Abstract
SAMHD1 is a cellular dNTPase that restricts lentiviral infection presumably by lowering cellular dNTP levels to below a critical threshold required for reverse transcription; however, lowering cellular dNTP levels may not be the sole mechanism of restriction. In particular, an exonuclease activity of SAMHD1 was reported to contribute to virus restriction. We further investigated the requirements for SAMHD1 restriction activity in both differentiated U937 and cycling HeLa cells. Using hydroxyurea treatment to lower baseline dNTP levels in HeLa cells, we were able to document efficient SAMHD1 restriction without significant further reduction in dNTP levels by SAMHD1. These results argue against a requirement for additional myeloid-specific host factors for SAMHD1 function but further support the notion that SAMHD1 possesses an additional dNTP-independent function contributing to lentiviral restriction. However, our own experiments failed to associate this presumed additional SAMHD1 antiviral activity with a reported nuclease activity.
Keywords: SAMHD1, HIV-1, SIV, dATP, dNTPase, nuclease, restriction
INTRODUCTION
Sterile alpha motif and HD domain protein 1 (SAMHD1) is a host factor contributing to the inefficient replication of HIV-1 in cells of myeloid lineage and other non-dividing cell types (Baldauf et al., 2012; Berger et al., 2011; Descours et al., 2012; Hrecka et al., 2011; Laguette et al., 2011). The SIVsm/HIV-2 Vpx proteins are however able to counteract SAMHD1 by targeting it for proteasomal degradation and viruses lacking this protein are restricted at the reverse transcription step in susceptible cell types (Goujon et al., 2007; Hrecka et al., 2011; Laguette et al., 2011; Sharova et al., 2008).
Early work on the SAMHD1 protein suggested it to be involved in regulating the innate immune response as mutations in SAMHD1 have been associated with Acardi-Goutieres Syndrome (AGS), a syndrome associated with increased production of interferon alpha (Dussaix et al., 1985; Rice et al., 2009). Accordingly, SAMHD1 knockout mice, while developmentally healthy, show increased expression of interferon stimulated genes (Behrendt et al., 2013; Rehwinkel et al., 2013). The main catalytic activity ascribed to SAMHD1 is its (d)GTP-dependent dNTPase activity with an active site located in the protein’s HD domain (Amie et al., 2013; Goldstone et al., 2011; Hansen et al., 2014; Ji et al., 2014; Ji et al., 2013; Koharudin et al., 2014; Powell et al., 2011; Zhu et al., 2013). This enzymatic activity allows SAMHD1 to degrade dNTPs to component nucleosides and free triphosphate in a single step. Therefore, dNTP degradation by SAMHD1 provides a counterpart to dNTP synthesis by ribonucleotide reductase (RNR) with both these proteins being carefully regulated to control the delicate dNTP balance in cells (Franzolin et al., 2013). It was thus suggested that SAMHD1 uses its dNTPase activity to restrict lentivirus (and other dNTP-dependent virus) infection by decreasing dNTP levels in susceptible cells to below the levels required for reverse transcription/replication (Hollenbaugh et al., 2013; Kim et al., 2012; Kim et al., 2013; Lahouassa et al., 2012).
Interestingly, SAMHD1 also possesses nucleic acid binding capability and has been reported in some studies to possess 3’–5’ exonuclease activity against ssRNA and viral genomes (Beloglazova et al., 2013; Goncalves et al., 2012; Ryoo et al., 2014; Tungler et al., 2013; White et al., 2013a). Indeed, several recent reports employing mutagenesis experiments to genetically separate the dNTPase and nuclease activities of the protein suggested that nuclease activity may be the main contributor to SAMHD1 restriction of lentiviruses (Choi et al., 2015; Ryoo et al., 2014). However, other studies have failed to detect nuclease activity associated with the SAMHD1 active site (Goldstone et al., 2011; Seamon et al., 2015) thus, leaving the question of the functional importance of a SAMHD1 exonuclease activity up for future investigations. Furthermore, while phosphorylation has been shown to negatively regulate SAMHD1 restriction ability (Cribier et al., 2013; Welbourn et al., 2013; White et al., 2013b) it is still unclear whether this modification might affect dNTPase activity, nuclease activity or possibly another characteristic of SAMHD1 not yet described (Arnold et al., 2015; Ryoo et al., 2014; Tang et al., 2015; Welbourn et al., 2013; White et al., 2013b; Yan et al., 2015). Therefore, whether lowering cellular dNTP levels is the sole mechanism of restriction used by SAMHD1 and whether or not this function is even necessary requires further investigation.
In this study we investigated the importance of cellular dNTP levels for virus restriction by SAMHD1. Unlike other restriction factors such as APOBEC3G that renders normally permissive HeLa cells restrictive for HIV-1, exogenous expression of SAMHD1 in HeLa cells shows little to no restrictive phenotype. It is possible that the continued synthesis of dNTPs in dividing HeLa does not allow SAMHD1 dNTPase activity to sufficiently lower the cellular dNTP pool for lentiviral restriction to occur. Alternatively, it cannot be ruled out that SAMHD1 exerts its antiviral effect in conjunction with additional host factor(s) not expressed in non-myeloid or dividing cell types. To address these questions, we employed SAMHD1 variants together with hydroxyurea treatment to modulate dNTP levels in HeLa cells. Hydroxyurea inhibits ribonucleotide reductase and thus reduces the cellular dNTP pool at the synthesis step (Nordlund and Reichard, 2006). Determination of cellular dATP levels confirmed that hydroxyurea dramatically reduced the baseline dNTP pool in treated HeLa cells with SAMHD1 exhibiting negligible additional effects on the dNTP pool. Using the hydroxyurea strategy of lowering baseline dNTP levels we were able to demonstrate SAMHD1 antiviral activity in HeLa cells whereas no additional antiviral activity was demonstrable in untreated cells. These results suggest that the lack of antiviral activity of SAMHD1 in normal HeLa cells is not due to the lack of additional cellular proteins but are due to the high dNTP levels in these cells. Thus, low cellular dNTP levels appear to be necessary for SAMHD1 restriction activity. However, our results also provide further evidence that SAMHD1 may possess an additional dNTP-independent function that contributes to lentiviral restriction but a contribution of a possible exonuclease activity could not be confirmed.
