanisms.
The SynRescue proteins also present an unusual opportunity to revisit the relationship between well-ordered structure and biological function. Moreover, because these sequences were devised de novo in the laboratory, we can ask whether uniquely folded 3-dimensional structures are essential for function in vivo in a system that is not biased by eons of evolutionary history. To address these questions, we investigated the biophysical properties of the SynRescue proteins, using both computational and experimental approaches. Results from circular dichroism, size exclusion chromatography, and NMR demonstrate that the SynRescue proteins are α-helical and relatively stable. Surprisingly, however, they do not form well-ordered structures. Instead, they form dynamic structures that fluctuate between monomeric and dimeric states. These findings show that well-ordered structure is not a prerequisite for function in vivo, and suggest that dynamic structures may have been important in the early evolution of protein function.
RESULTS
The SynRescue Proteins
For this investigation, we explored the biophysical and structural properties of seven SynRescue proteins: SynFes2 which rescues Δfes; SynGltA1 which rescues ΔgltA; SynIlvA1 which rescues ΔilvA; and SynSerB1, SynSerB2, SynSerB3, and SynSerB4 which rescue ΔserB. We compared their properties to three control proteins S824, S23, and WA20. The proteins S23 and S824 are sequences from the 2nd generation library (hence the ‘S’ prefix). We previously reported the solution NMR structure of S824, which confirmed that it folds into a 4-helix bundle, as designed 4. S23 was shown previously to be a monomeric molten globule α–helical protein 20.
S824 was the template sequence for the binary pattern and constant regions of the 3rd generation library 17. The SynRescue sequences are all members of the 3rd generation library, and they have between 42% and 51% sequence identity with S824. The protein WA20 is also a member of the 3rd generation library. We recently solved the crystal structure of WA20 to 2.2 Å, which revealed a 4-helix bundle comprising a domain swapped dimer 22. Figure 1 shows the sequences of the SynRescue proteins, the control proteins S23, S824, and WA20 (1B), and the experimentally determined structures of S824 (1C) and WA20 (1D).
Computational Structure Prediction
We performed computational structure prediction simulations for each of the SynRescue proteins and the control proteins S23, S824, and WA20, using the macromolecular modeling software Rosetta, which has been shown to accurately predict the structures of many small proteins (<150 residues) 24. The NMR solution structure of S824 has previously been solved (pdb code 1p68), and S824 is an extremely stable and well-ordered monomeric 4-helix bundle4. We attempted to computationally predict the structure of S824 as a positive control for Rosetta's ability to predict the structure of de novo sequences, not designed in Rosetta, and which have amino acid distributions that differ significantly from natural proteins (e.g. these sequences do not contain alanine or proline). Supplemental Figure 1 shows a plot of the root mean square deviation (RMSD) versus total Rosetta energy for the S824 structure prediction. In an ideal case, a single ‘folding funnel’ would be observed at low RMSD and low Rosetta energy 24, however the plot for S824 shows several funnels with approximately equal energies. While RMSD space is highly multidimensional, the lowest energy models in each funnel correspond to the different possible topologies of a 4-helix bundle. Although the Rosetta simulation samples the experimentally determined topology, the energy function is not able to accurately identify the correct structure of S824. For each of the four ‘folding funnels’, we used the experimentally determined NOE constraints from protein S824 to calculate the number of violations for each model structure. Only models with the same topology as the S824 NMR solution structure, left-handed 4-helix bundles (green funnel in Supplemental Figure 1), satisfied the NOE distance constraints. Models from the other three topologies have hundreds of long-range NOE distance violations, which confirms that the only structure compatible with these chemical shifts and NOE constraints is the experimentally determined structure.
We performed similar simulations for the SynRescue proteins, and like S824, they showed multiple funnels with similar energies. Investigation of the lowest energy models did not indicate which fold, if any, would be the true structure. (Supplemental Figure 2 shows the prediction results for the SynRescue sequences). We also performed structure prediction simulations for WA20. Since the X-ray crystal structure of WA20 is a homodimer, we used Rosetta's fold and dock protocol 25. The fold and dock structure prediction results for WA20 also showed multiple folding funnels with approximately equal energies. The lowest energy models in each funnel correspond to different arrangements of a helix-turn-helix homodimer. Again, the simulation sampled the experimentally determined topology, however the Rosetta energy function did not identify models with the topology of the X-ray crystal structure as the lowest energy models (Supplemental Figure 3).
These simulations demonstrate that for S824 and WA20, Rosetta's monomer and oligomer structure prediction methods sample the correct conformational space but the energy function does not identify the experimentally determined structure as having the lowest energy. This could occur for several reasons: (1) the sequences predicted here have features, which are not common in natural proteins or in Rosetta de novo designed proteins, such as they do not contain the amino acids alanine, proline, and cysteine, and have unusual amino acid distributions (e.g. over representation of histidine). Since many terms in the Rosetta energy function are trained on high-resolution X-ray crystal structures of natural proteins, and the Rosetta reference energy is trained specifically to recapitulate ‘natural’ amino acid distributions, the Rosetta energy function may not accurately represent the energies of these binary patterned proteins. (2) The actual physical energy differences between the structures sampled in the Rosetta simulations may be small and within the error of the Rosetta energy function. (3) In the cases of WA20 and the SynRescue proteins, we have not solved their NMR solution structures, thus, the Rosetta simulations may be correct in suggesting these sequences sample multiple topologies.
