Abstract
The two unrelated miRNAs, miR-143 and miR-145, co-expressed from the miR-143/145 cluster have been proposed to act as tumor suppressors in human cancer and therapeutic benefits of delivering miR-143 and miR-145 to tumors have been reported. In contrast, we found that tumor-specific deletion of miR-143/145 in an autochthonous mouse model of lung adenocarcinoma did not affect tumor development. This was consistent with the lack of endogenous miR-143/145 expression in normal and transformed lung epithelium. Surprisingly, miR-143/145 in the tumor microenvironment dramatically promoted tumor growth by stimulating the proliferation of endothelial cells. Loss of miR-143/145 in vivo led to derepression of the miR-145 target Camk1d, an inhibitory kinase, which when overexpressed prevents mitotic entry of endothelial cells. As a consequence, tumors in miR-143/145-deficient animals exhibited diminished neoangiogenesis, increased apoptosis and their expansion was limited by the tumor’s ability to co-opt the alveolar vasculature. These findings demonstrate that stromal miR-143/145 promotes tumorigenesis and cautions against the use of these miRNAs as agents in cancer therapeutics.
Keywords: miRNAs, non-small cell lung adenocarcinoma, tumor microenvironment, angiogenesis
INTRODUCTION
MicroRNAs (miRNAs) influence diverse cellular processes through their ability to modulate gene expression programs and dysregulation of miRNA expression has been linked to various disease states, including cancer (1, 2). In particular, miRNA profiling of tumors demonstrated that miR-143 and miR-145, two unrelated miRNAs co-expressed from the miR-143/145 cluster, show reduced expression compared to adjacent normal tissue in a variety of human tumors of epithelial origin (3-8). Functional studies have revealed that ectopic expression of these miRNAs promotes apoptosis, inhibits proliferation, and limits the tumor-forming capacity of diverse tumor cell types, suggesting a potential role for miR-143 and miR-145 in cell autonomous tumor suppression (9-12). Additionally, several reports indicate a therapeutic benefit of miR-143 or miR-145 delivery in subcutaneous and orthotopic models of pancreatic cancer and in colorectal carcinoma xenografts (13-15). In these overexpression studies, the tumor suppressive functions of miR-143 appear to be mediated through its ability to target the 3′ untranslated regions (3′UTRs) of several oncogenes, including K-ras and Erk5, whereas miR-145 represses the expression of Myc, Yes, Fascin, and Stat, among others (9, 11, 12, 16-18). Despite the existence of several independently generated miR-143/145 knockout mouse model, however, the tumor suppressive functions of endogenous miR-143 and miR-145 have not been tested in vivo (19-22).
Recent reports revealing the role of endogenous miR-143/145 in intestinal epithelial regeneration, pulmonary fibrosis, and skin wound healing have challenged the cell autonomous model for miR-143/145 function (23-25). These studies show that miR-143 and miR-145 are not expressed in cells of epithelial origin. Instead, upregulation of miR-143 and miR-145 in stress-activated myofibroblasts stimulates epithelial regeneration via paracrine mechanisms. To address the possibility that expression of the miR-143/145 cluster in the stroma may play an analogous, stimulatory role during the process of tumorigenesis, we have utilized the KrasLSL-G12D/+ (K) autochthonous mouse model of lung adenocarcinoma, a type of cancer, in which downregulation of the miR-143/145 cluster has been reported. Tumorigenesis in the K model is initiated in a subset of adult lung epithelial cells via intratracheal infection with viral Cre recombinase, which activates mutant KrasG12D from the endogenous locus (26), thus preserving its 3′UTR and the potential for regulatory interactions with miRNAs, including miR-143. Concomitant deletion of p53 in KrasLSL-G12D/+;p53F/F (KP) mice promotes the development of lung adenocarcinomas that closely recapitulate the pathophysiological features of the human disease (27).
Here, we demonstrate that the miR-143/145 cluster is not expressed in normal and transformed lung epithelium and does not play a cell-autonomous tumor suppressor role in lung tumorigenesis. Instead, we find that stromal miR-143/145 promotes tumor development in K mice. We report expression of these miRNAs in a subset of lung endothelial cells and identify a novel role for the miR-143/145 cluster in supporting tumor neoangiogenesis. This study highlights the utility of using autochthonous mouse models of cancer to understand the complex involvement of tumor stroma in cancer development.
RESULTS
Dissecting the proposed tumor suppressive functions of the miR-143/145 cluster using genetically engineered mouse models
To investigate the proposed tumor suppressive functions of miR-143 and miR-145 in vivo, we generated a conditional knockout for the miR-143/145 cluster (Supplementary Fig. S1A, B). miR-143/145-floxed (F) and null (−) alleles were obtained by crossing founders to FlpO or Deleter Cre transgenic animals, respectively (Supplementary Fig. S1A, C) (28, 29). Loss of mature miRNA expression in these animals was confirmed by northern blotting and miRNA Taqman assay (Supplementary Fig. S1D, E). As previously reported, mice lacking miR-143 and miR-145 were viable, born at Mendelian ratios, and exhibited predominantly age-related, smooth muscle-specific phenotypes, including loss of elasticity in the aorta, megacolon manifestation, and male infertility (Supplementary Fig. S1F, G) (19-22). Studies on ageing cohorts of littermate mice revealed that loss of expression of miR-143/145 alone or in the context of a sensitized p53 heterozygous background did not predispose to tumor development (Supplementary Fig. S1H-J) (30). Moreover, primary mouse embryonic fibroblasts (MEFs) lacking miR-143/145 did not exhibit increased proliferation rates (Supplementary Fig. 2SA, B). Finally, loss of these miRNAs failed to cooperate with oncogenic H-ras, c-Myc, or E1A in promoting oncogenic transformation of MEFs, assayed by the ability to form colonies at low densities, loss of contact inhibition, and anchorage-independent growth (data not shown). Together, these studies indicated that miR-143 and miR-145 did not appear to act as classical tumor suppressor genes.
Investigating the role of the miR-143/145 cluster in lung adenocarcinoma
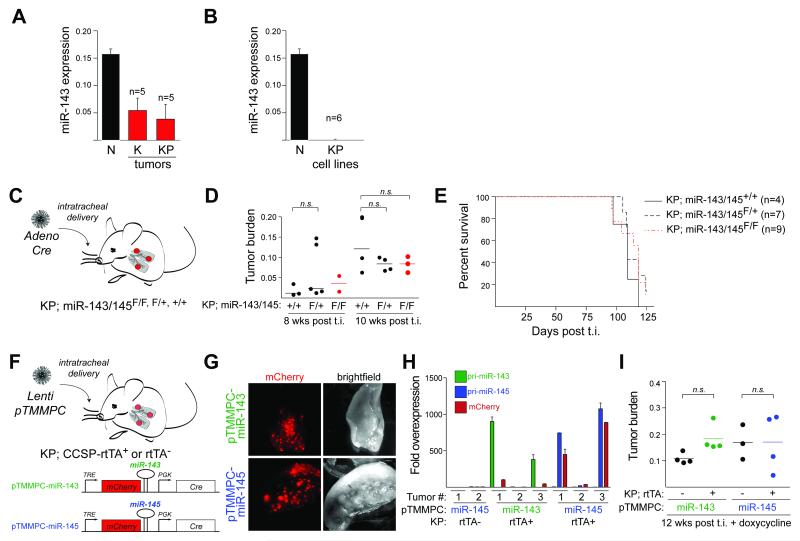
Next, we investigated the role of the miR-143/145 cluster in the KP mouse model of lung adenocarcinoma, a type of cancer, in which tumor suppressor activities of these miRNAs have been reported. Consistent with the human data (8), we found by quantitative RT-PCR (qRT-PCR) that miR-143 and miR-145 levels were significantly reduced in tumor tissue and tumor-derived cell lines compared to normal lung (Fig. 1A, B). To test directly whether loss of miR-143 and miR-145 affected tumorigenesis, we infected cohorts of KP;miR-143/145F/F, F/+ and +/+ mice with Cre recombinase to initiate lung tumors and induce concomitant tumor-specific deletion of miR-143 and miR-145 (Fig. 1C). Contrary to expectations, we found that mutation of the miR-143/145 cluster in KP tumors did not affect tumor burden at 8 and 10 weeks post tumor initiation and did not have a significant impact on overall survival (Fig. 1D, E). We concluded that loss of endogenous miR-143 and miR-145 expression did not promote lung adenocarcinoma development.