RESULTS AND DISCUSSION
SAMHD1 T592E decreases dATP levels in cells without causing significant restriction
We and others have shown that SAMHD1 phosphomimetics (T592E/T592D) were unable to restrict HIV infection in PMA-differentiated U937 cells yet still retained dNTPase activity in vitro (Welbourn et al., 2013; White et al., 2013b). While other recent studies have suggested phosphorylation (or phosphomimetics) might modulate dNTPase activity of SAMHD1 under certain conditions using recombinant protein (Arnold et al., 2015; Tang et al., 2015; Yan et al., 2015), only one group has reported cellular dNTP levels measured in cells under conditions where the SAMHD1 restrictive phenotype is lost due to phosphorylation (White et al., 2013b). We therefore wanted to confirm if the dNTPase activity we observed in vitro translated into a cellular effect and independently confirm the decreased cellular dNTP levels seen by others using a phosphomimetic mutant (White et al., 2013b). U937 cells were therefore transduced with lentiviral particles expressing SAMHD1 variants or empty vector, selected with puromycin, and differentiated with PMA. Parallel samples were used for western blot analysis, reporter virus infection, or dNTP isolation. All SAMHD1 variants were efficiently expressed (Fig 1A) and SAMHD1 WT and T592A were able to efficiently restrict HIV-1-GFP infection as compared to cells expressing the active site mutant (H206R/D207N [HD/RN]) or the empty vector control (Fig. 1B). As reported previously, the T592E protein was impaired in its ability to restrict HIV-1 infection (Fig 1B). Cellular dNTP extraction at time of infection showed that SAMHD1 T592E was indeed able to decrease cellular dATP levels to a similar extent as the wildtype and phosphoablative (T592A) proteins as compared to the much higher levels observed with the active site mutant or empty vector controls (Fig 1C). These results therefore independently confirm that SAMHD1 T592E is able to decrease cellular dNTP levels as efficiently as the WT protein, suggesting an additional mechanism of restriction by SAMHD1 may exist beyond nucleotide depletion.
Figure 1. Effect of SAMHD1 T592 phosphorylation on cellular dATP levels.
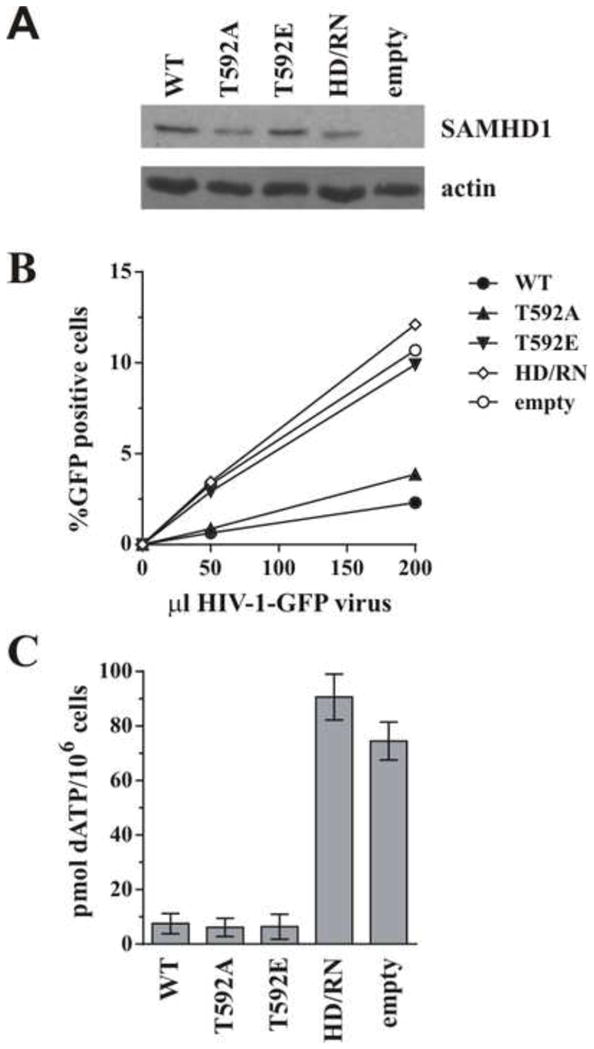
U937 cells were transduced with lentiviral particles encoding either WT SAMHD1, the indicated mutants, or an empty vector. Following puromycin selection, cells were differentiated overnight with 10 ng/ml PMA. A) Total cell extracts from differentiated U937 stable cell lines were separated by SDS-PAGE and subjected to immunoblotting for SAMHD1 and actin as indicated. B) Differentiated cells were infected with increasing amounts of VSV-G pseudotyped HIV-1-GFP (as described in (Welbourn et al., 2013)) and the percent infection (% GFP- positive cells) was determined by flow cytometry 48 h later. C) Cellular dNTP levels were isolated at time of infection and amounts of dATP present per million cells were determined using a polymerase based assay described in the Methods section. Results in panels A and B are representative of at least 3 independent experiments. Error bars in panel C represent the mean and standard deviation of quantitation from at least 3 independently generated cell lines.