Protein Expression and Purification
We expressed and purified the control proteins S23, S824, and WA20 and the seven SynRescue proteins. The control proteins S23, S824, and WA20 express and purify with high yield. However, some SynRescue sequences express and purify much more readily than others (see methods for details). In all cases, it was possible to generate pure protein (>95% by SDS-PAGE) at concentrations of at least 200 μM for biophysical and structural characterization.
The SynRescue Proteins Form α-Helical Secondary Structure
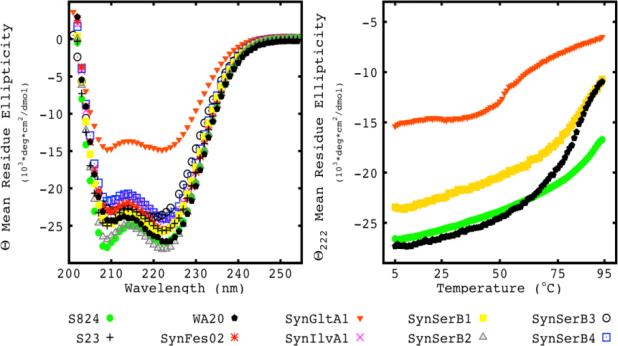
Circular dichroism (CD) measurements of the SynRescue and control proteins revealed canonical spectra with minima at 208 and 222 nm, thereby demonstrating that, as expected from their binary patterned design, the proteins are predominantly α-helical (Figure 2A).
Figure 2. The SynRescue proteins are helical and stable.
(A) Far-UV CD spectra. The control proteins S824 (green circle), S23 (plus sign) WA20 (black pentagon), and the rescue proteins SynFes02(red star), SynGltA1 (orange down triangle), SynIlvA1(purple X), SynSerB1 (yellow closed square), SynSerB2 (grey up triangle), SynSerB3 (black open circle), SynSerB4 (blue open square) display CD spectra consistent with α–helical structures, with prominent minima at 208 nm and 222 nm. (B) Thermal denaturation. The SynRescue proteins display a range of thermal stabilities. SynGltA1 (orange down triangle) has the lowest mid-point and SynSerB1 (yellow closed square) has one of the highest mid points. The control protein S824 (green closed circle) is shown for comparison as an extremely stable monomer. The dimer control protein WA20 (black pentagon) is shown and behaves similar to SynSerB1.
Most of the SynRescue proteins display similar levels of α-helical content, except for SynGltA1, which shows ~50% of the α-helical content of the other proteins. For helical proteins, the ratio of ellipticity at 222 nm relative to 208 nm indicates the amount of supercoiling. A 222/208 ratio greater than 1.0 is consistent with coiled-coil structures, whereas values between 0.9 and 1.0 indicate assemblies of non-supercoiled helices, and values less than 0.9 suggest independent helices 26. The control protein WA20 has a 222/208 ratio of 1.2 indicating it is supercoiled in solution, as expected from its crystal structure, where the domain swapped dimeric bundle is twisted by ~90° along its long axis. The protein S824 has a 222/208 ratio of 0.98 indicating it is not extensively supercoiled, consistent with the NMR structure. The SynRescue proteins also have 222/208 ratios of ~1, indicating they are not highly super coiled (Supplemental Table 1).
Thermal Stability
To assess the thermal stability of the SynRescue proteins, we monitored ellipticity at 222 nm as a function of temperature. The control proteins S824 (Figure 2B, green circle) and WA20 (Figure 2B, black pentagon) are thermostable, with unfolding midpoints of >100°C and 80°C respectively. All of the SynRescue proteins are also stable, with denaturation midpoints between 50-90°C (Supplemental Table 1). The thermal denaturations for SynSerB1 (yellow filled squares) and SynGltA1 (orange filled triangles) are shown in figure 2B and are representative of the extremes of the SynRescue proteins. The denaturation curves of the SynRescue proteins have a range of cooperativities, with some being barely cooperative (SynFes2) and others modestly cooperative (SynIlvA1).
Thermal denaturations of the SynRescue and control proteins were thermodynamically reversible: After cooling to the original temperature, followed by a period of equilibration, ellipticity at 222 nm regained 95%-100% of the original native values. Although all of the samples display thermodynamic reversibility, the kinetics of refolding differed among the various sequences. Protein S824, which is known to form a well-ordered monomeric 4-helix bundle 4, refolded relatively rapidly with its renaturation curve nearly superimposable on its denaturation curve. In contrast, WA20, which is known from crystallography to form a domain swapped dimer 22, refolded more slowly, with its renaturation lagging behind the original denaturation curve. The SynRescue proteins displayed delayed renaturation, similar to that observed for the WA20 dimer (Supplemental Figure 4).