Figure 1. Lack of cell-autonomous role for the miR-143/145 cluster in lung adenocarcinoma.
(A, B) Taqman analysis of mature miR-143 expression in normal lung (N), (A) dissected KrasG12D/+ (K) and KrasG12D/+;p53−/− (KP) lung tumors or (B) cell lines derived from KrasG12D/+;p53−/− (KP) lung tumors. Data show mean±SEM.
(C) Schematic of the experimental design, showing the method for tumor initiation (t.i.) via intratracheal infection with adenoviral Cre recombinase (Adeno Cre).
(D) Quantification of the tumor burden of mice in (C). Mean and p-values using unpaired t-test are indicated (n=2-5 mice).
(E) Survival curve of mice in (C) (n=4-9 mice).
(F) Schematic of the experimental design to test the effects of miR-143/145 overexpression.
(G) Fluorescence detection of mCherry and brightfield microscopy of lungs.
(H) qRT-PCR analysis of individual tumors dissected at 12 weeks post t.i.. Data show mean±SEM of technical replicates.
(I) Quantification of tumor burden of mice in (F). Mean and p-values using unpaired t-test are indicated (n=3-4 mice).
Given the apparent controversy with published data, we reasoned that these miRNAs may exhibit tumor suppressive activities in the context of forced overexpression through their ability to target critical oncogenes, as previously reported. To test this, we cloned miR-143 and miR-145 into the 3′UTR of mCherry in the pTMMPC lentiviral vector, which drives doxycycline-regulated induction of mCherry and concomitant miRNA expression (Fig. 1F) (31). This bifunctional pTMMPC vector also constitutively expresses Cre recombinase, required to initiate tumorigenesis in this model. We generated pTMMPC-miR-143 and pTMMPC-miR-145 lentivirus and infected cohorts of KP;CCSP-rtTA+ or control CCSP-rtTA− mice (Fig.1F) (32). All mice were fed doxycycline diet at tumor initiation. At 12 weeks post tumor initiation, we confirmed transgene expression by mCherry fluorescence in whole lungs and qRT-PCR of individual tumors (Fig. 1G, H). Low levels of miR-143 and miR-145 observed in two of the KP;CCSP-rtTA+ tumors likely reflected repressive features of the lentiviral integration sites or were a consequence of loss of activity from the CCSP promoter. Despite evidence that the majority of KP;CCSP-rtTA+ tumors expressed high levels of either miR-143 or miR-145, there was no significant difference in overall tumor burden between rtTA+ and rtTA− cohorts, indicating that forced miRNA expression did not influence tumor development in this model (Fig. 1I). Similarly, exogenous introduction of miR-143 and miR-145, individually or in combination, did not affect the viability or alter the proliferation rates in KP cell lines (Supplementary Fig. S2C, D). Finally, expression of miR-143/145 at endogenous levels did not appear to target K-ras or RREB1, two previously proposed targets relevant to the tumor suppressive functions of miR-143/145 (Supplementary Fig. S2E, F) (11). In summary, these data did not support previous studies, which have reported significant cell autonomous tumor suppressive activities of miR-143 and miR-145 in tumor cell lines in vitro and in vivo.
Expression of miR-143/145 in tumor stroma promotes lung tumorigenesis
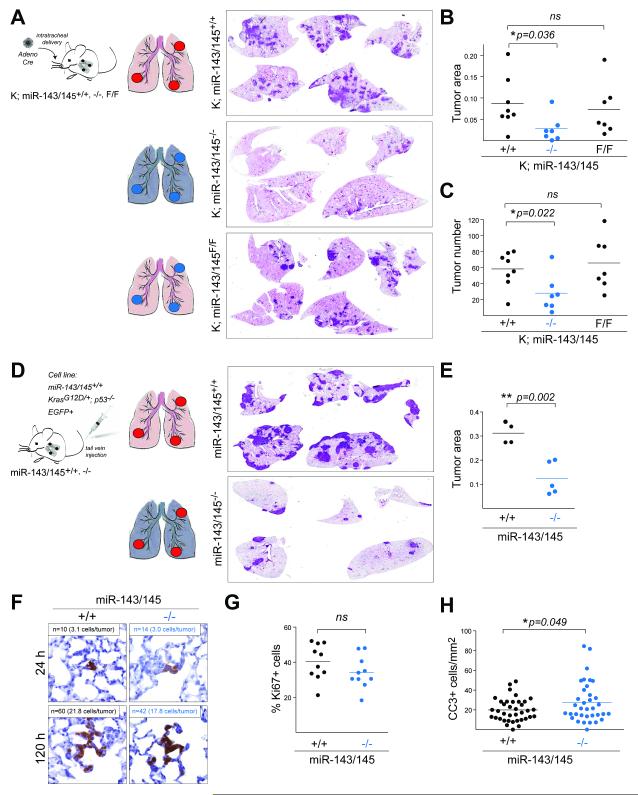
Since the miR-143/145 cluster has been shown to promote epithelial regeneration through a non-cell autonomous mechanism, we examined the role of stromal miR-143 and miR-145 in the context of lung cancer development (20). To this end, we initiated tumorigenesis using adenoviral Cre recombinase in cohorts of K;miR-143/145+/+, F/F, and −/− mice and assayed tumor burden at 12 weeks (Fig. 2A). Consistent with the data shown in Fig. 1D, the tumor burden of K;miR-143/145F/F animals, which have tumor-specific miRNA deletion, was indistinguishable from control animals (Fig. 2A-C). Surprisingly, however, histological analysis of K;miR-143/145−/− animals, which have organism-wide deletion of the cluster, revealed a dramatically reduced tumor burden (Fig. 2A). There was a two-fold decrease in tumor area and a three-fold reduction in the number of tumors per section (Fig. 2B, C). These data indicated that systemic miR-143/145 expression played an unanticipated role in promoting tumor development.
Figure 2. Stromal miR-143/145 promotes lung tumorigenesis.
(A) (Left) Experimental setup. Color coded schematic of the status of the miR-143/145 locus in tumor cells vs lung stroma (miR-143/145+/+: red; miR-143/145−/−: blue). (Right) H&E staining of lung sections at 12 weeks post t.i..
