Abstract
Exome sequencing has recently been elevated to the standard of care for genetic diagnostic testing, particularly for genetically diverse and clinically heterogeneous disorders. This review provides a clinically oriented discussion of the next-generation sequencing technology that makes exome sequencing possible and how such technology is applied to the diagnosis of Mendelian disease, including clinically significant de novo variation, interpretation of variants of uncertain clinical significance, the future potential for genetic assessments of disease risk, and the substantial benefits in diagnostic efficiency. Important caveats are also discussed, including the implications of incidental or secondary findings detected during exome sequencing, and the relationship of exome sequencing to other methods of clinical genomic testing, such as chromosomal microarray and genome sequencing. Overall, the widespread adoption and use of exome sequencing in routine clinical practice is expected to improve diagnosis rates and reduce test costs, while leading to improvements in patient outcomes and a renewed emphasis on disease management.
Keywords: exome, genome, neurogenetics, genetic testing, genetics, sequencing
The challenge of modern genetic testing
Within the last decade, it has become essentially impossible to ignore the impact of genetic disease on the practice of neurology and psychiatry. Virtually all of the major common neurologic and psychiatric disorders have been shown to possess some degree of significant heritability,1 and the number of rare familial or private Mendelian causes of both common and rare phenotypes has risen considerably. A current search of the Online Mendelian Inheritance in Man database2 reveals over 2100 disorders with a neurologic phenotype and an established genetic etiology. Such daunting numbers constitute a significant challenge for the physician in general or specialty practice, as they give a sense of the truly vast diagnostic dilemma that patients currently face. Further, while most general or subspecialty physicians should not be expected to perform as geneticists, the need for complex genetic testing in a potentially large segment of the clinical populace demands a new paradigm in clinical evaluation. To meet this challenge, the incorporation of new technology allowing for rapid and inexpensive (next-generation) DNA sequencing has enabled vast improvements in the rates of genetic diagnosis with considerable savings to the patient and the physician in terms of cost and time. The most clinically relevant application of next-generation sequencing in current practice is in the sequencing of the exome, the approximately 1% of the human genome that codes for proteins and where the majority of mutations currently known to cause genetic disease are found.1,3–5 In this review we will discuss the methodology and use of next-generation sequencing in routine practice, emphasizing awareness of important clinical caveats and highlighting the advantages this technology brings to the diagnostic evaluation in complex hereditary disease.
Next-generation sequencing
Published by Frederick Sanger in 1977,6 Sanger (also known as chain-termination) sequencing has been the most commonly used DNA sequencing method for decades and is still considered to be the gold standard methodology in molecular diagnostic laboratories. Sanger sequencing, accompanied by the automated capillary sequencing instrument, was the main technology used in the Human Genome Project7 completed in 2001.8 It took roughly $2.7 billion and 13 years to complete the Human Genome Project primarily because Sanger sequencing has significant limitations, including the number of samples that can be sequenced in parallel and a high cost per base sequenced (~$10/kb). These limitations stimulated efforts to develop less-expensive and higher-throughput sequencers, and a new chapter in DNA sequencing technology began in 2005 when the first next-generation sequencing (NGS) instrument by 454 Life Sciences (purchased by Roche in 2007) was introduced to the world.9 The following year, Solexa (later purchased by Illumina) released the Genome Analyzer, and Agencourt (initially purchased by Applied Biosystems, then by Life Technologies) released the SOLiD (Sequencing by Oligo Ligation Detection).10 Other companies, such as Complete Genomics, Pacific Biosciences, and Helicos Biosciences, also released NGS instruments, but the first three companies currently market the most widely used systems with ongoing improvements in read length, sequencing speed, and accuracy. NGS technology has now advanced to the point where the entire human genome can be sequenced in approximately 2 days for under $3000 (~ $0.001/kb), and this cost continues to decline. Table 1 illustrates the versatility of next-generation sequencing for diagnostic testing, while Table 2 summarizes the specifications and applications of the current leading NGS systems in the clinical marketplace.
Table 1.
Comparison of DNA-sequencing technologies for genetic diagnosis
| Diagnostic test | Single-gene / multi-gene panel |
Next-generation panel | Exome sequencing | Genome sequencing |
|---|---|---|---|---|
| Sequencing method | Sanger | Next-generation | Next-generation | Next-generation |
|
Approximate number of
genes tested |
1 or < 30 (panel) | 100–1000 | 20,000 | 20,000, including noncoding sequence and structural variation |
|
Estimated clinical cost
per gene sequenced |
$1000 | $1.00 or less | Varies by laboratory ($0.25 or more) |
Varies by laboratory ($0.50 or more) |
Table 2.