NMR Spectroscopy
To probe the structural properties of the SynRescue proteins, we recorded their 1H15N HSQC NMR spectra (Figure 3). In such spectra, a mono-disperse, well-folded protein is expected to show a cross peak for each backbone NH, and a pair of cross peaks for each asparagine and glutamine side chain. The control monomeric protein, S824, yields such a spectrum, with abundant and well-resolved peaks (Figure 3A). In contrast, a molten globule protein, which is compact, but dynamic, would be expected to produce a spectrum with limited chemical shift dispersion. The control monomeric protein, S23, yields a spectrum consistent with the molten globule state (Figure 3B). The control protein WA20, which forms a dimer in solution and in its x-ray crystal structure, has spectra consistent with a molecule undergoing exchange between multiple states on the time scale of the NMR experiment. The peaks of WA20's 1H15N HSQC are broad, low intensity, and poorly resolved.
Figure 3. 1H15N HSQC NMR spectra indicate the SynRescue proteins are dynamic.
(A) The spectrum of the well folded de novo protein S824 shows intense peaks with unique chemical shifts for each backbone NH and asn and gln side chain NH. (B) The spectrum of the control molten globule S23 shows numerous peaks but with many overlapping chemical shifts. (C) The spectrum of the extended dimer WA20 shows numerous broad, low intensity peaks consistent with a structure undergoing intermediate exchange. (D-E) The spectra of SynIlvA1 and SynSerB4 show numerous broad, low intensity peaks indicating they are dynamic. (F) The spectrum of SynGltA1 shows ~⅕ the expected backbone peaks indicating it is primarily unfolded or extremely dynamic.
The 1H15N HSQC NMR spectra of the SynRescue proteins resemble neither the well-folded nor molten globule monomeric control proteins but instead resemble the spectra of WA20. The spectra of the SynRescue proteins show peaks with low intensity and broad linewidths. In some cases, the SynRescue spectra have numerous broad, low intensity backbone NH peaks (e.g. SynIlvA1 & SynSerB4 in Figure 3D, 3E), while in other cases the spectra display relatively few broad, low intensity peaks (SynGltA1 in Figure 3F and SynSerB1, SynSerB2, SynSerB3 and SynFes2 in Supplemental Figure 5). These spectra are consistent with dynamic structures undergoing exchange on a range of intermediate timescale. Given the nature of the 1H15N HSQC spectra and the slow reversibility of refolding observed in the thermal denaturations, we considered the possibility that the SynRescue proteins might be undergoing exchange between monomeric and oligomeric states on an intermediate timescale.
In some cases, it is has been possible to assign or partially assign the chemical shifts of proteins undergoing exchange on an intermediate time scale. However considering the quality of the SynRescue 1H15N HSQC spectra and similar data quality in other experiments traditionally used in backbone and side chain assignment (1H13C HSQC, HNCA, HNCO, HNCACB, HNCACO were collected for several SynRescue proteins, data not shown), we concluded that it would not be possible to assign or even partially assign the backbone or side chain chemical shifts of the SynRescue proteins using traditional methods and the conditions tested. While we could not determine the structures of the SynRescue proteins by solution NMR, we still wanted to investigate the oligomeric state of the SynRescue proteins. Therefore we probed the solution state of these proteins by size exclusion chromatography.
Size Exclusion Chromatography
The oligomeric states of the SynRescue proteins were assessed by size exclusion chromatography (SEC). Because SEC is a non-equilibrium method, the apparent molecular weight of a protein undergoing monomer/oligomer exchange on an intermediate timescale will be influenced by the time spent on the column, and by the flow rate and size of the column. Therefore, we measured the apparent molecular weights of the de novo proteins using columns of three different sizes: (i) a S75 5/150 analytical column with a 3 mL bed volume; (ii) a S75 10/300 semi-preparative column with a 24 mL bed volume; and (iii) a S75 26/600 preparative column with a 318 mL bed volume.
The well-folded monomer S824, the molten globule monomer S23, and the domain swapped dimer WA20 provide appropriate controls for this experiment. The molecular weights of S824 and S23, calculated from their amino acid sequences, are both 11.9 kDa. In SEC experiments, both proteins run at ~12 kDa on all three columns, confirming that these proteins exist in solution as mono-disperse monomeric globular structures (Figure 4A and Table 1).
Figure 4. Apparent Molecular Weights of the SynRescue Proteins.
(A) The monomeric control protein, S824, has apparent molecular weights of ~12 kDa on three size exclusion columns from smallest to largest: S75 5/150 (blue), S75 10/300 (green), and S75 26/600 (red). (B) The dimer control protein, WA20, has apparent molecular weights of 31 kDa (5/150), 25 kDa (10/300), and 20 kDa (26/600). The SynRescue proteins SynFes2 and SynGltA1 are presented as representatives of the extremes of the SynRescue protein's behaviors. SynFes2 has apparent molecular weights of 28 kDa (5/150), 24 kDa(10/300) and 20 kDa (26/600), respectively, and SynGltA1 where the apparent molecular weight on all three columns is ~ 18 kDa.
Table 1.