(B-C) Quantification of (B) tumor area and (C) tumor number of experiment described in (A). Mean and p-values using unpaired t-test are indicated (n=7-8 mice).
(D) (Left) Experimental setup. Color coding as in (A). (Right) H&E staining of lung sections at 1 month post t.v.i.
(E) Quantification of tumor area of mice in (D). Mean and p-value using unpaired t-test are indicated (n=4-5 mice).
(F) IHC detection of EGFP-positive cells at indicated time-points post t.v.i. of mice in (D). Number of tumor nodules per section (n) and the number of EGFP-positive cells per tumor nodule are indicated.
(G) Fraction of Ki67-positive cells in lung tumors from mice in (D). Mean and p-value using unpaired t-test are indicated (n=10 tumors).
(H) Quantification of cleaved caspase 3 (CC3)-positive cells per tumor area of lung sections from mice in (D). Mean and p-value using unpaired t-test are indicated (n>35 tumors).
Similar reduction of tumor burden in the lung was observed when a KP;miR-143/145+/+;EGFP+ cell line was introduced via tail vein injection (t.v.i.) in congenic miR-143/145+/+ or miR-143/145−/− animals (Fig. 2D). In this transplant model, tumor cells preferentially colonize the lungs, where they develop aggressive lung adenocarcinomas with short latency. Histological analysis at 4 weeks post t.v.i. revealed that formation of tumor nodules in the lungs of miR-143/145-deficient animals was significantly impaired relative to wild-type, littermate controls (Fig. 2D, E). Time-course experiments, combined with immunohistochemistry (IHC) detection of tumor cells using the EGFP marker, indicated that absence of miR-143/145 did not affect the colonization of the lungs (quantified as tumor nodules/section) and only moderately limited the initial expansion of the tumor nodules (quantified by number of cells/tumor nodule) (Fig. 2F). Surprisingly, even at late timepoints, when the tumor burden was significantly lower in knockout animals, there was no apparent difference in proliferation rates (Fig. 2G). Instead, there was a small but significant increase in the incidence of apoptosis in tumors developing in miR-143/145-deficient animals, indicating that systemic loss of these miRNAs imposed stress that limited tumor expansion (Fig. 2H).
Together, these experiments provided genetic evidence that systemic expression of miR-143 and/or miR-145 supported tumor cell survival in the lung through a non-cell autonomous mechanism, pointing to a potential role of these miRNAs in the tumor microenvironment. This was specific to the lung as the rate of subcutaneous growth of the KP;miR-143/145+/+;EGFP+ cell line was indistinguishable between miR-143/145-proficient and –deficient animals, indicating functional differences between the stroma of the lung and the tumor microenvironment at subcutaneous transplant sites (Supplementary Fig. S3A, B).
In an effort to elucidate the underlying mechanism, we performed IHC analysis of the infiltration pattern and morphological characteristics of tumor-associated cell types but failed to detect significant miRNA-dependent differences (Supplementary Fig. S4A, B). In addition, immunofluorescence (IF) analysis of lung tumors showed no apparent differences in IGF signaling, assayed by the pattern and intensity of pIGFR-Y116 staining, contrasting a previously proposed model for the contribution of fibroblast-derived miR-143/145 expression to epithelial regeneration in the colon (Supplementary Fig. S4C) (20). We hypothesized that miR-143 and miR-145 promoted lung tumor development through a novel mechanism.
Rare endothelial cells in the lung stroma express miR-143/145
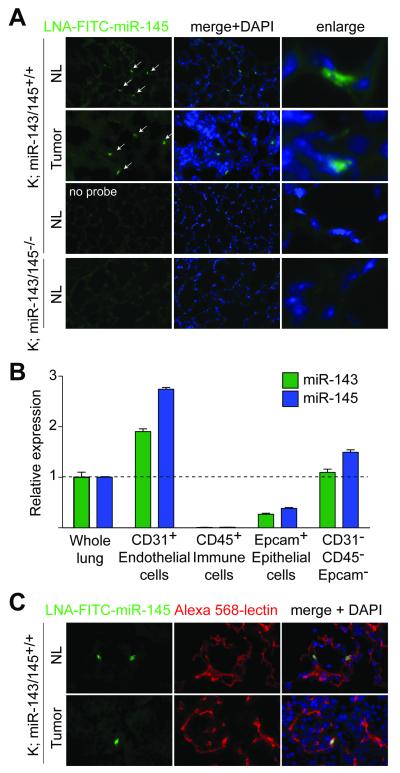
To identify the stromal compartments that expressed miR-143/145, we performed miRNA fluorescence in situ hybridization (FISH) in sections of normal and tumor-bearing lungs. As a positive control, we confirmed that miR-143 and miR-145 were abundant in the submucosa and muscularis mucosa layers in the colon and found them highly expressed in the smooth muscle lining of bronchioles and arteries in the lung (Supplementary Fig. S5A) (28, 29). The lung epithelium appeared largely devoid of miRNA signal with the exception of rare miRNA positive cells found in the alveolar walls as well as scattered throughout KP tumors (Fig. 3A). The staining was specific to miR-145 as there was no signal in sections where the probes were omitted or in miR-143/145-deficient tissues (Fig. 3A).
Figure 3. Pattern of expression of the miR-143/145 cluster in normal and tumor-bearing lungs.
(A) miRNA-FISH analysis of miR-145 expression pattern in normal (NL) or tumor-bearing lungs. DNA was counterstained with DAPI. No probe as negative control.
(B) Taqman analysis of mature miR-143 and miR-145 expression levels in sorted cell populations from K; miR-143/145+/+ lungs 16 weeks post t.i. Data show mean±SEM of technical replicates and were confirmed in a biological replicate.
(C) miRNA-FISH analysis of miR-145 and fluorescence detection of lectin in NL and tumor-bearing lungs in K; miR-143/145+/+ mice, sacrificed 16 weeks post t.i. DNA was counterstained with DAPI.
To determine the identity of the miR-143/145-expressing cells, we performed fluorescence-activated cell sorting (FACS) of dissociated lungs using cell surface markers to isolate the prevalent cell types. This strategy allowed us to evaluate miRNA levels in endothelial cells (CD31+, also known as PECAM1), immune cells (CD45+, also known as PTPRC), epithelial cells (Epcam+), and in the triple-negative fraction (CD31-CD45−Epcam−), thought to include smooth muscle cells, fibroblasts, pericytes, and activated neutrophils (Supplementary Fig. S5B, C). Taqman probes specific to the mature miR-143 and miR-145 sequences revealed that, relative to total lung, this cluster was depleted in immune and epithelial cells and enriched in endothelial cells and the triple-negative fraction (Fig. 3B). miRNA-FISH detection of miR-145 in normal lung and lung tumors showed close association of miRNA-positive cells with the vasculature, marked by fluorescently-labeled lectin, further pointing to endothelial identity (Fig. 3C). We concluded that the epithelial compartment of the lung was devoid of miR-143 and miR-145 expression, providing an explanation for the lack of tumor suppressive activities of these miRNAs during lung adenocarcinoma development. Instead, we observed that endothelial cells in vivo are enriched for the miR-143/145 cluster.