Specifications of NGS systems commonly used in clinical laboratories.
| HiSeq2500 | HiSeq XTen | MiSeq (v2 chemistry) |
Ion PGM Ion 316 or 318 chip |
|
|---|---|---|---|---|
| Read length | 100 bp X 2a | 150 bp X 2 | 150 bp X 2 or 250 bp X 2 |
200 bp or 400 bp |
| Run time | 27 hb | 3 daysb | 1 day or 1.5–2 days |
4–7 h |
| Data generated | 100–120 Gbb | 1.6–1.8 Tbb | 4.5–5 Gb or 7.5–8.5 Gb |
300 Mb–1 Gb |
| Common application |
Exome sequencing | Genome sequencing | Gene panel sequencing |
Gene panel or somatic mutation hot-spot genotyping |
Read length can be up to 250 bp X 2 (machine run time: 60 h)
Per dual flow cell on 1 HiSeq system
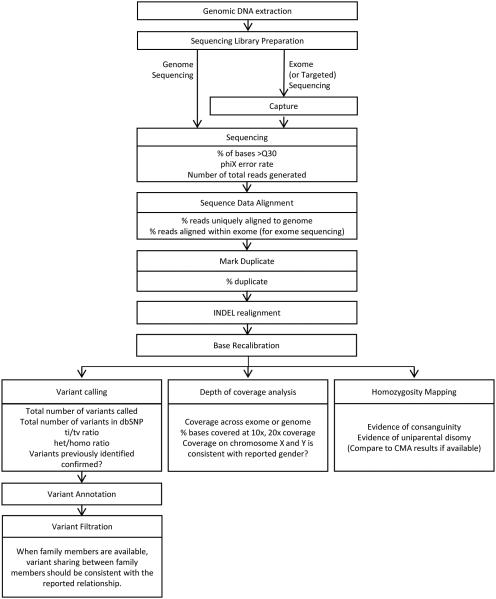
Typical NGS library preparation involves generating small fragments of DNA to which universal adaptors are ligated in order to simultaneously amplify the smaller oligonucleotides and sequence the fragments. For genome sequencing, genomic DNA can be randomly sheared to generate small fragments (Fig. 1). However, both exome sequencing and targeted sequencing require an additional step of capturing the genomic regions of interest. When NGS became available, a technology that allows simultaneous amplification of multiple genomic loci became crucial. Earlier attempts used target-specific primers to amplify selected regions, enrich for the amplified products, and further amplify the enriched products with universal primers.11–13 However, these methods were labor intensive and subject to failure due to primer multiplexing. To overcome these challenges, a method using short oligonucleotide microarray-based capture was developed.14–16 This evolved into an in-solution hybridization method using RNA- or DNA-based baits, and is currently the most commonly used method in commercially available capture kits. Different versions of capture kits vary by probe design and, accordingly, depth of coverage.17 In general, GC-rich regions are harder to capture and sequence, and hence many first exons do not get sequenced to comparable levels as other exons. Probe designs have further evolved, and the most suitable capture kit should be selected depending on the specific need. For example, some of the capture kits target only the coding regions of the genome, while others also target the untranslated regions (UTRs). Additionally, some of the kits focus on covering clinical genes (i.e., those known to cause rare Mendelian disorders) better than genes that are not associated with any human disease.
Figure 1.
Typical workflow for next-generation sequencing, analysis, and analytical quality control metrics.