Apparent molecular weights of the SynRescue and control proteins.
| Construct | MWAA (kDa) | MW 5/150 (kDa) | MW 10/300 (kDa) | MW 26/600 (kDa) |
|---|---|---|---|---|
| S23 | 11.9 | 12.3 | 12.4 | 12.5 |
| S824 | 11.9 | 12.3 | 12.4 | 12.5 |
| WA20 | 12.5 | 31.4 | 25.2 | 20.6 |
| SynFes02 | 12.5 | 27.8 | 23.5 | 20.6 |
| SynGltA1 | 12.5 | 18.6 | 18.2 | 18.4 |
| SynIlvA1 | 12.6 | 26.0 | 23.1 | 19.5 |
| SynSerB1 | 12.6 | 28.8 | 24.6 | 19.8 |
| SynSerB2 | 12.3 | 27.0 | 22.0 | 18.6 |
| SynSerB3 | 12.7 | 28.8 | 21.8 | 20.1 |
| SynSerB4 | 12.6 | 27.5 | 24.0 | 17.4 |
The apparent molecular weights of the SynRescue and control proteins were determined using three size exclusion columns: an analytical S75 5/150 (MW5/150), a semi-preparative S75 10/300 (MW10/300), and a large preparative S75 26/600 (MW26/600). Comparison of the expected monomer molecular weight (MWAA) as calculated from the amino acid sequence, with the experimentally determined apparent molecular weights shows that the SynRescue proteins have apparent molecular weights that are consistent with the formation of weakly associated dimers similar to the known dimer WA20.
The other control protein, WA20, has a covalent molecular weight of 12.5 kDa, calculated from its amino acid sequence. The crystal structure of WA20 shows a domain swapped dimer, and the expected molecular weight of this dimer would be 25 kDa. However, the dimer seen in the crystal structure is elongated. This is because turn 1 and turn 3 of the intended design did not form, and instead continue helix 1 into helix 2, and helix 3 into helix 4 (Figure 1A & 1C). This causes WA20 to be shaped more like a rod than a sphere. Since SEC separates proteins based on their hydrodynamic radii 27, the rod shaped structure of WA20 would be expected to run through SEC columns with an apparent molecular weight that is larger than would be observed for a more spherical 25 kDa protein.
On the smallest SEC column (5/150), WA20 runs with an apparent molecular weight of 31.4 kDa, which is ~2.5 times its expected monomer molecular weight, and is consistent with its elongated structure and larger hydrodynamic radius. However, on the medium sized column (10/300), the apparent molecular weight of WA20 is shifted to 25.2 kDa, which is ~2 times its covalent molecular weight. Finally, using the largest column (26/600), the apparent molecular weight of WA20 is further shifted to 20.6 kDa, which is only 1.7 times WA20's covalent monomer weight. These results suggest that during the longer runs on the larger SEC column, WA20 dissociates from its dimeric structure.
Figure 4 compares the apparent molecular weights – on all three SEC columns – of the control proteins S824 and WA20, with representative SynRescue proteins, SynFes2 and SynGltA1. (The other SynRescue proteins display behaviors between the extremes of SynFes2 and SynGltA1, and are summarized in Table 1.) The apparent molecular weight of SynFes2 is highly dependent on the column size, running at 27.8 kDa, 23.5 kDa, and 20.6 kDa for the small, medium, and large columns, respectively. On the smallest column, the apparent molecular weight of SynFes2 is 2.3 times its monomer weight. We interpret this as indicating that SynFes2 forms an extended dimer similar to WA20. This assumption is strengthened by the finding that the apparent molecular weights of SynFes2 on the medium and large columns are similar to those of WA20.
For SynGltA1, the situation is somewhat different. The apparent molecular weight of SynGltA1 does not depend on column size; it runs at ~18 kDa on all three columns. We interpret this to indicate that SynGltA1 forms either a very weakly associating dimer or that it forms an extended monomer. It seems unlikely that SynGltA1 forms a canonical 4-helix bundle (similar to S824 in figure 1B) because we do not observe an apparent molecular weight consistent with that structure.
We also evaluated the apparent molecular weight of the control proteins and the SynRescue proteins as a function of protein concentration on the analytical S75 5/150. We tested the proteins at the same concentration used in the NMR, ≥200 μM, and also diluted to 30 μM. In the concentration range tested, the apparent molecular weight was independent of protein concentration.
The results of the SEC experiments for the remaining SynRescue proteins are summarized in Table 1. All together, we take these results to indicate that the SynRescue proteins form extended helical monomers that assemble into extended dimer structures similar to the crystal structure of WA20. Most importantly, these data, together with NMR spectra, indicate that the SynRescue proteins do not form well-folded or molten globule monomeric structures like S824 or S23. Instead, the SynRescue proteins appear to fluctuate between monomeric and dimeric α-helical bundles similar to WA20.
DISCUSSION
We investigated the biophysical and structural properties of several de novo proteins that were shown previously to provide activities capable of sustaining the growth of living cells. We determined that the SynRescue proteins are α-helical and thermostable, and that they denature reversibly. However, 1H15N HSQC NMR experiments demonstrate that their structures are dynamic and undergo kinetic exchange on an intermediate timescale. Size exclusion chromatography indicates that the SynRescue proteins do not form long-lived monomeric structures, but instead form extended dimers that are kinetically unstable on the time scale of the chromatography experiments.
The SynRescue proteins are members of a 3rd generation library of binary patterned sequences designed to form α-helical bundles. The crystal structure of another protein from this same library, WA20, was solved recently and shown to form two extended α-helical hairpins, which intertwine to form a domain swapped dimer (Figure 1D) 22. The sequences of the SynRescue proteins are 31% - 52% identical to WA20 and they behave similarly in CD, NMR, and size exclusion chromatography. Therefore, we suggest that the transient dimeric structures observed for the SynRescue proteins resemble the extended dimer seen in the X-ray crystal structure of WA20 (Figure 1C) or a related structure with a different arrangement of helices similar to the models produced by Rosetta's Fold and Dock structure prediction protocol (Supplemental Figure 3), or perhaps they sample a range of these structures as monomers and dimers.