Lack of SAMHD1 restriction activity in dividing cells with high dNTP levels
Although SAMHD1 is present in many actively dividing cell types, the restrictive effect has generally only been observed in differentiated/non-dividing cells (Baldauf et al., 2012; Berger et al., 2011; Descours et al., 2012; Hrecka et al., 2011; Laguette et al., 2011; St Gelais et al., 2012). There are several possible explanations, including a lowering of set point dNTP levels upon cell differentiation, the presence of a cell-type specific co-factor, or modulation of SAMHD1 restriction activity via phosphorylation at T592 in dividing cells (Cribier et al., 2013; Welbourn et al., 2013; White et al., 2013b). We therefore investigated how important low cellular dNTP levels are for lentiviral restriction by SAMHD1. Dividing HeLa cells that express only low levels of endogenous SAMHD1 were transduced to either express empty vector, SAMHD1 WT, HD/RN or the phosphoablative T592A mutant (Fig 2A). Upon infection with increasing doses of WT or Vpx-defective SIV-GFP, neither WT SAMHD1 nor the “active” T592A mutant showed significant restriction activity; in fact, efficient infection was seen with all SAMHD1 variants tested (Fig 2B). This is consistent with the lack of SAMHD1 restriction activity observed toward an HIV-1 reporter in 293T cells or undifferentiated U937 cells (Arnold et al., 2015; St Gelais et al., 2014). While SAMHD1 WT and T592A were able to slightly decrease dATP levels compared to the active site mutant and empty vector control (Fig 2C), the resulting levels were still quite high, presumably due to continued dNTP synthesis in these actively dividing cells. Similarly, St Gelais et al. also reported only modest decreases in dNTP levels by the WT SAMHD1 protein in cycling cells (St Gelais et al., 2012). This lack of restriction activity in cycling HeLa cells with high dNTP levels, even with the constitutively active T592A non-phosphorylated variant, suggests that low dNTP levels may indeed be required for SAMHD1 restriction activity.
Figure 2. Effect of SAMHD1 on lentiviral infection in dividing cells.
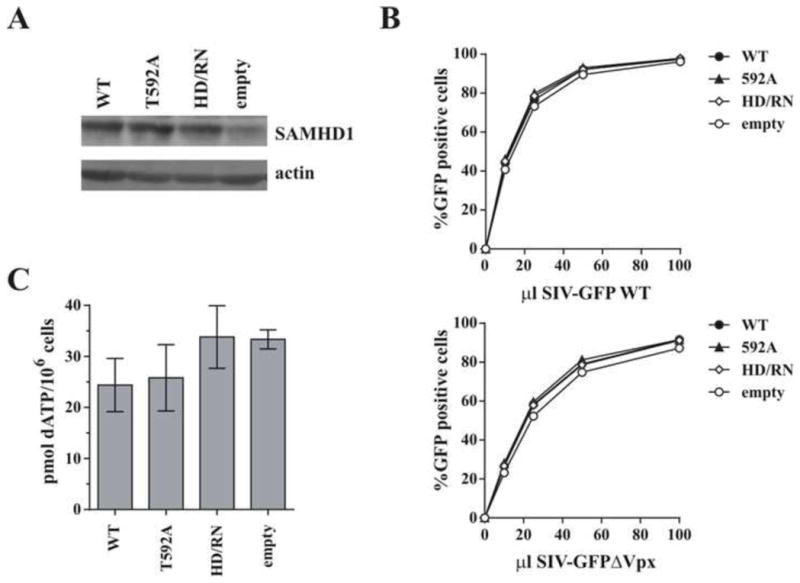
HeLa cells were transduced with lentiviral particles encoding either WT SAMHD1, the indicated SAMHD1 mutants, or an empty vector and selected with puromycin for 48 h. A) Total cell extracts were separated by SDS-PAGE and subjected to immunoblotting for SAMHD1 and actin as indicated. B) Cells were infected with increasing volumes of VSV-G-pseudotyped SIV-GFP (White et al., 2013a), with or without Vpx, and the percent infection (% GFP-positive cells) was determined by flow cytometry 48 h later. C) Cellular dNTPs were isolated at time of infection and the amount of dATP present per million cells was determined using a polymerase-based assay. Results in panels A and B are representative of at least 3 independent experiments. Error bars in panel C represent the mean and standard deviation of quantitation from at least 3 independently generated cell lines.