Reduced endothelial proliferation in miR-143/145-deficient mice
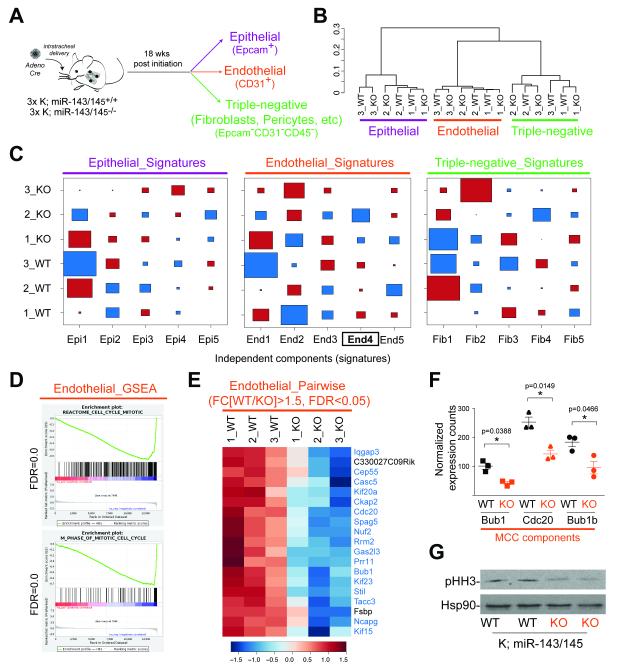
To elucidate the pathway that promoted tumor development downstream of miR-143/145, we performed global gene expression analysis in sorted epithelial (Epcam+), endothelial (CD31+) and triple-negative (CD31−CD45−Epcam−) cell populations, isolated 18 weeks post tumor initiation from three K;miR-143/145−/− mice and three K;miR-143/145+/+ littermate controls (Fig. 4A). Unsupervised clustering of RNA sequencing (RNA-seq) data segregated the three fractions as distinct populations (Fig. 4B). Independent Component Analysis (ICA) elucidated in an unbiased manner five statistically significant signatures within the dataset, establishing the level of purity and specificity of each sorted fraction (Supplementary Fig. S5D, E; Supplementary Table 1) (Bhutkar, A. et al, in preparation).
Figure 4. Mitotic defect in miR-143/145-deficient tumor endothelium.
(A) Schematic of the sorting strategy
(B) Unsupervised clustering of RNA-seq of samples in (A).
(C) Hinton plot of ICA of samples from (A). Columns represent the expression pattern of each signature. Red squares represent positive correlation and blue represents negative correlation with the signature; the size of the square reflects the degree of correlation. Statistically significant signature highlighted in bold.
(D) GSEA of the top 2%-ranked genes in Signature_End4 from (C) showing mitotic signature (FDR=0.0).
(E) Heatmap of relative expression of differentially expressed genes from pair-wise comparison; mitotic factors highlighted in blue.
(F) Normalized expression counts of three mitotic checkpoint components (MCC) Bub1, Cdc20, and Bub1b in experiment described in (A). Mean±SEM (n=3 mice) and p-values using Mann-Whitney test are shown.
(G) Immunoblot analysis of pHH3-Ser10 in whole cell lysates of CD31+ endothelial cells sorted from tumor-bearing lungs of indicated genotypes as described in (A). Lanes represent samples collected from independent animals. Hsp90 was used as a loading control.
Next, we investigated the differences between miR-143/145-deficient and – proficient samples. ICA did not detect biologically relevant differences between wild-type and null epithelial and triple-negative populations but highlighted one statistically significant signature that distinguished miR-143/145−/− endothelial samples from controls (Signature End4, p<0.05, Fig. 4C, Supplementary Table 2). GSEA of the top ranked genes within this signature identified gene sets related to the mitotic cell cycle and revealed a strong depletion of mitotic genes in miR-143/145-deficient endothelial cells compared to wild-type controls (Fig. 4D). This finding was supported by pair-wise comparison of differentially expressed genes between knockout and wild-type endothelial samples, which showed that 17 out of the 19 significantly repressed genes (FC[KO/WT]<0.67, p<0.05) were either mediators of mitosis or factors, whose levels peak in M-phase of the cell cycle (Fig. 4E and Supplementary Table 3 and 4). Genes showing consistently reduced levels in miR-143/145-deficient endothelial samples compared to controls included three essential components of the mitotic checkpoint complex (Bub1, Bub1b, and Cdc20) as well as the well-established marker of proliferation Ki67 (Fig. 4F and Supplementary Table 3). Finally, immunoblot analysis of CD31+ endothelial cells isolated from tumor-bearing lungs of knockout animals showed diminished levels of the mitotic marker, phosphorylated histone H3 (pHH3-Ser10), compared to wild-type controls (Fig. 4G). Collectively, these data pointed to a mitotic defect in the endothelium of tumor-bearing miR-143/145-deficient lungs and suggested that loss of these miRNAs may impair the process of tumor angiogenesis.
The miR-143/145 cluster promotes tumor neoangiogenesis
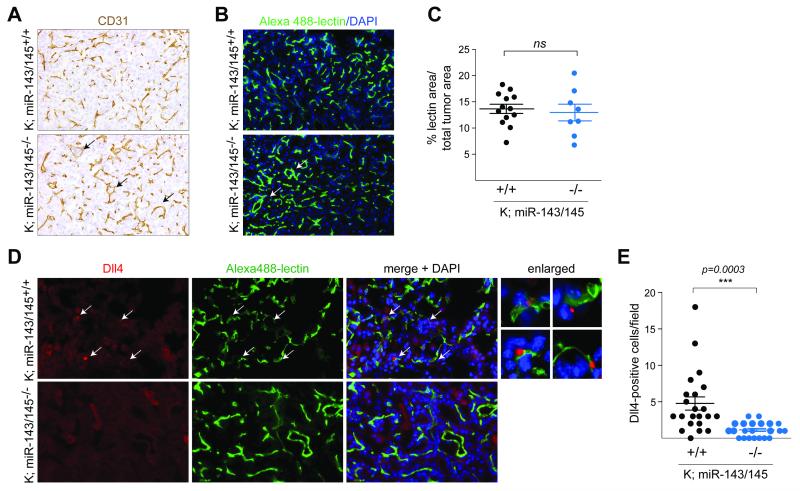
Based on the reduction of the mitotic index, we expected that the tumor vascularization in K;miR-143/145−/− animals would be grossly impaired. However, quantification of the vascular area using the endothelial marker CD31 or fluorescently labeled lectin did not reveal a miR-143/145-dependent difference (Fig. 5A-C). Instead, we noted honeycomb structures in K;miR-143/145−/− tumors, reminiscent of the normal alveolar architecture, whereas tumor vessels in K;miR-143/145+/+ mice appeared short and stubby (Fig. 5A, B, arrows). This pattern suggested that tumors in null animals may have expanded by co-opting the normal lung vasculature, rather than by sprouting new vessels (33). In agreement with this model, we observed reduced numbers of Dll4-positive tip cells, a marker of vessel sprouting and active neoangiogenesis in K;miR-143/145−/− animals compared to control K;miR-143/145+/+ mice (Fig. 5D, E; 4.4-fold reduction) (34). We propose that impaired activation of the angiogenic switch is the mechanism by which loss of miR-143/145 expression inhibits tumor development.
Figure 5. The miR-143/145 cluster promotes tumor neoangiogenesis.
(A) CD31 IHC staining in lung tumors at 12 weeks post t.i. Arrows highlight honeycomb structures.