Analysis of next-generation sequencing data
The data analysis workflow for exome (or genome) sequencing is shown in Figure 1. Although the overall workflow is generally standard across different laboratories, it is important to note that key differences in bioinformatic algorithms, software versions, and other proprietary methods of analysis can (and do) vary; accordingly, a percentage of rare variant calls can differ between any two laboratories, leading to differing technical reports and, possibly, alternate clinical interpretations (discussed further below). The general bioinformatics pipeline for analyzing NGS data involves aligning the short sequence reads to the reference genome, marking potential PCR duplicates, fine tuning the alignment, and then calling the variants. Once the variants are identified, they are annotated with enriched information about the variant and its gene collected from databases such as OMIM (Online Mendelian Inheritance of Man database; http://www.omim.org/), HGMD (Human Gene Mutation Database; http://www.hgmd.org/), EVS (Exome Variant Server database; http://evs.gs.washington.edu/EVS/), and ExAC (Exome Aggregation Consortium database; http://exac.broadinstitute.org/), among others. (More comprehensive lists of these and other recommended databases are available from the American College of Medical Genetics and Genomics (ACMG).18) Annotated variants are then filtered according to the inheritance pattern (if known, or if parents and/or other family members were sequenced simultaneously with the patient), minor allele frequency of the variants, and evidence of association with rare human disorders. Sequencing unaffected parents along with the patient allows identification of de novo variants and phased compound heterozygous variants, significantly reducing the number of variants that have to be examined and increasing the diagnostic rate, particularly for recessive disorders.4
Quality control of next-generation sequencing data
Quality control (QC) is especially important in clinical laboratories and for NGS testing; accordingly, multiple QC checkpoints exist throughout preparation of the sequencing library, the actual sequencing, and the data analysis and interpretation. Even though the test is highly complex, the results are extremely consistent across samples within a test center, as long as the same protocol is followed throughout. In the rare instances of a data problem, it is relatively easy to identify samples that are outliers,4 and most laboratories repeat such samples. Common practice for handling outlier samples would be to acquire a new blood sample and reprocess the entire test. Some of the common QC checkpoints are shown in Figure 1.
Clinical variant reporting and challenges
Variant reporting for single-gene testing is typically straightforward, with most laboratories choosing to report detection of all pathogenic variants, variants of uncertain clinical significance, and benign variants. In these cases, the number of variants detected is usually relatively small (often 1–5 per gene). However, as the size of the target increases, the likelihood of finding additional variants increases as well. Therefore, for next-generation sequencing–based panels, exomes, and genomes, with the aim of providing a more meaningful clinical report, laboratories will often choose not to report most of the variants they detect (including all benign variants, as well as most of the variants of uncertain clinical significance) and only highlight the ones which they deem most likely to be causal for a patient’s clinical indications. A number of considerations may affect variant interpretation, including the suspected mode of inheritance, the class of the potential mutation, the penetrance and expressivity of the associated disease, and various other factors.1 For those select variants of uncertain clinical significance that are reported, each of them may be shown later to be causal, contributory, or not relevant at all through additional clinical or research studies. How laboratories perform the process of narrowing down a list of variants from approximately 20,000 (the total number of coding variants that would likely be detected from an exome sequence) to the one or two causal variants that will be reported is based on each laboratory’s specific bioinformatic analysis pipeline. Because a single best method for identifying a potentially pathogenic variation or relevant variants of uncertain clinical significance (VUS) has not yet been established, these pipelines can be quite distinct; as a result, the final clinical reports can vary widely for such variants. It would not be unexpected to see one laboratory reporting a rare variant that another laboratory may not even detect or may have excluded from further consideration. This does not reflect on the quality of a given laboratory or the validity of its findings, only on the methodological differences and complexities inherent to the interpretation of these forms of genomic testing. Laboratories should thus be transparent regarding their analytical methods to aid clinicians and geneticists in detailed interpretation of the full results.
For those variants that are detected and deemed to be reportable, many laboratories have chosen to confirm each variant using an orthogonal method (typically Sanger sequencing) out of concerns over reporting false-positive results, while other groups have chosen to only confirm low-quality variants before reporting.19 Once the relevant variants have been confirmed or are of suitable quality, they are often reported in a hierarchical manner, with the most relevant variants presented at the top of the report. Laboratories are encouraged to communicate clearly regarding whether a particular reported variant is known or likely to be pathogenic (and is therefore explanatory for a patient’s condition), or whether it is a VUS (and additional work should be performed by the ordering clinician in order to strengthen its causality, such as sequencing the variant in additional affected family members). At this stage, the ordering clinician may also wish to consult with a clinical geneticist or other genetics specialist (e.g., neurogeneticist, etc.) to assist with interpretation of the findings and/or to create an appropriate management plan for the patient.