Lowering dNTP levels in HeLa cells reveals an additional antiviral effect of SAMHD1
To explore whether SAMHD1 has additional antiviral effect above and beyond lowering dNTP levels, we artificially lowered dNTP levels in HeLa cells to see if under such conditions SAMHD1 can exert any extra restriction activity over low dNTPs alone. To do so, HeLa cell lines expressing SAMHD1 WT, HD/RN, or an empty vector were generated (Fig 3A) and treated for 4h prior to infection with hydroxyurea to block de novo dNTP synthesis by inhibiting ribonucleotide reductase (Nordlund and Reichard, 2006). Figure 3B shows dATP levels for dNTPs extracted at time of infection. Similar to Figure 2, in the absence of hydroxyurea, WT SAMHD1 decreased dATP levels less than 2-fold (Fig 3B), which was not sufficient depletion to restrict infection by wildtype or Vpx-defective SIV-GFP (Fig 3C). In contrast, hydroxyurea treatment reduced dATP levels by about 10-fold even in the absence of SAMHD1, resulting in globally 3–5 fold lower infection rates (compare percentage of infection in Fig 3C and 3D). Importantly, in the presence of hydroxyurea, expression of SAMHD1 WT resulted in an additional 2–3 fold reduction in infection by SIV-GFPΔVpx over empty vector containing cells (Fig 3D & E) even though hydroxyurea treatment of control cells was able to decrease dATP to levels approaching those observed when SAMHD1 was also expressed (Fig 3B). Significantly, this extra restriction was not consistently observed with the SIV-GFP WT virus that encodes a functional Vpx to counteract SAMHD1. Slightly lower levels of dATP were consistently observed in the presence of hydroxyurea when WT SAMHD1 was present (Fig 3B). While it cannot be formally ruled out that this difference contributes to the additional restriction measured, it seems unlikely this small difference could account for the magnitude of the effect on viral infection observed. Taking into consideration the results presented in Fig 1, these data therefore provide further evidence that SAMHD1 may possess an additional dNTP-independent function that can contribute to lentiviral restriction at low dNTP levels.
Figure 3. Effect of SAMHD1 on lentiviral infection in hydroxyurea-treated cells.
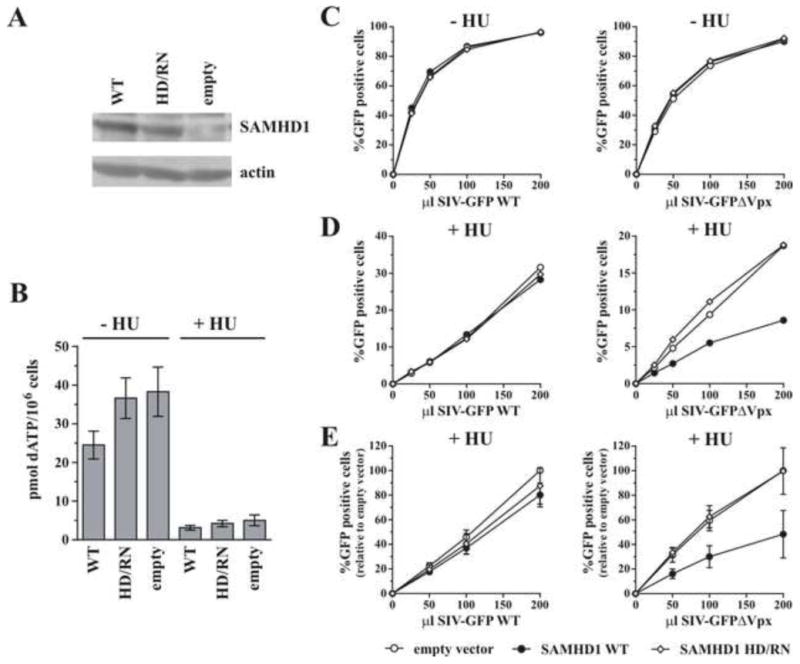
HeLa cells were transduced with lentiviral particles encoding either WT SAMHD1, the indicated SAMHD1 mutants, or an empty vector and selected with puromycin for 48 h. Cells were treated +/− 1mM hydroxyurea (HU) for 4 h prior to infection or dNTP isolation (in continued presence or absence of HU). A) Total cell extracts were separated by SDS-PAGE and subjected to immunoblotting for SAMHD1 and actin as indicated. B) Cellular dNTPs were isolated at time of infection and the amount of dATP present per million cells was determined using a polymerase-based assay. Data are presented as mean and standard deviation of quantitation from at least 3 independently generated cell lines. C, D & E) Cells were infected with increasing volumes of VSV-G pseudotyped SIV-GFP, with or without Vpx, and the percent infection (% GFP-positive cells) was determined by flow cytometry 24 h later. Results in panels A, C, and D show representative results from one of at least 3 independent experiments. Panel E represents the mean and standard deviation from 3 independent experiments performed as in panel D. The maximum amount of infection determined in the empty vector sample was defined as 100% for each experiment. All other data points were normalized accordingly.
No SAMHD1 nuclease activity is detected against single-stranded RNA
Several recent reports have suggested nuclease activity as an important SAMHD1 mechanism of action for viral restriction (Choi et al., 2015; Ryoo et al., 2014). To test for potential nuclease activity, SAMHD1 was isolated by immunoprecipitation from PMA-differentiated U937 cells that had been transduced with lentiviral particles encoding Flag-tagged WT SAMHD1, SAMHD1 HD/RN, or empty vector. Of note, the HD/RN mutant was included here because this dNTPase active site has been reported to be necessary for the SAMHD1 nuclease function (Beloglazova et al., 2013). Eluates containing immunoprecipitated SAMHD1 (Fig 4A) were incubated with a 32P-labeled 20-nucleotide single stranded RNA probe (Ryoo et al., 2014). Reaction products were separated on denaturing urea-PAGE and visualized by autoradiography. As shown in Figure 4B, no SAMHD1-associated nuclease activity was seen as no increase in degradation products of the radiolabeled probe was observed with the isolated WT SAMHD1 protein over the no protein (mock) control or eluates from empty vector or HD/RN mutant containing cells. In contrast, an RNase A exonuclease positive control quantitatively degraded the RNA probe. These data are consistent with a recent paper that also failed to detect SAMHD1 active-site associated nuclease activity (Seamon et al., 2015). Degradation of viral genomic RNA by SAMHD1 nuclease activity as a relevant mechanism of restriction is also hard to reconcile with a report that restriction can be relieved even when SAMHD1 degradation by Vpx is delayed until 24h post-infection (Hofmann et al., 2013).