(B) Fluorescence detection of lectin in lung tumors at 12 weeks post t.i. DNA was counterstained with DAPI. Arrows highlight honeycomb structures.
(C) Quantification of lectin in samples from (B). Mean±SEM and p-value using unpaired t-test are indicated.
(D) IF analysis of Dll4 staining and fluorescence detection of lectin in mice from (B). DNA was counterstained with DAPI. Arrows in merged images and enlarged panels indicate Dll4-positive tip cells that are closely associated with the vasculature.
(E) Quantification of Dll4-positive cells of images in (D). Mean±SEM and p-value using unpaired t-test are indicated.
Identification of relevant miRNA targets
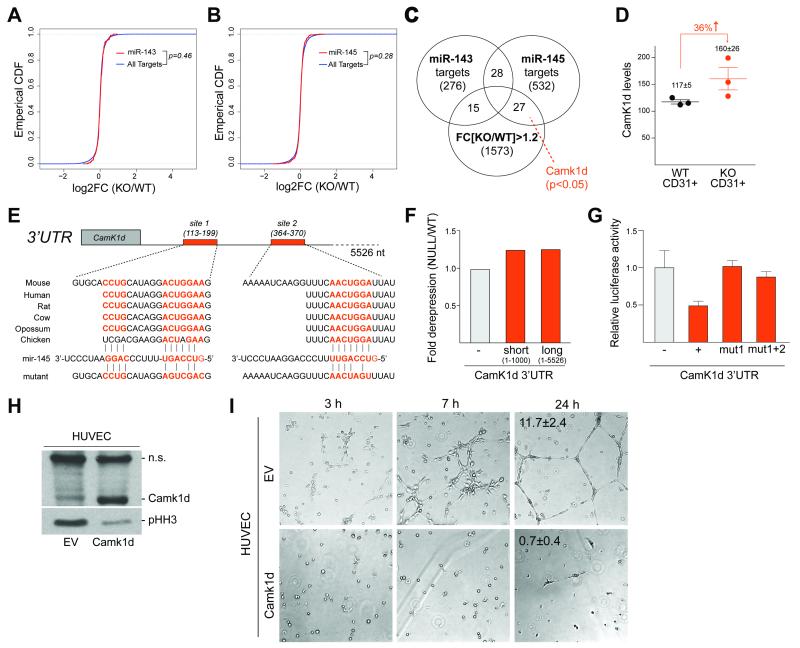
Finally, we addressed the mechanism by which miR-143/145 regulated mitotic entry in endothelial cells. To identify biologically relevant miRNA targets, we analyzed the endothelial cells expression data focusing on conserved miR-143 and miR-145 targets predicted by Targetscan (35). Using several independent approaches, we did not detect global patterns that distinguished between wild-type and null endothelial samples (Fig. 6A, B and data not shown). However, at the gene level, we noted that the conserved miR-145 target, Camk1d (Ca2+/calmodulin kinase 1d) was consistently upregulated in null samples compared to wild-type controls (Fig. 6C). Camk1d showed a 36% increase in expression in null samples relative to wild-type controls (p<0.05, Fig. 6D and Supplementary Table 3). Camk1d is a poorly characterized member of the Ca2+/calmodulin-dependent family of serine/threonine kinases, which have been implicated in the regulation of a wide range of cellular processes, including control of cellular proliferation and angiogenesis (36, 37). Notably, the 3′UTR of Camk1d contains two conserved miR-145 sites. We confirmed that Camk1d is a direct target of miR-145 using two independent approaches. Loss of endogenous miR-145 led to derepression of luciferase activity when the luciferase gene was cloned prior to the 3′UTR of Camk1d (Fig. 6E, F). In addition, site-directed mutagenesis of one or both miR-145 sites within the 3′UTR of Camk1d abolished repression of luciferase activity (Fig. 6E, G).
Figure 6. Derepression of the miR-145 target, Camk1d, impairs angiogenesis.
(A-B) CDF plots of (A) miR-143 and (B) miR-145 target expression fold change in miR-143/145-deficient endothelial cells relative to wild-type control in RNA-seq from Fig. 4A.
(C) Venn diagram of the overlap of conserved miR-143 targets, conserved miR-145 targets, and genes, derepressed in miR-143/145−/− endothelial samples relative to miR-143/145+/+ controls (mean FC[KO/WT]>1.2). Only Camk1d showed significant differential expression (p<0.05). p-values determined using Krushal Wallis test.
(D) Normalized expression counts of Camk1d in RNA-seq of sorted CD31+ endothelial cells isolated from tumor-bearing mice in Fig. 4A. Data show mean±SEM (n=3 mice).
(E) Schematic of the sequence, evolutionary conservation, and mutational analysis of the two miR-145 target sites in the 3′UTR of Camk1d.
(F) Bar graph of luciferase reporter assay in miR-143/145-deficient cells relative to wild-type controls transfected with no (−), short (1-1000 nt ) or long (full-length 1-5526) Camk1d 3′UTR constructs.
(G) Bar graph of luciferase reporter assay in miR-143/145-proficient cells transfected with constructs expressing luciferase with no 3′UTR (−), wild-type Camk1d 3′UTR (+), mutant Camk1d 3′UTR at site 1 (mut1) or mutant 3′UTR at sites 1 and 2 (mut1+2).
(H) Immunoblot analysis of Camk1d and pHH3 levels in HUVECs, infected with empty vector (EV) or human Camk1d lentiviral construct. Non-specific band on the Camk1d immunoblot was used as a loading control.
(I) Brightfield microscopy at indicated time-points of tube formation assay with HUVECs infected with EV or human Camk1d lentiviral construct. Numbers indicate tubes per image field and show mean±SEM (n=5, unpaired t-test p=0.0059).
Finally, we tested whether increased expression of Camk1d affected mitotic progression in endothelial cells using lentiviral delivery of Camk1d in human umbilical vein endothelial cells (HUVECs). Camk1d overexpression led to diminished levels of pHH3-Ser10, recapitulating the in vivo phenotype of miR-143/145 deficiency (Fig. 6H). In addition, ectopic Camk1d dramatically impaired the ability of HUVECs to form capillary-like structures in a tube formation assay, an approach used to model the process of angiogenesis in vitro (Fig. 6I). Together, these results pointed to Camk1d as a potential mediator of the mitotic defects observed in miR-143/145-deficient endothelial cells. However, we cannot exclude that derepression of additional miR-143 and miR-145 targets may contribute to the observed phenotype.
DISCUSSION
Our in vivo analysis of miR-143/145 function in a mouse model of lung adenocarcinoma provides genetic evidence that stromal miR-143 and miR-145 play a role in promoting tumor development. This conclusion contrasts with a large number of previous reports, which have implicated these miRNAs in cell autonomous tumor suppression. The basis for the majority of these studies has been the observation that expression of the miR-143/145 cluster is reduced in tumor tissues and tumor-derived cell lines compared to normal samples. Here, we provide evidence that the reported differential expression is not likely due to effects in the transformed cells per se but reflects the cell-type specificity of miR-143/145 expression and can be attributed to the cellular composition of the samples tested. Using two independent approaches in vivo, we demonstrate low levels of mature miR-143 and miR-145 in epithelial cells and higher levels in smooth muscle cells, fibroblasts, and endothelial cells. These data suggest that the apparent reduction of miR-143/145 expression in tumors can be explained by their largely epithelial composition (20). These findings also shed light on why tumor-specific depletion of these miRNAs did not affect tumorigenesis in the K and KP models.