Utility of clinical exome sequencing in neurologic and psychiatric disease
In recent years, exome sequencing has been effectively utilized to identify novel genetic causes of a variety of diseases, including neurological and psychiatric disorders.20 This has proven to be a powerful technique for gene discovery. However, the more immediately clinically accessible role for exome sequencing is its use as an unbiased approach to identify mutations in known genes. Several studies, predominantly from large diagnostic sequencing laboratories, have examined the usefulness of exome sequencing in establishing diagnoses in patients with suspected underlying genetic disorders. This work has demonstrated a particularly effective diagnosis rate in patients with neurological disease, as shown in Table 3. The greatest advantage is already being seen with patients presenting with heterogeneous clinical phenotypes that could result from a wide variety of genetic causes, where exome sequencing has been endorsed as the standard of care.21,22 The unbiased nature of exome sequencing reduces the impact of disease variability on genetic testing strategies. Clinicians often struggle with such phenotypic diversity and are challenged to select appropriate genetic testing for complex cases, which could, for example, represent either the typical phenotype of an extremely rare genetic disorder or an unusual presentation of a more common genetic condition. Exome sequencing solves this conundrum by weighing all disease genes equally and allowing all identified variants to be assessed simultaneously in a clinical context. In this way, the true phenotypic variability of genetic disease can be assessed, and there are already examples in the literature of patients identified with pathogenic variation that would never have been considered for single-gene testing based on published phenotypes.23–25 This emphasizes an important role for clinical input in bioinformatic analysis when assessing the likely contributions of novel genetic variation to disease in a particular patient.4,24 This is of particular relevance, as establishing a correct genetic diagnosis leads to altered management in numerous ways ranging from changes to medical therapy, implementation of disease-specific preventive care, initiation of reproductive and genetic counseling, and participation in research and/or clinical trials.24,26,27
Table 3.
Rates of diagnosis for neurological disorders using clinical exome sequencing
Exome sequencing improves detection of de novo genetic disorders
Dominant and recessive inheritance patterns have long been appreciated for the transmission of single-gene traits. The methods developed for human disease gene discovery were directed towards identifying disease genes fitting those patterns. Because of this confluence of ideas and tools, the majority of disease-gene associations discovered before the advent of clinical exome/genome sequencing matched dominant, sex-linked, or recessive models.
However, there have long been tantalizing clues that sporadic genetic diseases, those occurring due to de novo DNA variants arising in an affected offspring, play major roles in medical genetics. One classic example is de novo gain-of-function variants in the FGFR3 gene, which cause achondroplasia (MIM: 9300204).28,29 This disease–gene association was identified by studying familial forms of the condition, which was possible because achondroplasia is not genetically lethal. For severely disabling or sterilizing conditions, such familial forms do not exist, rendering the classic tools of molecular genetics ineffective.
It is also worth noting that sporadic de novo genetic disease can be indistinguishable from recessive disease when studying family histories. If there is a negative or non-informative family history with only a single affected individual, either model is plausible. In the case of affected siblings, recessive inheritance is more likely, although sporadic de novo disorders have been seen due to parental germline mosaicism in some cases.30 For extremely rare conditions, it may not be possible to determine the correct genetic mechanism in the absence of molecular genetic evidence.
Exome or genome sequencing of proband–mother–father trios enables, for the first time, genome-wide detection of single-gene de novo variants. This has led to an explosion of new disease–gene associations, beginning with the molecular characterization of the previously confounding Kabuki syndrome.31 Clinical cohort studies from across the world have clearly demonstrated that sporadic de novo gene mutations are a major cause of genetic disease.4,5,32,33 Next-generation sequencing has also allowed for the identification of novel gene-associated syndromes, such as CHD834 and KAT6A.35 Although de novo mutations in these genes cause neurological symptoms and dysmorphic features, neither causes a sufficiently specific syndrome to robustly allow for identification of cases without molecular testing. These disease–gene discovery efforts translate immediately to clinical molecular genetic testing, providing constant updates to the knowledge base available to analysts interpreting the results of clinical genome or exome results.
Much like autosomal recessive disorders that can be caused by homozygosity or compound heterozygosity, dominant diseases should be thought of as caused by heterozygous variants either inherited from an affected parent or arising de novo in a proband. For dominant conditions with severe impact on fitness, including those with congenital or early onset neurological symptoms, de novo mutation may be the only mechanism observed.
Incidental and secondary findings detected during exome sequencing
Variants may be detected during the course of clinical exome/genome sequencing that are not directly relevant to the primary clinical indications of the patient (so called “incidental” findings). The ACMG has issued guidelines36 addressing these variants, which initially recommended that clinical laboratories report any known or suspected pathogenic variants (such as novel truncating variants) detected in any one of a list of 56 genes without regard to various factors, such as patient age or indication for testing. However, these guidelines have since been revised37 to allow patients to opt out of receiving these types of results, and the term incidental findings has been formally replaced by secondary findings, which reflects a desire for laboratories to actively search for these types of variants during the course of clinical testing (as opposed to stumbling upon these variants while reviewing the sequencing data). While some laboratories have chosen to adhere closely to the guidelines, other clinical laboratories have chosen to include additional genes, beyond those recommended in the guidelines, in their incidental/secondary variant analyses, and others have even chosen not to actively search for these variants at all, keeping them as true incidental findings. Therefore, physicians who wish to order exome/genome sequencing need to be aware of the possibility that incidental/secondary findings will appear on the report if a patient has opted to receive them and are encouraged to closely review the consent form of the providing laboratory to understand their specific approach to incidental/secondary variant reporting. Most of the recommended genes involve autosomal dominant cancer predisposition syndromes (such as the BRCA1 and BRCA2 genes that predispose an individual to breast and ovarian cancer); as an example, in a patient with a pathogenic incidental/secondary variant reported in the BRCA1 gene, referral to an oncologist may be necessary.