Given the tendency of the 3rd generation sequences to sample dimeric states, we wished to understand which features in the design of the 3rd generation library promote this dimerization. We were particularly curious about this because the design of the 3rd generation library was inspired by the sequence of S824 (from a 2nd generation library), which formed a well-ordered monomeric 4-helix bundle with a disperse 1H15N HSQC NMR spectrum and a persistent structure that was readily solved by NMR 4.
We have identified three features that may have favored the formation of extended (double length) α-helical hairpins that assemble into domain swapped dimers. In each case, ‘negative design’ might have prevented extension of the helices and the resulting dimerization 1. These three features of negative design are summarized as follows:
-
(i)
Breaking the Hydrophobic Register: The underlying premise of the binary patterning strategy is that matching the sequence periodicity of polar and nonpolar residues with the structural periodicity of the desired secondary structure will direct a chain to form amphiphilic secondary structures that bury hydrophobic side chains in the protein core. For α-helices, this requires placing nonpolar residues every 3 or 4 positions to match the helical repeat of 3.6 residues/turn. If this periodicity continues throughout a designed sequence, then one might expect the entire sequence to form one long amphiphilic helix. In particular, if the last nonpolar residue of one helix and the first nonpolar residue of the next helix are 3, 4, or 7 residues apart, then the two helices may form a single long helix with a continuous hydrophobic face. To avoid this possibility, one can use negative design to break this periodicity, offset the hydrophobic face of the helix, and disfavor the continuation of long helices. This feature of negative design was not incorporated into the 3rd generation library: Thus, the sequences of the SynRescue proteins and WA20 have 7 residues from the last nonpolar residue of helix 1 (Trp23) to the first nonpolar residue of helix 2 (Leu30). Likewise, there are 7 residues from the last nonpolar residue of helix 3 (Leu75) to the first nonpolar residue of helix 4 (Val82). Since these sequences do not offset the hydrophobic register of an idealized amphiphilic α-helix, perhaps it is not surprising that the crystal structure of WA20 shows that helices 1 & 2, and helices 3 & 4 form continuous double length helices. We presume the SynRescue sequences form similar extended helices in their dimeric structures.
-
(ii)
Preventing Favorable Buried Polar Interactions: Another premise of the binary patterning strategy is that polar residues avoid burial. Therefore in our libraries, polar residues are used only in positions designed to be on the solvent exposed faces of helices or in inter-helical loops. However, if these loops do not form at the expected locations and the helices continue through the intended loop sequences, then some of these polar residues will be on the buried faces of the extended helices. This is observed in the crystal structure of the WA20 dimer. Moreover, as shown in figure 1C, the sequences that were designed to form loops between helices 1 & 2 and between helices 3 & 4 pack against one another in the domain swapped dimer. In the structure of WA20, the burial of these polar residues is enabled by a favorable electrostatic interaction between His26 and Glu78. Similarly, all the SynRescue proteins studied here have charged and/or hydrogen bonding groups at positions 26 and 78 that could be satisfied by the formation of extended dimer structures similar to WA20. These residues at positions 26 and 78 are shown in bold in Figure 1A. These favorable buried polar interactions, which presumably stabilize the dimeric structure, could be prevented by using negative design to place like charges at these sites (e.g. K/R26 and K/R78).
-
(iii)
Interrupting Helix Propensity: Another way to use negative design to prevent the helices from extending through the intended loops would be to include helix breaking residues in the loops. The control proteins, S23 and S824, contain two glycines in each of the relevant loops. Glycine is well known as a helix breaker, and the structure of S824 shows the intended loops at these locations. Thus, S23 and S824 are both monomeric. In contrast, protein WA20 has no glycines at these positions, and its crystal structure shows helices that continue through the intended loop sequences. Likewise, the SynRescue proteins rarely have glycines in these regions, and are presumed to form extended dimers similar to WA20.
While the features described above may have caused the SynRescue proteins to adopt less ordered structures, which vacillate between monomeric and dimeric states, this diminished order has not prevented the possibility of biological function. Quite the contrary; more than 20 different sequences from the 3rd generation library provide life-sustaining activities in E. coli: These sequences enable cell growth in strains that cannot grow in their absence 16. These findings demonstrate that a well-ordered structure is not a prerequisite for biological function.
For natural proteins, structural biologists had long assumed that ordered structures are essential for biological function. However this assumption arose, in part, from a bias that developed because the only protein structures that had been observed were those that ‘held still’ long enough for their structures to be determined by crystallography or NMR. More recently, as new methods have been developed to study dynamic structures, it is becoming clear that many proteins essential for life are indeed dynamic and/or intrinsically disordered 14; 15.
Advances in protein engineering provide additional compelling evidence that well-defined structures are not required for activity – even for high levels of enzyme catalysis. Most notably, Hilvert and co-workers demonstrated that an engineered version of chorismate mutase exists as a dynamic molten globule, yet retains kcat and Km values similar to the wild type enzyme 28.