In addition to clarifying the cell type specificity of expression, several lines of evidence presented here challenge the view of miR-143 and miR-145 as universal tumor suppressors. We observed lack of predisposition of miR-143/145 knockout mice to tumor development, no effect of miR-143/145 loss on the proliferation rate or transformation capacity in MEFs, and no tumor suppressive consequences of miR-143 and miR-145 overexpression in tumor cells in vitro and in vivo. Collectively, these data suggest that the previously reported tumor suppressive activities of miR-143 and miR-145 are likely to be context-specific.
Instead, we present evidence that stromal miR-143/145 plays a pro-tumorigenic role in vivo and promotes lung adenocarcinoma progression. This was specific to the lung tumor microenvironment but not at sites of subcutaneous transplants, revealing organ-specific requirements. The unexpected ability of miR-143/145 to support tumor growth is consistent with a previous study, which examined the role of the cluster in the context of epithelial regeneration following colonic injury (20). Our analysis further revealed a novel lung-specific mechanism by which stromal miR-143/145 supports tumor growth through the stimulation of mitotic progression of endothelial cells. We propose that a failure to activate the angiogenic switch in miR-143/145-deficient mice leads to increased apoptotic rates and limited tumor expansion. A similar function in promoting pathological angiogenesis has previously been described for a number of miRNAs, including miR-132 and miR-93 (38, 39).
Interestingly, deletion of the miR-143/145 cluster did not lead to a global derepression of miR-143 or miR-145 targets in endothelial cells. Of note, miR-143 has two additional family members expressed in the mouse genome, miR-1721 and miR-4770, which may provide redundancy in the regulation of miR-143 targets. In addition, we imagine that the small changes in gene expression induced by miRNA deficiency, while physiologically relevant, may be difficult to detect in vivo. Alternatively, lack of an apparent miRNA signature may be explained by the observation that only a subset of endothelial cells express miR-143/145 and thus expression analysis of bulk endothelial populations may not allow for target identification. Detection of miR-143/145 expression in a small subset of endothelial cells could be explained by transcriptional control by factors such as KLF2, a known mediator of endothelial activation (40) or by their transfer from smooth muscle cells to endothelial cells, as recently shown in vitro (41). In the context of these in vivo limitations, we identified only one miR-145 target, Camk1d as significantly derepressed in the miR-143/145-deficient endothelium. The targets of Camk1d in mammalian cells have not been identified and it remains to be determined how Camk1d overexpression impairs tumor neoangiogenesis in vivo. Interestingly, a recent study in S. pombe reported a role for the yeast homolog of this kinase in the phosphorylation and inhibition of Cdc25 (42). This provides a potential explanation for the mitotic defect observed in miR-143/145-deficient endothelial cells, as in mammalian cells the Cdc25c phosphatase mediates the critical step that drives progression into mitosis. In support of this model, exogenous overexpression of Camk1d in HUVECs resulted in defective mitosis and reduced tube formation in an angiogenesis assay in vitro. These findings implicate miR-145 as a regulator of neoangiogenesis at least in part through the repression of a kinase in control of a key regulatory signal transduction pathway.
In summary, this study elucidates a non cell-autonomous requirement for miR-143/145 expression in lung cancer development and identifies a novel role for these miRNAs as key modulators of endothelial function in cancer biology. This work highlights the power of using defined genetic systems and autochthonous mouse models of cancer to dissect the in vivo contribution of stroma-specific factors to tumor progression. The model presented here has general implications for the role of cell-type specific miRNAs in influencing critical steps in tumorigenesis and the potential to harness these regulatory networks for therapeutic purposes.
MATERIALS AND METHODS
Mouse strains and treatments
All animal work was conducted in accordance with a protocol approved by the MIT Committee on Animal Care. The miR-143/145 targeting construct was electroporated into V6.5 ES cells. Correctly targeted clones were used to generate miR-143/145targ/+ mice by tetraploid complementation and mice were back-crossed to pure C57BL/6 background using Taconic Speed Congenics. KrasLSL-G12D/+, p53F/F, and CCSP-rtTA mice have previously been described (26, 27, 32).
Lung tumorigenesis was initiated as described in (43) by intratracheal infection of mice with Cre-expressing adenovirus (Ad5-CMV-Cre, University of Iowa Gene Transfer Core) or with Cre- and miR-143- or miR-145-expressing pTMMPC lentiviruses (31).
For histological analyses, tissues were fixed in 4% formalin, embedded in paraffin, and sections were stained with hematoxylin and eosin (H&E). Tumor area was determined using NIS-Elements imaging software.
For IF and miRNA-FISH analysis, mice, untreated or injected intravenously with an Alexa Fluor 568-conjugated Isolectin IB4 conjugate (100 μg in 100 μl, Life Technologies) for 10 minutes were perfused with PBS, followed by perfusion with 2% paraformaldehyde (PFA). After an overnight fixation in 4% PFA at 4°C, lung and control organs were placed in 30% sucrose overnight at 4°C with shaking. Lungs were inflated with an OCT:PBS (1:1) solution and embedded in OCT (Tissue-Tek). Frozen sections (10 μm thick) were cut on a CryoStar NX70 (Thermo Scientific).
Cell lines and treatments
We isolated primary MEFs from littermate E13.5 embryos in our laboratory and maintained the cells in DMEM supplemented with 15% FBS, pen/strep, L-glutamine, non-essential amino acids, and β-mercaptoethanol. miR-143/145F/F;CreER and control MEFs were isolated October 2010; miR-143/145F/F;KrasFSF-G12D/+;CreER and control MEFs were isolated August 2011. All MEFs were authenticated by genotyping and RNA expression analysis.
We derived the KrasG12D/+;p53−/− cell line (LG1233) in our laboratory in May 2012 from individual tumors dissected from KrasLSL-G12D/+;p53F/F C57BL/6 mice at 10 week after intratracheal administration of Ad5-CMV-Cre. The cell lines used have not been authenticated. The cells were transduced with a lentivirus co-expressing GFP and luciferase and selected for GFP expression by FACS. Following passaging in C57BL/6J immunocompetent mice, individual lung tumors were re-isolated. The cells with highest GFP expression levels were selected by FACS (LG1233S1P1) and used for intravenous and subcutaneous transplantation by injecting 105 cells in 100 μl PBS.
Primary miRNA sequences cloned downstream of the tetracycline response element (TRE) in the pSLIK lentiviral expression system (44) were introduced in KrasG12D/+;p53−/− cell lines. Cells were selected with hygromycin and treated with 1 μg/ml doxycycline to induce miRNA expression.