Clinical interpretation of variants of uncertain significance
Genomic sequencing methods, while benefiting from an unbiased and comprehensive evaluation of all genes in the genome, are challenged by the frequent identification of sequence variation that has never been previously observed. Previously observed variation can be easily annotated with regard to a likelihood of pathogenicity, given that it is either rare or common in control populations (either non-diseased or having alternate symptoms/phenotypes) and has either been seen in similar patients before or not. Novel variation could represent a rare polymorphism or a disease variant, and this determination cannot be made by viewing the sequence in isolation.38 Bioinformatic analysis may be helpful, but, as this is still an inexact science,39–41 the findings can most often only be used as support for other lines of interpretation. Segregation of such variation within a family can be helpful in assessing pathogenicity, but caveats such as penetrance, expressivity, genetic mosaicism, and other factors may still limit clear identification. The same holds true for observed variation seen only rarely in control populations (i.e., at frequencies of 1% or less), and one cannot confidently exclude such variation as being wholly benign.38,42 The much overused term “variant of uncertain clinical significance” is often ascribed to such genetic changes and is therefore of limited value as a descriptor. In the vast majority of cases, it is most clinically useful for laboratories (or interpreting bioinformaticists) to establish a line of evidence either supporting or refuting a given variant as pathogenic, thus enabling clinicians to perform additional clinical or research evaluations to confirm the status, if such options exist for the disease in question. Additionally, as more genomic sequencing and variant validation is performed, the status of these uncertain variants may change, and periodic reassessment of exome sequencing results (at the request of the clinician) should be considered for patients without a definitive diagnosis.
Exome sequencing may aid efforts to assess genetic risk of disease
It is clear that common variants with modest effects can increase the risk for certain diseases, and that rare variants can cause Mendelian conditions with phenotypes similar to, or indistinguishable from, common traits. As one example, researchers found that familial and sporadic Alzheimer’s disease (AD) have similar functional connectivity perturbations,43 which highlights a degree of indeterminacy in distinguishing the monogenic and polygenic phenotypes. It was also demonstrated recently that common variants affect the age of onset of AD.44 Thus, for a hypothetical patient presenting with typical onset AD and no family history, these findings––combined with the appreciation of sporadic de novo genetic disease––raise the complex possibility that the patient may actually carry a de novo monogenic mutation predisposing him/her to early onset AD but with a common variant profile that leads to delayed disease onset. Currently, genetic testing for AD is typically restricted to familial or early onset cases, so our knowledge of the probability of the scenario described above is lacking. However, as access to next-generation gene-panel and exome sequencing improves, it may become prudent to incorporate such testing into common practice for sporadic diseases such as AD in order to account for such scenarios.
For many neurological disorders, the scope of the genetic contribution to disease is uncertain. Establishing the degree of heritability for a condition is a necessary first step to set the stage for genetic analysis, but this can be a challenge, especially for adult-onset conditions where truly negative individuals are difficult to find. For example, estimates of the heritability of non-familial amyotrophic lateral sclerosis (ALS) have been based on relatively small-scale twin studies and thus include a wide range of possible heritability scores.45 Studying a cohort of over 1200 sporadic cases of ALS enabled estimation of a more precise heritability of 17–25%.46 This helps to contextualize genome-wide association studies, such as a recent meta-analysis of 61,000 ALS cases that reported significant common variants at three distinct loci and predicted that common variants account for ~12% of ALS heritability.47 This suggests that the relatively small remainder of heritability may be caused by rare variants. Efforts to detect such variants by genome or exome sequencing are sure to follow in the coming years. Integrating common variant genotypes with rare variant sequences will be challenging but may lead to improved cause attribution and risk prediction.