Fluctuating or dynamic structures may have also played an important role in the early evolution of proteins. Jensen postulated that early in the history of life on earth, proteins did not have well-defined specific activities. He suggested that primordial proteins had low levels of activity and low specificity. Instead of the highly specialized enzymes that we see in modern organisms, Jensen suggested that primordial proteins were promiscuous generalists. Broad specificity would have been advantageous at the early stages of molecular evolution because it would “maximize the catalytic versatility of an ancestral cell that functioned with limited enzyme resources” 29. While Jensen's discussion of primordial proteins focused primarily on function, rather than structure, it seems reasonable to assume that nonspecific promiscuous functions would have been facilitated by nonspecific promiscuous structures. While it is not possible to go back in time to perform structural measurements and/or assay the biological fitness of primordial proteins, the de novo sequences in our libraries may in fact resemble the sequences that existed in the early history of life on earth.
Indeed, one of the de novo sequences described in the current study, SynIlvA1, has now been shown to be dynamic, both in terms of structure and function. The structural dynamics of SynIlvA1 are illustrated by the experiments described above, and recently we reported the functional promiscuity of SynIlvA1, which was originally selected for its ability to rescue the isoleucine auxotroph ΔilvA, but also rescues Δfes, which is essential for the assimilation of iron 30. These observations suggest that dynamic proteins may not merely be ‘acceptable’ structures for biological function, but may in fact play key roles in evolutionary trajectories from multifunctional generalists to highly active specialists.
METHODS
Computational Simulations using Rosetta
Protein structure prediction simulations were performed using the Rosetta macromolecular modeling software fragment assembly protocol 24. Briefly, this protocol combines three residue and nine residues fragments (from high-resolution crystal structures) using a reduced centroid model of the protein, coarse-grained energy functions, and a Monte Carlo search procedure, followed by an all-atom high-resolution structure refinement step. The three and nine residue fragments are chosen based on sequence similarity and predicted secondary structure of the target protein sequence. Fragments were generated using the Robetta fragment server http://robetta.bakerlab.org/ and simulations were performed on a Princeton University Dell/SGI computer cluster with 10,304 cores. Sample command lines are given in the supplemental information.
To predict the structure of suspected oligomers, we used the Rosetta fold and dock protocol which has been used to predict the structure of protein oligomers 25. We used the protocol to predict the structures of the proteins studied here under the assumption that they were symmetric homodimers with C2 symmetry. The fold and dock protocol essentially performs the standard Rosetta ab initio simulation, while simultaneously docking monomers A and A’ in a symmetric complex, allowing translation and rotation in the x, y, and z directions. Sample command lines are given in the supplemental information.
Protein Expression and Purification
The genes for the proteins studied here are in a modified pCA24N vector 16. The vector contains the chloramphenicol resistance gene, Chloramphenicol Acetyl Transferase (CAT), an IPTG inducible T5 promoter, and a ribosome binding site upstream from the gene of interest. The gene of interest is between a 5’ Nde1 site at the initiator methionine, and a 3’ BsrG1 site that cleaves in the last four amino acids followed by a stop codon. Amino acid sequences for the constructs S824, S23, WA20, SynIlvA1, SynFes2, SynGltA1, SynSerB1, SynSerB2, SynSerB3, and SynSerB4 are listed in the supplemental information and with their European Nucleotide Archive accession number.
Proteins were expressed in E. coli BL21 (DE3) pLysS cells. Cells were grown in 1L LB with 30 μg/ml chloramphenicol at 37°C to an OD600 between 0.4-0.6, and induced with 100 μM IPTG for 12-16 hours at 18°C. Cells were recovered by centrifugation at 5000 × g for 30 minutes. Cell pellets were resuspended in 50 mM sodium phosphate with 200 mM sodium chloride (pH 7.4), and lysed by passing through an Emulsiflex C3 homogenizer at 15,000 psi for three cycles. Cell lysates were clarified by centrifugation at 7,000 × g for 30 minutes. The supernatant was filtered using 0.22 μm PES membrane syringe filters.
Proteins were purified using immobilized metal affinity chromatography (IMAC). While, our constructs do not contain a canonical histidine tag, they do contain a high percentage of histidines, on average 15%, and are readily purified using IMAC with a modified buffer system. The running buffer does not contain imidazole and is 50 mM sodium phosphate and 200 mM sodium chloride at pH 7.4 and the elution buffer is 50 mM sodium phosphate, 200 mM sodium chloride, and 500 mM imidazole at pH 7.4. The IMAC purification was performed as follows; filtered supernatant was applied to a 5mL HisTRAP column (GE Healthcare) equilibrated in running buffer without imidazole. The column was washed with 5 column volumes of running buffer. A second wash step of 5 column volumes with 10% elution buffer, removes proteins non-specifically bound to the column, with the primary contaminating protein being chloramphenical acetyl transferase. The proteins of interest were then eluted using 75% elution buffer. Eluted fractions were pooled, typically 10 mLs, and further purified by size exclusion chromatography on a HiLoad Superdex 75 26/600 column (GE Healthcare). Purity of proteins from this two-step procedure was >95% as assessed by SDS-PAGE (Supplemental Figure 6).