HUVEC cells (ATCC CRL-1730, authenticated by short tandem repeat analysis by ATCC and not re-authenticated by our lab) were serum starved for 16 hours. 2×105 cells were seeded on growth-factor reduced Matrigel (Corning)-coated wells, and tube formation was imaged by brightfield microscopy at indicated time-points.
miRNA Fluorescence in situ hybridization (miRNA FISH)
Cryostat sections were dried and fixed in 4% PFA for 10 minutes. Sections were blocked in Hybridization buffer (50% formamide, 5xSSC, 1xDenhards, 500 μg/ml yeast tRNA in H20) for 30 minutes at 54°C and incubated with FITC-labeled miR-143 or miR-145-specific miRCURY LNA Detection Probes (Exiqon, 38515-04 and 38517-04) diluted in Hybridization buffer for 2 hours at 50°C in shaking waterbath. Following three washes with 0.1xSSC at 58°C and one wash with 2xSSC at RT, sections were incubated with 3% H2O2 for 10 minutes and washed three times for 3 minutes in TN buffer (0.1M Tris-HCl, pH 7.5; 0.15M NaCl). The FITC label was amplified by blocking for 30 minutes with Blocking buffer (2 mg/ml BSA; 2% sheep serum in TN buffer) and incubating for 30 minutes with HRP-conjugated anti-FITC antibody (Dako, P5100) diluted in Blocking buffer, followed by three 5-minute washes with Washing buffer (TN with 0.3% Triton X-100). Signal amplification was performed by incubating slides with FITC-tyramide (TSA-Fluorescien System, Perkin Elmer) for 10 minutes. Three 5-minute washes with TNT were performed and the slides were mounted in ProLong Gold Mounting Medium (Invitrogen). Images were visualized under a Nikon microscope and processed in Photoshop.
Fluorescence Activated Cell Sorting (FACS)
Lungs were minced using a razor blade and incubated in dissociation solution containing DNAse (40 U/ ml, Roche) and Collagenase IV (1,000 U/ml, Worthington) rotating at 37°C for 1 hour. Following red blood cell lysis, cells were blocked for 15 minutes at 4°C and stained with the following primary antibodies for 25 minutes at 4°C: CD31-APC (BD Pharmingen), CD45-PE-Cy7 (BD Pharmingen), Epcam-FITC (Biolegend). DAPI (Invitrogen) was used to exclude dead cells. FACS sorting was carried out on a BD FACSAria (BD Biosciences) prior to RNA sequencing, qRT-PCR, or immunoblot analyses.
RNA isolation and quantitative RT-PCR
RNA was isolated with the RNeasy Mini Kit (Qiagen), reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and used for qPCR using KAPA SYBR Fast Master Mix (Kapa Biosystems, KK4604). The following primers were used: GAPDH F- agcttgtcatcaacgggaag, R- tttgatgttagtggggtctcg; pri-miR-143 F- gaagatgtcctgcagccgtca, R- tctagggacaacctatctagtcc; pri-miR-145 F- tggatctgtctccctccca, R- ttacctccgacgaaccgcctcc; mCherry F- cacctacaaggccaagaagc, R- tggtgtagtcctcgttgtgg; Kras F- ggtggttggagctggtggcg, R- cctgtaggagtcctctatcgtagggt; RREB1 F- cgaggactgctgcgccacaat, R- acaccaggcctgagagattcga; CD31 F- cggtgttcagcgagatcc, R- actcgacaggatggaaatcac; CD45 F- catccatcctcgtccactg, R- attgaacaaggcacagaacaac; SPC F- ccactggcatcgttgtgtat, R- gccatcttcatgatgtagcagt; SMA F- ctctcttccagccatctttcat, R- tataggtggtttcgtggatgc. Expression levels were calculated relative to GAPDH and normalized to control samples.
Analysis of mature miRNA levels was performed by isolating RNA using the miRNeasy Mini Kit (Qiagen) and performing miR-143 and miR-145-specific Taqman MicroRNA Assays (Life Technologies) using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and Taqman Fast Universal PCR Master Mix (Life Technologies). Expression levels were calculated relative to U6 snRNA (Life Technologies).
Immunoblotting
Cells were collected, counted and lysed in 2x Laemmli buffer. Samples were heated to 95°C for 7 minutes and passed through an insulin syringe. Immunoblotting was performed using the following antibodies: Camk1d (Cell Signaling, 3365), pHH3 (Cell Signaling, 9701), Hsp90 (BD Biosciences, 610418).
Immunofluorescence (IF)
Frozen sections were dried, rehydrated in PBS, and preincubated with 1xPNB blocking buffer (Perkin Elmer). Sectioned were incubated overnight at 4°C with the following antibodies diluted in 1xPNB: CD31 (Dianova, DIA-310), Dll4 (R&D Systems, AF1389), and pIGFR Y116 (Abcam, ab39398), followed by 1-hour incubation at RT with Alexa dye-tagged secondary antibodies (Life Technologies). DAPI (Invitrogen) was used to label the nuclei. Slides were mounted in ProLong Gold Mounting Medium (Invitrogen). Images were visualized under a Nikon microscope. Quantitation of lectin was performed using ImageJ.
Immunohistochemistry (IHC)
Mice were sacrificed by CO2 asphyxiation and lungs were inflated with 10% zinc formalin (Polysciences Inc.), fixed overnight in zinc formalin at RT, then transferred to 70% ethanol until paraffin embedding. IHC was performed on 5 μm thick sections using ABC Vectastain kit (Vector Labs) with antibodies to CD31 (Dianova, DIA-310), GFP (Cell signaling 2956), CD3 (Abcam, ab5690), B220 (Biolegend, 103202), MPO (Thermo Scientific, #RB-373-A0), IbaI (Wako), cleaved caspase 3 (Cell signaling 9661), KI67 (Vector Labs, VP-RM04) and SMA (Sigma Aldrich, A5228). The staining was visualized with DAB (Vector Labs, SK-4100) and slides were counterstained with hematoxylin. For the apoptosis and proliferation analysis, quantitation was performed using ImageJ to determine the percentage of Ki67-positive cells and the number of cleaved caspase 3-positive cells per tumor area.
Luciferase assay
Full-length (5526 nt) or a shorter wild-type or mutant segment (1000 nt) of the 3′UTR of mouse Camk1d was cloned at the 3′ of Firefly luciferase in the pGL3-Promoter Luciferase Reporter Vector (Promega). Empty or 3′UTR-containing pGL3 were co-transfected with Renilla luciferase-expressing vector (pRL-TK, Promega) in MEFs using Attractene transfection reagent (Qiagen). Luciferase activity was assayed at 48 hours post transfection using Dual Luciferase Reporter Assay System (Promega). Firefly luciferase signal was normalized to Renilla luciferase signal.
RNA-seq analysis
Illumina HiSeq 2000 40-nt single-ended reads were mapped to the UCSC mm9 mouse genome. Independent Component Analysis (ICA), targeted pair-wise differential expression analysis, and gene set enrichment analysis (GSEA) were performed as described in (45). All primary RNA-seq data are available at Gene Expression Omnibus (GSE71877).
Supplementary Material
STATEMENT OF SIGNIFICANCE.
This study shows that miR-143/145 expressed from the tumor microenvironment stimulates neoangiogenesis and supports tumor expansion in the lung, demonstrating a surprising role for the putative tumor suppressor miRNA cluster in promoting tumorigenesis. We propose inhibition of miR-143/145 as a therapeutic avenue to modulate tumor neoangiogenesis.
ACKNOWLEDGEMENTS
We thank Jesse Zamudio, Francisco Sanchez-Rivera, and Tuomas Tammela for critical review of the manuscript. We are grateful to the Koch Institute Swanson Biotechnology Center (SBC) for technical support, specifically Aurora Burds O’Connor from the Rippel ES cell and Transgenics Facility and Kathleen Cormier at the Hope Babette Tang (1983) Histology Facility. We thank Roderick Bronson for pathological analysis.