Figure 4. SAMHD1 in vitro nuclease assay.
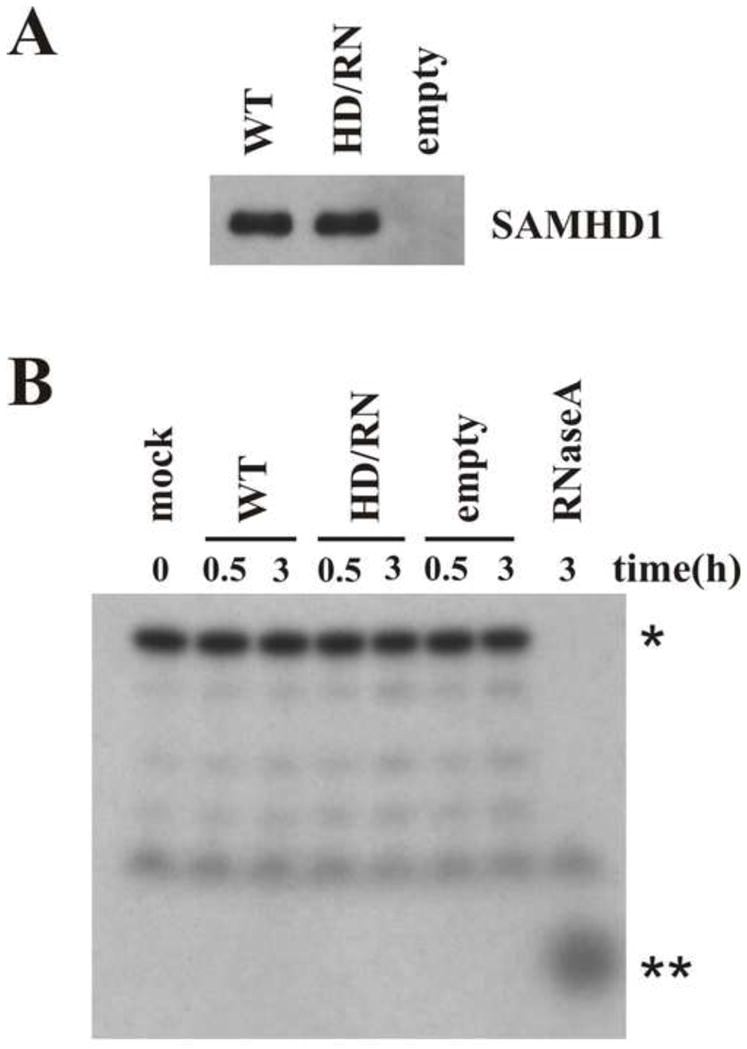
U937 cells were transduced with lentiviral particles encoding 3xFlag-SAMHD1 (WT or HD/RN) or an empty vector. After puromycin selection, cells were differentiated overnight with PMA and cell extracts were used for immunoprecipitation using Flag beads followed by elution with 3xFlag peptide. A) A portion of the eluate was separated by SDS-PAGE and subjected to immunoblotting with SAMHD1-specific antiserum. B) The remaining SAMHD1-containing eluate was incubated at 37°C with a 20 nucleotide RNA probe and the reaction products were separated on a 15% denaturing urea polyacrylamide gel as described in Materials and Methods. * indicates the full length undigested probe and ** indicates the migration of the products obtained after complete digestion with the RNaseA positive control.
CONCLUSIONS
Overall, the results presented here are consistent with low dNTP levels being necessary but not sufficient for full SAMHD1 restriction activity. While several recent studies have now shown phosphorylation to modulate SAMHD1 dNTPase activity in vitro (Arnold et al., 2015; Tang et al., 2015; Yan et al., 2015), this modification still allows residual activity and our data indicate that SAMHD1 T592E is still able to lower dATP levels in PMA-U937 cells similar to the WT protein without causing restriction. We were also unable to confirm SAMHD1 nuclease activity, leaving the question of what additional SAMHD1 activity is required for full restriction open for future investigation. Interestingly, the fact that hydroxyurea treatment in Figure 3D rendered HeLa cells susceptible only to wildtype SAMHD1 would argue against a myeloid-specific cofactor required for SAMHD1 restriction and would further suggest that the HD/RN dNTPase active site mutant has also lost its dNTP-independent function. Therefore, the identification of all factors involved in SAMHD1 restriction of lentiviruses, whether other co-factors are involved or whether different mechanisms of action are at play under different conditions or in different cell types, should remain a subject for future investigations.
MATERIALS AND METHODS
Cell culture and transfections
HeLa and 293TN cells were grown in Dulbecco’s modified Eagles medium (DMEM) containing 10% fetal bovine serum. U937 cells were maintained in RPMI 1640 media containing 10% fetal bovine serum. U937 cells were differentiated by overnight treatment with 10 ng/ml of phorbol-12-myristate-13-acetate (PMA, Sigma). Cells were then immediately infected, harvested for western blot analysis, or harvested for dNTP isolation and quantitation. For generation of pCDH-SAMHD1 lentiviral transduction particles, 293TN cells (Systems Biosciences) were transfected using LipofectAMINE PLUS™ (Invitrogen Corp. Calsbad CA) following the manufacturer’s recommendations. For each 25 cm2 flask, 0.67 μg of pCDH plasmid was used together with 6.7 μl of pPACKH-1 packaging mix (Systems Biosciences). Lentiviral particle-containing supernatants were collected after 48 h, clarified by filtration through a 0.45 μm filter and stored at −80°C. GFP-HIV-1, SIV-GFP-WT, and SIV-GFP-ΔVpx reporter viruses were produced from 293TN cells as described (e.g. (White et al., 2013a)).