The proteins S824, S23, WA20, SynIlvA1, SynFes2, and SynSerB1 expressed and purified in high yield giving >30 mg/L expression culture. SynGltA1 and SynSerB2 expressed well, but purified with lower yield giving ~10 mgs/L of culture. SynSerB3 did not express at significant levels and was difficult to purify giving ~1 mg/L of culture. SynSerB4 had modest expression and did not purify in high yield giving ~5 mgs/L of culture. It is interesting that SynSerB1 and SynSerB3 behaved so differently, given that their amino acid sequences are 94% identical, with only six contiguous residues being different (Figure 1). Additionally, the SynRescue proteins were prone to precipitation at protein concentrations above 200 μM, especially at sodium chloride concentrations below 100 mM.
Circular Dichroism Spectroscopy
CD data were collected on a Chirascan Circular Dichroism spectrometer (Applied Photophysics). Far-UV CD spectra were collected using a 1 mm pathlength cuvette and protein concentrations of ~30 μM in 50 mM sodium phosphate and 100 mM sodium chloride at pH 7.4. Thermal denaturation experiments were performed by monitoring the α-helical CD signal at 222 nm, as the temperature was increased/decreased at 1°C/min from 5°C to 95°C and then back to 5°C. Thermal denaturation curves were fit to a two state model of unfolding using gnuplot (see supplemental information for details).
Nuclear Magnetic Resonance Spectroscopy
NMR spectra were collected on an 800 MHz AVANCE III HD spectrometer (Bruker) with a 5 mM cryoprobe. Proteins were in 90% H20/10% D20 with 50 mM sodium phosphate and 200 mM sodium chloride (pH 6.8). One dimensional proton spectra were collected using WATERGATE solvent suppression 31. Two dimensional 1H,15N-HSQC were collected on uniformly labeled 15N samples using the ‘hsqcfpf3gpphwg’ pulse sequence from the Bruker library modified to use excitation sculpting water suppression. Labeled samples were grown as described previously, except that prior to induction, cultures were centrifuged and transferred to a minimal media containing 1.0 g/L of 15N ammonium chloride. NMR samples had concentrations ≥200 μM, as protein solubility allowed. All spectra were processed and visualized using TopSpin (Bruker) and CCPNMR 32.
Size Exclusion Chromatography Experiments
A Superdex 75 5/150 column (GE Healthcare) was used for analytical size exclusion chromatography. A set of standard proteins of BSA (66 kDa), Carbonic Anhydrase (29 kDa), Cytochrome C horse heart (12.4 kDa), and Aprotinin (6.5 kDa) were run on the column to measure elution volume, resolution, and sensitivity. Blue dextran (~2,000 kDa) was used to identify the column void volume. These data were used to generate a standard curve of the ratio of elution volume over void volume versus Log10(molecular weight). The same was done for the Superdex 75 10/300 and Superdex 75 26/600 columns. The size exclusion chromatography experiments were performed using the same samples concentrated for the 1H,15N HSQC experiments, with concentrations of ≥200 μM and also at dilutions of 30μM both in 50 mM sodium phosphate and 200 mM sodium chloride at pH 6.8 giving similar results. The injection volumes were 1000 μL, 500 μL, and 100 μL for the 26/600, the 10/300, and the 5/150 columns. The flow rates were 2.6 mL/min, 1.0 mL/min, and 0.5 mL/min for the 26/600, the 10/300, and the 5/150 columns. Molecular weights were calculated from elution volumes by rearranging the standard curve equation for the S75 5/150 to be MW=10^(−2.1362*(EV/3.0)/0.48+7.3678), S75 10/300 to be MW=10^(−1.5068*(EV/24)/0.32+6.5893), and S75 26/600 to be MW=10^(−1.0806* (EV/318)/0.37+6.0493)).
Supplementary Material
Research Highlights.
Well-ordered protein structure is commonly assumed to be an essential feature of protein function.
Here we investigated the biophysical and structural properties of a family of de novo designed proteins that provide life-sustaining functions in E. coli.
We discovered that the de novo proteins tested here do not form well-ordered structures and instead form dynamic dimer structures.
These results highlight the importance of protein dynamics in protein function and also suggest that dynamic structures may have been important in early evolution.
Acknowledgements
We thank Dr. Istvan Pelczer and Ken Conover from the Princeton University Chemistry Department NMR facility for helpful discussions on NMR pulse sequences and results. We also thank the Princeton University Research Computing center for access to the Tiger cluster. We also thank Ann Mularz and Katherine Digianantonio for helpful discussions on this research and manuscript. This work was funded by NSF grants MCB-1050510 and MCB-1409402 to MHH and a NIH F32 fellowship (1F32GM106622) to GSM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
GSM, JBG, and MHH designed the research. GSM and JBG performed the experiments. GSM, JBG and MHH analyzed the data. GSM, JBG, and MHH wrote the paper.