GRANT SUPPORT
This work was supported by NIH (P01-CA42063-26), the Howard Hughes Medical Institute and the Ludwig Center for Molecular Oncology at MIT. This work was partially supported by the Cancer Center Support (Core) Grant P30-CA14051 from the National Cancer Institute. N.D. was supported by a Damon Runyon Fellowship Award. V.G. is supported by a Jane Coffin Childs Memorial Fund Postdoctoral Fellowship. T.J. is the David H. Koch Professor of Biology and a Daniel K. Ludwig Scholar at MIT.
ABBREVIATIONS
- miRNAs
MicroRNAs
- 3′UTRs
3′ untranslated regions
- K
KrasLSL-G12D/+
- KP
KrasLSL-G12D/+; p53F/F
- F
floxed
- -
null
- MEFs
mouse embryonic fibroblasts
- qRT-PCR
quantitative RT-PCR
- t.v.i.
tail vein injection
- IHC
immunohistochemistry
- IF
immunofluorescence
- FISH
fluorescence in situ hybridization
- FACS
fluorescence-activated cell sorting
- ICA
Independent Component Analysis
- RNAseq
RNA sequencing
- HUVECs
human umbilical vein endothelial cells
- H&E
hematoxylin and eosin
- PFA
paraformaldehyde
- TRE
tetracycline response element
Footnotes
CONFLICT OF INTEREST STATEMENT
There are no conflicts to disclose
REFERENCES
- 1.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–79. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 2.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 3.Michael MZ, SM OC, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–91. [PubMed] [Google Scholar]
- 4.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 5.Sempere LF, Liu X, Dmitrovsky E. Tumor-suppressive microRNAs in Lung cancer: diagnostic and therapeutic opportunities. ScientificWorldJournal. 2009;9:626–8. doi: 10.1100/tsw.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77:12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 7.Papaconstantinou IG, Manta A, Gazouli M, Lyberopoulou A, Lykoudis PM, Polymeneas G, et al. Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas. 2013;42:67–71. doi: 10.1097/MPA.0b013e3182592ba7. [DOI] [PubMed] [Google Scholar]
- 8.Shen H, Shen J, Wang L, Shi Z, Wang M, Jiang BH, et al. Low miR-145 expression level is associated with poor pathological differentiation and poor prognosis in non-small cell lung cancer. Biomed Pharmacother. 2015;69:301–5. doi: 10.1016/j.biopha.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–92. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 10.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer. 2009;45:2197–206. doi: 10.1016/j.ejca.2009.04.039. [DOI] [PubMed] [Google Scholar]
- 11.Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, et al. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010;24:2754–9. doi: 10.1101/gad.1950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clape C, Fritz V, Henriquet C, Apparailly F, Fernandez PL, Iborra F, et al. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS One. 2009;4:e7542. doi: 10.1371/journal.pone.0007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borralho PM, Simoes AE, Gomes SE, Lima RT, Carvalho T, Ferreira DM, et al. miR-143 overexpression impairs growth of human colon carcinoma xenografts in mice with induction of apoptosis and inhibition of proliferation. PLoS One. 2011;6:e23787. doi: 10.1371/journal.pone.0023787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibrahim AF, Weirauch U, Thomas M, Grunweller A, Hartmann RK, Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71:5214–24. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 15.Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10:1470–80. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Zeng H, Guo Y, Liu P, Pan H, Deng A, et al. miRNA-145 inhibits non-small cell lung cancer cell proliferation by targeting c-Myc. J Exp Clin Cancer Res. 2010;29:151. doi: 10.1186/1756-9966-29-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, et al. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer. 2010;102:883–91. doi: 10.1038/sj.bjc.6605570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gregersen LH, Jacobsen AB, Frankel LB, Wen J, Krogh A, Lund AH. MicroRNA-145 targets YES and STAT1 in colon cancer cells. PLoS One. 2010;5:e8836. doi: 10.1371/journal.pone.0008836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–47. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–8. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–10. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–78. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang S, Cui H, Xie N, Icyuz M, Banerjee S, Antony VB, et al. miR-145 regulates myofibroblast differentiation and lung fibrosis. FASEB J. 2013;27:2382–91. doi: 10.1096/fj.12-219493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chivukula RR, Shi G, Acharya A, Mills EW, Zeitels LR, Anandam JL, et al. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell. 2014;157:1104–16. doi: 10.1016/j.cell.2014.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gras C, Ratuszny D, Hadamitzky C, Zhang H, Blasczyk R, Figueiredo C. miR-145 Contributes to Hypertrophic Scarring of the Skin by Inducing Myofibroblast Activity. Mol Med. 2015;21:296–304. doi: 10.2119/molmed.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–8. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 28.Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb Symp Quant Biol. 1997;62:159–68. [PubMed] [Google Scholar]
- 29.Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–40. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 30.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 31.Xue W, Dahlman JE, Tammela T, Khan OF, Sood S, Dave A, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A. 2014;111:E3553–61. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tichelaar JW, Lu W, Whitsett JA. Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J Biol Chem. 2000;275:11858–64. doi: 10.1074/jbc.275.16.11858. [DOI] [PubMed] [Google Scholar]
- 33.Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3:401–10. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 34.Hellstrom M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–80. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 35.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Verploegen S, Lammers JW, Koenderman L, Coffer PJ. Identification and characterization of CKLiK, a novel granulocyte Ca(++)/calmodulin-dependent kinase. Blood. 2000;96:3215–23. [PubMed] [Google Scholar]
- 37.Colomer J, Means AR. Physiological roles of the Ca2+/CaM-dependent protein kinase cascade in health and disease. Subcell Biochem. 2007;45:169–214. doi: 10.1007/978-1-4020-6191-2_7. [DOI] [PubMed] [Google Scholar]
- 38.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–14. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fang L, Deng Z, Shatseva T, Yang J, Peng C, Du WW, et al. MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-beta8. Oncogene. 2011;30:806–21. doi: 10.1038/onc.2010.465. [DOI] [PubMed] [Google Scholar]
- 40.Hergenreider E, Heydt S, Treguer K, Boettger T, Horrevoets AJ, Zeiher AM, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14:249–56. doi: 10.1038/ncb2441. [DOI] [PubMed] [Google Scholar]
- 41.Climent M, Quintavalle M, Miragoli M, Chen J, Condorelli G, Elia L. TGFbeta Triggers miR-143/145 Transfer From Smooth Muscle Cells to Endothelial Cells, Thereby Modulating Vessel Stabilization. Circ Res. 2015;116:1753–64. doi: 10.1161/CIRCRESAHA.116.305178. [DOI] [PubMed] [Google Scholar]
- 42.Cisneros-Barroso E, Yance-Chavez T, Kito A, Sugiura R, Gomez-Hierro A, Gimenez-Zaragoza D, et al. Negative feedback regulation of calcineurin-dependent Prz1 transcription factor by the CaMKK-CaMK1 axis in fission yeast. Nucleic Acids Res. 2014;42:9573–87. doi: 10.1093/nar/gku684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–72. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI, et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci U S A. 2006;103:13759–64. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li CM, Gocheva V, Oudin MJ, Bhutkar A, Wang SY, Date SR, et al. Foxa2 and Cdx2 cooperate with Nkx2-1 to inhibit lung adenocarcinoma metastasis. Genes Dev. 2015;29:1850–62. doi: 10.1101/gad.267393.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.