Antibodies
A polyclonal antibody to human SAMHD1 (SAM416) was described previously (Welbourn et al., 2012). A polyclonal antibody to actin was purchased from Sigma-Aldrich, Inc. (St. Louis MO; Cat#A-5060) and used as a loading control.
Plasmids
The pCDH-SAMHD1 lentiviral expression constructs (WT, T592A, T592E) were described previously (Welbourn et al., 2013). The SAMHD1 active site mutant (H206R/D207N, HD/RN) was generated by quikchange mutagenesis as described for the other mutants (Welbourn et al., 2013). To generate N-terminally Flag-tagged lentiviral expression constructs, the SAMHD1 sequence (WT, HD/RN) was first PCR-amplified from pcDNA-SAMHD1 constructs using primers containing EcoRI and Xba1 restriction sites and inserted into p3xFlagCMV7.1 (Sigma-Aldrich, Inc., St. Louis MO). The full Flag-SAMHD1 sequence was then amplified by PCR using primers containing Bmt1 and BamH1 restriction sites and inserted into pCDH-CMV-MCS-EF1-puro (Systems Biosciences).
Immunoblotting
For immunoblot analysis of cell-associated proteins, whole cell lysates were prepared as follows: Cells were washed once with PBS, suspended in PBS and mixed with an equal volume of sample buffer (4% sodium dodecyl sulfate, 125 mM Tris-HCl, pH 6.8, 10% glycerol and 0.002% bromophenol blue). Proteins were solubilized by heating 10–15min at 95°C with occasional vortexing to shear cellular DNA. Cell lysates were subjected to SDS-PAGE; proteins were transferred to PVDF membranes and reacted with appropriate antibodies as described in the text. Membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (GE healthcare, Piscataway NJ) and proteins were visualized by enhanced chemiluminescence (ECL, GE healthcare, Piscataway NJ).
U937-based HIV-1 restriction assay
HIV-1 restriction assays were performed essentially as described (Welbourn et al., 2013; White et al., 2013a). Monocytic U937 cells were transduced with pCDH-SAMHD1 lentiviral particles and selected with 0.4 μg/ml puromycin for approximately one week. 6×104 cells were then differentiated overnight in a 24-well plate using 10ng/ml of PMA. The next day, the differentiated cells were washed and infected with increasing amounts of HIV-1-GFP as indicated in the text. Fourty-eight hours later, the percentage of infected cells was determined by flow cytometry for GFP. Cells were also seeded/differentiated in parallel and harvested for dNTP isolation at time of infection.
Infection of SAMHD1 expressing HeLa cells and hydroxyurea treatment
HeLa cells were transduced with pCDH-SAMHD1 lentiviral particles and selected with 3 μg/ml puromycin for 48 h. Cells were treated +/− 1 mM hydroxyurea (HU, Sigma) for 4 h prior to infection with increasing amounts of reporter virus or dNTP isolation (in continued presence or absence of HU). Twenty-four hours later, the percent infected cells was analyzed by flow cytometry for GFP.
dATP quantitation
dNTPs were isolated from cells essentially as described (Diamond et al., 2004). Cells were harvested, counted, washed twice in PBS and the cell pellet suspended in ice cold 65% methanol (100 μl/106 cells). The solution was vortexed for 2 min, boiled for 3min and then centrifuged for 6min at 16,000xg. The clarified supernatant was then evaporated to dryness. Pellets were suspended in H2O (100 μl/106 cells) and 2–5 μl extract used for quantitation. dATP levels were quantified using the method from Sherman and Fyfe (Sherman and Fyfe, 1989) as modified by Ferraro et al. (Ferraro et al., 2010) using 32P-dTTP as the probe. In brief, 2–5 μl of dNTP extract was incubated with 40 mM TrisHCl, pH 7.4, 10 mM MgCl2, 5 mM dithiothreitol, 0.25 μM oligonucleotide, 1.5 μg RNaseA, 0.25 μM labeled dTTP (α-32P dTTP, Perkin Elmer, diluted 1 in 30 with unlabeled dTTP) and 0.025 units Klenow polymerase for 1 h at 37°C. 15 μl of the reaction mixture was then spotted on DE81 paper (GE healthcare), washed 3 times 10 min with 5% Na2HPO4, once with H2O and once with absolute ethanol. The bound radioactivity was determined by scintillation counting. These conditions gave a linear standard curve from 0.125 pmol to 4 pmol dATP and the amount of dATP in the extract is expressed as pmol/106 cells.
Nuclease Assay
SAMHD1 proteins used in nuclease assays were isolated from U937 cells transduced to express 3xFlag-SAMHD1 (WT or HD/RN) or empty vector. Cells were differentiated overnight with 10 ng/ml PMA, lysed in 50 mM Tris pH 8.0, 150 mM NaCl, 1% Triton X-100 for 20 min at 4°C and clarified at 10,000xg for 10 min at 4°C. Cleared lysates were then incubated for 2 h with anti-Flag conjugated agarose beads (EZview Red ANTI-FLAG M2 affinity gel, Sigma-Aldrich) at 4°C. The samples were then washed twice with wash buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% Triton X-100) and twice with assay buffer (1x PBS, 2 mM DTT, 10% glycerol, 0.01% NP-40). Bound proteins were eluted from the beads using 200 ng/μl 3xFlag peptide (ApexBio) in assay buffer. Nuclease assays were performed using a 32P-labeled 20nt RNA probe as described in (Ryoo et al., 2014). Immunoprecipitated proteins and the probe were incubated at 37°C in assay buffer for up to 3 h. As a positive control, a sample was treated in parallel with RNaseA (40 μg/ml). Formamide loading buffer was added, the samples heated to 65°C for 5min and an aliquot of each sample was separated on a 15% denaturing urea polyacrylamide gel (SequaGel, National Diagnostics) prior to autoradiography.