In addition to the discovery of many new association loci, common variant studies published in 2014 highlight the complex interconnectivity of neurological diseases. Clinical observation and pedigree analysis have long shown that schizophrenia, depression, and bipolar disorder often run in the same families. Genetic analysis has revealed a subtlety within these relationships not previously appreciated. One study, surveying immune-related genetic loci associated with multiple sclerosis, found that these loci also influence risk for schizophrenia but not bipolar disorder.48 This finding suggests that there are genetic subtypes of schizophrenia with an immunological underpinning that do not strongly overlap with bipolar disorder. This type of finding would not be possible without the molecular pathways pinpointed through genome-wide association studies. The impact of rare variants on schizophrenia risk remains murky, as microarray analysis and exome sequencing studies remain somewhat inconclusive.49–51 This appears to be a condition where common variants play a very large role in heritability; but our understanding of the complex interplay between these variants remains in the early stages. Further sequencing efforts may help expand knowledge of clinical and risk-associated variation.
Polygenic risk modeling is another promising area that has already yielded some interesting results. Functional imaging has been used to identify specific changes in brain activity strongly linked to polygenic schizophrenia-risk scores in both patients and controls,52 demonstrating that common variants can affect subclinical neurological traits that put individuals at risk for developing clinical disorders. It is hoped that future work may establish specific clinical correlations between such variation and disease, so that it may provide diagnostic utility.
Cost and relative value of clinical exome sequencing
While ordering a simple molecular test (e.g., fragile X CGG repeat allele sizing for intellectual disability) are relatively inexpensive, full gene sequencing tests (e.g., CACNA1A gene for epilepsy) are often on the order of $1000 or more, and panels/exomes/genomes are currently on the order of $5000–10,000 or even more. Rather than discuss the cost of an individual test, we wish to focus on the overall cost to the healthcare system. When an ordering clinician is uncertain about the most likely relevant causal gene in a given patient, if one laboratory is able to offer a gene panel or exome for ~$5000, while another laboratory is able to offer a single full gene sequencing test for $1000, the value of expanded genomic testing becomes clear once the first five genes in the clinical suspicion list are not found to harbor any pathogenic variants. However, this does not take into account the frequent waiting time between each individual test result (often 1–2 months) and the cost to the patient (both financially and emotionally) for each recurrent clinic visit. Ordering five sequential genetic tests in five subsequent clinic visits may result in the patient going almost a year without a clear answer. One group has estimated that if a diagnosis is not determined during an initial genetics consultation an average of roughly $25,000 is spent on subsequent molecular testing in order to arrive at a diagnosis.53 Therefore, if comparing the overall potential cost versus the cost of an individual genomic sequencing test, the value of exome sequencing to an individual patient, insurance provider, and/or institution becomes more obvious.
The relationship among and advantages of exome sequencing and other diagnostic testing methods
Sanger sequencing is still considered to be the gold standard sequencing methodology, especially in clinical laboratories, and many genetic tests are based on it. However, many of these tests are gradually being replaced by NGS, as this is much more efficient for sequencing multiple genetic loci in parallel and, hence, is cost and labor effective. In addition to higher sequencing cost, time and resources spent on primer design for individual target loci, optimizing and running PCR and analyzing the sequencing traces are not insignificant, and thus genotyping one variant as a clinical test currently costs several hundred dollars. Still, there will be areas where Sanger sequencing will continue to be used. First, even though some laboratories have shown that it is possible to empirically determine the quality threshold to only require Sanger validation on low-quality variants while maintaining clinically acceptable sensitivity and specificity,19 most clinical labs perform Sanger validation on all reportable variants. Second, as more exome sequencing is being performed, test volume requesting genotyping of a single variant identified in the proband in other family members is increasing, and for those cases Sanger sequencing will be the best fit. Lastly, there are patients with rather obvious suspected genetic disorders where testing a single gene would be more cost- and time- efficient than performing exome sequencing. Those single-gene tests will have to be done by Sanger sequencing until clinical exome sequencing cost and turnaround time decrease even further. However, it is not uncommon to find cases where negative results by Sanger sequencing were overturned by clinical exome sequencing, as Sanger methods can be more susceptible to common lab errors such as sample swaps, especially when the parent samples are sequenced simultaneously. Additional ways of broadly integrating Sanger sequencing, NGS panels, and exome sequencing to improve diagnosis have also been suggested.54
Chromosomal microarray (CMA), including array-based comparative genomic hybridization (aCGH) and single-nucleotide polymorphism (SNP) array, is a commonly ordered clinical test for genetic disorders such as developmental delay (including intellectual disability), autism, and congenital anomalies.55 Because of its higher resolution than G-banded karyotyping and higher diagnostic rate, CMA is often ordered as a first-tier genetic test.55 Common clinical cutoffs for identifying and reporting copy number variants (CNVs) from a genome-wide CMA are on the order of ~50 Kb, a resolution not high enough to detect smaller CNVs that might span, for example, only one exon. However, lowering the resolution would reveal more benign variants and clinical specificity would be compromised. This same reasoning applies to detecting CNVs using exome-sequencing. Technically, it is possible to identify exon-level CNVs using exome-sequencing data, but the noise variance is even higher than for CMA data, as gene density is not uniform across the genome, which is why exome sequencing has not yet replaced CMA. However, there have been reports where single-exon CNVs have been identified from exome-sequencing data by clinical laboratories and confirmed by alternate methods.4,5 More recently, microarrays focusing on identifying exon-level CNVs in clinically relevant genes have been developed, but it has yet to be seen how these will be utilized clinically.