REFERENCES
- 1.Hecht MH, Richardson JS, Richardson DC, Ogden RC. De novo design, expression, and characterization of Felix: a four-helix bundle protein of native-like sequence. Science. 1990;249:884–91. doi: 10.1126/science.2392678. [DOI] [PubMed] [Google Scholar]
- 2.Regan L, DeGrado WF. Characterization of a helical protein designed from first principles. Science. 1988;241:976–8. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- 3.Harbury PB, Plecs JJ, Tidor B, Alber T, Kim PS. High-resolution protein design with backbone freedom. Science. 1998;282:1462–7. doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 4.Wei Y, Kim S, Fela D, Baum J, Hecht MH. Solution structure of a de novo protein from a designed combinatorial library. Proc Natl Acad Sci U S A. 2003;100:13270–3. doi: 10.1073/pnas.1835644100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–8. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 6.Murphy GS, Sathyamoorthy B, Der BS, Machius MC, Pulavarti SV, Szyperski T, Kuhlman B. Computational de novo design of a four-helix bundle protein--DND_4HB. Protein Sci. 2015;24:434–45. doi: 10.1002/pro.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koga N, Tatsumi-Koga R, Liu G, Xiao R, Acton TB, Montelione GT, Baker D. Principles for designing ideal protein structures. Nature. 2012;491:222–7. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang PS, Oberdorfer G, Xu C, Pei XY, Nannenga BL, Rogers JM, DiMaio F, Gonen T, Luisi B, Baker D. High thermodynamic stability of parametrically designed helical bundles. Science. 2014;346:481–5. doi: 10.1126/science.1257481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–21. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–5. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 11.Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, St Clair JL, Gallaher JL, Hilvert D, Gelb MH, Stoddard BL, Houk KN, Michael FE, Baker D. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–13. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, 3rd, Hilvert D, Houk KN, Stoddard BL, Baker D. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–91. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. A Three-Dimensional Model of the Myoglobin Molecule Obtained by X-Ray Analysis. Nature. 1958;181:662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- 14.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oldfield CJ, Dunker AK. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu Rev Biochem. 2014;83:553–84. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 16.Fisher MA, McKinley KL, Bradley LH, Viola SR, Hecht MH. De novo designed proteins from a library of artificial sequences function in Escherichia coli and enable cell growth. PLoS One. 2011;6:e15364. doi: 10.1371/journal.pone.0015364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradley LH, Kleiner RE, Wang AF, Hecht MH, Wood DW. An intein-based genetic selection allows the construction of a high-quality library of binary patterned de novo protein sequences. Protein Eng Des Sel. 2005;18:201–7. doi: 10.1093/protein/gzi020. [DOI] [PubMed] [Google Scholar]
- 18.Patel SC, Bradley LH, Jinadasa SP, Hecht MH. Cofactor binding and enzymatic activity in an unevolved superfamily of de novo designed 4-helix bundle proteins. Protein Sci. 2009;18:1388–400. doi: 10.1002/pro.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamtekar S, Schiffer JM, Xiong H, Babik JM, Hecht MH. Protein design by binary patterning of polar and nonpolar amino acids. Science. 1993;262:1680–5. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 20.Wei Y, Liu T, Sazinsky SL, Moffet DA, Pelczer I, Hecht MH. Stably folded de novo proteins from a designed combinatorial library. Protein Sci. 2003;12:92–102. doi: 10.1110/ps.0228003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Go A, Kim S, Baum J, Hecht MH. Structure and dynamics of de novo proteins from a designed superfamily of 4-helix bundles. Protein Sci. 2008;17:821–32. doi: 10.1110/ps.073377908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arai R, Kobayashi N, Kimura A, Sato T, Matsuo K, Wang AF, Platt JM, Bradley LH, Hecht MH. Domain-swapped dimeric structure of a stable and functional de novo four-helix bundle protein, WA20. J Phys Chem B. 2012;116:6789–97. doi: 10.1021/jp212438h. [DOI] [PubMed] [Google Scholar]
- 23.Cherny I, Korolev M, Koehler AN, Hecht MH. Proteins from an unevolved library of de novo designed sequences bind a range of small molecules. ACS Synth Biol. 2012;1:130–8. doi: 10.1021/sb200018e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradley P, Misura KM, Baker D. Toward high-resolution de novo structure prediction for small proteins. Science. 2005;309:1868–71. doi: 10.1126/science.1113801. [DOI] [PubMed] [Google Scholar]
- 25.Das R, Andre I, Shen Y, Wu Y, Lemak A, Bansal S, Arrowsmith CH, Szyperski T, Baker D. Simultaneous prediction of protein folding and docking at high resolution. Proc Natl Acad Sci U S A. 2009;106:18978–83. doi: 10.1073/pnas.0904407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau SY, Taneja AK, Hodges RS. Synthesis of a model protein of defined secondary and quaternary structure. Effect of chain length on the stabilization and formation of two-stranded alpha-helical coiled-coils. J Biol Chem. 1984;259:13253–61. [PubMed] [Google Scholar]
- 27.Erickson HP. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online. 2009;11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vamvaca K, Vogeli B, Kast P, Pervushin K, Hilvert D. An enzymatic molten globule: efficient coupling of folding and catalysis. Proc Natl Acad Sci U S A. 2004;101:12860–4. doi: 10.1073/pnas.0404109101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jensen RA. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–25. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 30.Smith BA, Mularz AE, Hecht MH. Divergent evolution of a bifunctional de novo protein. Protein Sci. 2015;24:246–52. doi: 10.1002/pro.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single- quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–5. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 32.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–96. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.