SAMHD1 T592E mutant decreases dATP levels in cells without causing significant restriction.
Lack of SAMHD1 restriction activity in dividing cells with high dNTP levels
Lowering dNTP levels in HeLa cells reveals an additional antiviral effect of SAMHD1.
No SAMHD1 nuclease activity is detected against single-stranded RNA
Acknowledgments
We thank Sandra Kao, Eri Miyagi, Chia-Yen Chen, Haruka Yoshii-Kamiyama and Sayaka Sukegawa for helpful discussions and critical reading of the manuscript and Alisha Buckler-White and Ron Plishka for sequence analysis. This work was supported in part by the Intramural Research Program of the NIH, NIAID. SW was supported by a postdoctoral fellowship from the Canadian Institutes of Health Research (C.I.H.R) and an Intramural AIDS Research Fellowship from NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amie SM, Bambara RA, Kim B. GTP is the primary activator of the anti-HIV restriction factor SAMHD1. J Biol Chem. 2013;288:25001–25006. doi: 10.1074/jbc.C113.493619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold LH, Groom HC, Kunzelmann S, Schwefel D, Caswell SJ, Ordonez P, Mann MC, Rueschenbaum S, Goldstone DC, Pennell S, Howell SA, Stoye JP, Webb M, Taylor IA, Bishop KN. Phospho-dependent Regulation of SAMHD1 Oligomerisation Couples Catalysis and Restriction. PLoS Pathog. 2015;11:e1005194. doi: 10.1371/journal.ppat.1005194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendt R, Schumann T, Gerbaulet A, Nguyen LA, Schubert N, Alexopoulou D, Berka U, Lienenklaus S, Peschke K, Gibbert K, Wittmann S, Lindemann D, Weiss S, Dahl A, Naumann R, Dittmer U, Kim B, Mueller W, Gramberg T, Roers A. Mouse SAMHD1 has antiretroviral activity and suppresses a spontaneous cell-intrinsic antiviral response. Cell Rep. 2013;4:689–696. doi: 10.1016/j.celrep.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, Yakunin AF. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J Biol Chem. 2013;288:8101–8110. doi: 10.1074/jbc.M112.431148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Sommer AF, Zwarg J, Hamdorf M, Welzel K, Esly N, Panitz S, Reuter A, Ramos I, Jatiani A, Mulder LC, Fernandez-Sesma A, Rutsch F, Simon V, Konig R, Flory E. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7:e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Ryoo J, Oh C, Hwang S, Ahn K. SAMHD1 specifically restricts retroviruses through its RNase activity. Retrovirology. 2015;12:46. doi: 10.1186/s12977-015-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier A, Descours B, Valadao ALC, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by Cyclin A2/CDK1 Regulates Its Restriction Activity toward HIV-1. Cell Rep. 2013;3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Descours B, Cribier A, Chable-Bessia C, Ayinde D, Rice G, Crow Y, Yatim A, Schwartz O, Laguette N, Benkirane M. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4+ T-cells. Retrovirology. 2012;9:87–87. doi: 10.1186/1742-4690-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond TL, Roshal M, Jamburuthugoda VK, Reynolds HM, Merriam AR, Lee KY, Balakrishnan M, Bambara RA, Planelles V, Dewhurst S, Kim B. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J Biol Chem. 2004;279:51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussaix E, Lebon P, Ponsot G, Huault G, Tardieu M. Intrathecal synthesis of different alpha-interferons in patients with various neurological diseases. Acta Neurol Scand. 1985;71:504–509. doi: 10.1111/j.1600-0404.1985.tb03235.x. [DOI] [PubMed] [Google Scholar]
- Ferraro P, Franzolin E, Pontarin G, Reichard P, Bianchi V. Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 2010:38. doi: 10.1093/nar/gkp1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzolin E, Pontarin G, Rampazzo C, Miazzi C, Ferraro P, Palumbo E, Reichard P, Bianchi V. The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc Natl Acad Sci U S A. 2013;110:14272–14277. doi: 10.1073/pnas.1312033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- Goncalves A, Karayel E, Rice GI, Bennett KL, Crow YJ, Superti-Furga G, Burckstummer T. SAMHD1 is a nucleic-acid binding protein that is mislocalized due to aicardi-goutieres syndrome-associated mutations. Human mutation. 2012;33:1116–1122. doi: 10.1002/humu.22087. [DOI] [PubMed] [Google Scholar]
- Goujon C, Riviere L, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology. 2007;4:2–2. doi: 10.1186/1742-4690-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen EC, Seamon KJ, Cravens SL, Stivers JT. GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state. Proc Natl Acad Sci U S A. 2014;111:E1843–1851. doi: 10.1073/pnas.1401706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Norton TD, Schultz ML, Polsky SB, Sunseri N, Landau NR. Inhibition of CUL4A Neddylation causes a reversible block to SAMHD1-mediated restriction of HIV-1. J Virol. 2013;87:11741–11750. doi: 10.1128/JVI.02002-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbaugh JA, Gee P, Baker J, Daly MB, Amie SM, Tate J, Kasai N, Kanemura Y, Kim DH, Ward BM, Koyanagi Y, Kim B. Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathog. 