Genome sequencing is already being used as a clinical test in some laboratories (see discussion below). However, the analysis is mostly limited to the coding region of the genome (i.e., equivalent to the exome), as we lack understanding of the vast majority of variants in intronic or intergenic regions. The advantage of performing genome, as opposed to exome, sequencing would be the ability to more sensitively identify structural variants such as translocations, inversions, and CNVs, and to identify variants in regulatory regions that are most likely not captured and sequenced by exome sequencing. Genome sequencing also produces more uniform coverage than exome sequencing, because the sequencing library preparation for the former is not as complex, with no capture and fewer PCR cycles (Fig. 1).
Clinical genome sequencing
As recently demonstrated by three independent groups, clinical exome sequencing is extremely powerful for detecting disease-causing, protein-coding variants in the clinical setting.4,5,22,24,33 Presently, approximately 95% of single nucleotide (SNVs) and small insertion/deletion (indels) variants known to cause monogenic genetic disease are detectable via exome sequencing. Although this percentage will likely decrease as the field of human genetics research delves deeper into intronic and other non-coding forms of DNA variation, at this time genome sequencing has only a marginal potential to improve the detection of disease-causing SNVs and indels. This potential is typically not met; genome sequencing to exome-level coverage is cost prohibitive, meaning that the benefit of detecting intronic variants by genome sequencing is balanced by the cost of poorer coverage of coding sequences. Thus, genome sequencing is not simply a “deluxe” version of exome sequencing and that, in addition to increased cost, is likely an important reason why it has not been as readily adopted as a clinical test, compared to exome sequencing.
Genome sequencing does have great potential in that, theoretically, it can detect structural variants. The data generated for genome sequencing holds clues that can be used to detect medium-scale insertions/deletions (20–50,000bp), a blind spot lying between what can be covered by exome and chromosomal microarray analyses. The degree to which blind spot variants are causative of monogenic disease is very poorly understood, as the methods to detect them are presently in the incubation stage. Though some successes have been made in structural variant identification using genome sequencing,56–58 there is as yet no consensus for what tool or set of tools should be used to ensure a high level of technical validity.59 Generally speaking, the methods currently available for structural variant identification lack sufficient sensitivity and specificity, and require time- and resource-consuming confirmatory testing, which prevent such analysis from being of routine clinical utility. Newly available microarrays allow for the identification of exon-level deletions and duplications,60 providing testing to cover part, but not all, of the blind spot. It remains to be seen if such testing is of sufficient yield to merit widespread clinical adoption. As researchers coalesce around methods for converting genome-sequence data into reliable structural variant detection by genome sequencing, it will become an increasingly attractive complement to exome sequencing, and an eventual likely replacement for chromosomal microarray analysis.
Conclusions and future directions
Recent advances in technology have enabled the rapid and efficient sequencing of DNA, which is revolutionizing the concept of genetic testing. Previously, clinicians were charged with the judicious use of single-gene testing that, although effective in specifically defined phenotypes, often proved less useful in disorders with variable or inconsistent phenotypes. As the number of available individual genetic tests has risen,1 so has the tendency for patients to receive extensive or unwarranted genetic testing, resulting in increased cost-of-care burden on the patient.61 The advent of cheap and efficient sequencing has enabled the unbiased testing of all protein-coding genes simultaneously, vastly improving cost efficiency and accelerating time to diagnosis––to the benefit of patients. Genomic testing methods, particularly exome sequencing, have become the new standard of care for genetic testing,21,22 and it is vital that the medical community incorporate this technology into routine practice. While questions still remain regarding the comprehensiveness of variant identification and analysis, as well as the precision of subsequent bioinformatic assessment and the need for functional validation of novel variation, there is no debate as to the advantages of exome sequencing over other genetic testing methodologies. As its clinical use becomes more widespread, the time to diagnosis for many patients should further decrease, hopefully allowing for a shift in the current clinical emphasis on diagnosis to much needed efforts aimed at improving disease treatment and management.