2013;9:e1003481. doi: 10.1371/journal.ppat.1003481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Tang C, Zhao Q, Wang W, Xiong Y. Structural basis of cellular dNTP regulation by SAMHD1. Proc Natl Acad Sci U S A. 2014;111:E4305–4314. doi: 10.1073/pnas.1412289111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Wu Y, Yan J, Mehrens J, Yang H, DeLucia M, Hao C, Gronenborn AM, Skowronski J, Ahn J, Xiong Y. Mechanism of allosteric activation of SAMHD1 by dGTP. Nat Struct Mol Biol. 2013;20:1304–1309. doi: 10.1038/nsmb.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA. Tight Interplay among SAMHD1 Protein Level, Cellular dNTP Levels, and HIV-1 Proviral DNA Synthesis Kinetics in Human Primary Monocyte-derived Macrophages. J Biol Chem. 2012;287:21570–21574. doi: 10.1074/jbc.C112.374843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ET, White TE, Brandariz-Nunez A, Diaz-Griffero F, Weitzman MD. SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. J Virol. 2013;87:12949–12956. doi: 10.1128/JVI.02291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koharudin LM, Wu Y, DeLucia M, Mehrens J, Gronenborn AM, Ahn J. Structural basis of allosteric activation of sterile alpha motif and histidine-aspartate domain-containing protein 1 (SAMHD1) by nucleoside triphosphates. J Biol Chem. 2014;289:32617–32627. doi: 10.1074/jbc.M114.591958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nature immunology. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- Powell RD, Holland PJ, Hollis T, Perrino FW. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem. 2011;286:43596–43600. doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Maelfait J, Bridgeman A, Rigby R, Hayward B, Liberatore RA, Bieniasz PD, Towers GJ, Moita LF, Crow YJ, Bonthron DT, Reis E Sousa C. SAMHD1-dependent retroviral control and escape in mice. EMBO J. 2013;32:2454–2462. doi: 10.1038/emboj.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BCJ, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim SY, Seo D, Kim J, White TE, Brandariz-Nunez A, Diaz-Griffero F, Yun CH, Hollenbaugh JA, Kim B, Baek D, Ahn K. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med. 2014;20:936–941. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seamon KJ, Sun Z, Shlyakhtenko LS, Lyubchenko YL, Stivers JT. SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res. 2015;43:6486–6499. doi: 10.1093/nar/gkv633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova N, Wu Y, Zhu X, Stranska R, Kaushik R, Sharkey M, Stevenson M. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 2008;4:e1000057. doi: 10.1371/journal.ppat.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman PA, Fyfe JA. Enzymatic assay for deoxyribonucleoside triphosphates using synthetic oligonucleotides as template primers. Anal Biochem. 1989;180:222–226. doi: 10.1016/0003-2697(89)90420-x. [DOI] [PubMed] [Google Scholar]
- St Gelais C, de Silva S, Amie SM, Coleman CM, Hoy H, Hollenbaugh JA, Kim B, Wu L. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology. 2012;9:105–105. doi: 10.1186/1742-4690-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Gelais C, de Silva S, Hach JC, White TE, Diaz-Griffero F, Yount JS, Wu L. Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J Virol. 2014;88:5834–5844. doi: 10.1128/JVI.00155-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C, Ji X, Wu L, Xiong Y. Impaired dNTPase Activity of SAMHD1 by Phosphomimetic Mutation of T592. J Biol Chem. 2015 doi: 10.1074/jbc.M115.677435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tungler V, Staroske W, Kind B, Dobrick M, Kretschmer S, Schmidt F, Krug C, Lorenz M, Chara O, Schwille P, Lee-Kirsch MA. Single-stranded nucleic acids promote SAMHD1 complex formation. J Mol Med (Berl) 2013;91:759–770. doi: 10.1007/s00109-013-0995-3. [DOI] [PubMed] [Google Scholar]
- Welbourn S, Dutta SM, Semmes OJ, Strebel K. Restriction of Virus Infection but Not Catalytic dNTPase Activity Is Regulated by Phosphorylation of SAMHD1. J Virol. 2013;87:11516–11524. doi: 10.1128/JVI.01642-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welbourn S, Miyagi E, White TE, Diaz-Griffero F, Strebel K. Identification and characterization of naturally occurring splice variants of SAMHD1. Retrovirology. 2012;9:86–86. doi: 10.1186/1742-4690-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Carlos Valle-Casuso J, Amie S, Nguyen L, Kim B, Brojatsch J, Diaz-Griffero F. Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology. 2013a;436:81–90. doi: 10.1016/j.virol.2012.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. The Retroviral Restriction Ability of SAMHD1, but Not Its Deoxynucleotide Triphosphohydrolase Activity, Is Regulated by Phosphorylation. Cell Host Microbe. 2013b;13:441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Hao C, DeLucia M, Swanson S, Florens L, Washburn MP, Ahn J, Skowronski J. CyclinA2-Cyclin-dependent Kinase Regulates SAMHD1 Protein Phosphohydrolase Domain. J Biol Chem. 2015;290:13279–13292. doi: 10.1074/jbc.M115.646588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Gao W, Zhao K, Qin X, Zhang Y, Peng X, Zhang L, Dong Y, Zhang W, Li P, Wei W, Gong Y, Yu XF. Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nature communications. 2013;4:2722. doi: 10.1038/ncomms3722. [DOI] [PubMed] [Google Scholar]