Acknowledgements
This work was supported by the National Institute for Neurological Disorders and Stroke (R01NS082094 to BLF).
Footnotes
Conflicts of interest
The authors have no conflicts of interest with regard to this manuscript.
References
- 1.Fogel BL, Geschwind DH. Clinical Neurogenetics. In: Daroff R, Fenichel G, Jankovic J, Mazziotta J, editors. Neurology in Clinical Practice. Elsevier; Philadelphia, PA: 2012. pp. 704–734. [Google Scholar]
- 2.OMIM . McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; Baltimore, MD: accessed February, 2015. [Google Scholar]
- 3.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, Das K, Toy T, Harry B, Yourshaw M, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, Ward P, Braxton A, Wang M, Buhay C, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300:286–290. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 9.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 11.Dahl F, Gullberg M, Stenberg J, Landegren U, Nilsson M. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahl F, Stenberg J, Fredriksson S, Welch K, Zhang M, Nilsson M, Bicknell D, Bodmer WF, Davis RW, Ji H. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc Natl Acad Sci U S A. 2007;104:9387–9392. doi: 10.1073/pnas.0702165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fredriksson S, Baner J, Dahl F, Chu A, Ji H, Welch K, Davis RW. Multiplex amplification of all coding sequences within 10 cancer genes by Gene-Collector. Nucleic Acids Res. 2007;35:e47. doi: 10.1093/nar/gkm078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 15.Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 16.Lee H, O'Connor BD, Merriman B, Funari VA, Homer N, Chen Z, Cohn DH, Nelson SF. Improving the efficiency of genomic loci capture using oligonucleotide arrays for high throughput resequencing. BMC Genomics. 2009;10:646. doi: 10.1186/1471-2164-10-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark MJ, Chen R, Lam HY, Karczewski KJ, Euskirchen G, Butte AJ, Snyder M. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strom SP, Lee H, Das K, Vilain E, Nelson SF, Grody WW, Deignan JL. Assessing the necessity of confirmatory testing for exome-sequencing results in a clinical molecular diagnostic laboratory. Genet Med. 2014;16:510–515. doi: 10.1038/gim.2013.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabbani B, Tekin M, Mahdieh N. The promise of whole-exome sequencing in medical genetics. J Hum Genet. 2014;59:5–15. doi: 10.1038/jhg.2013.114. [DOI] [PubMed] [Google Scholar]
- 21.Miyatake S, Matsumoto N. Genetics: Clinical exome sequencing in neurology practice. Nat Rev Neurol. 2014;10:676–678. doi: 10.1038/nrneurol.2014.213. [DOI] [PubMed] [Google Scholar]
- 22.Gomez CM, Das S. Clinical exome sequencing: the new standard in genetic diagnosis. JAMA Neurol. 2014;71:1215–1216. doi: 10.1001/jamaneurol.2014.2015. [DOI] [PubMed] [Google Scholar]
- 23.Lu JT, Campeau PM, Lee BH. Genotype-phenotype correlation--promiscuity in the era of next-generation sequencing. N Engl J Med. 2014;371:593–596. doi: 10.1056/NEJMp1400788. [DOI] [PubMed] [Google Scholar]
- 24.Fogel BL, Lee H, Deignan JL, Strom SP, Kantarci S, Wang X, Quintero-Rivera F, Vilain E, Grody WW, Perlman S, et al. Exome sequencing in the clinical diagnosis of sporadic or familial cerebellar ataxia. JAMA Neurol. 2014;71:1237–1246. doi: 10.1001/jamaneurol.2014.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerreiro R, Bras J, Hardy J, Singleton A. Next generation sequencing techniques in neurological diseases: redefining clinical and molecular associations. Hum Mol Genet. 2014;23:R47–53. doi: 10.1093/hmg/ddu203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava S, Cohen JS, Vernon H, Baranano K, McClellan R, Jamal L, Naidu S, Fatemi A. Clinical whole exome sequencing in child neurology practice. Ann Neurol. 2014;76:473–483. doi: 10.1002/ana.24251. [DOI] [PubMed] [Google Scholar]
- 27.Iglesias A, Anyane-Yeboa K, Wynn J, Wilson A, Truitt Cho M, Guzman E, Sisson R, Egan C, Chung WK. The usefulness of whole-exome sequencing in routine clinical practice. Genet Med. 2014;16:922–931. doi: 10.1038/gim.2014.58. [DOI] [PubMed] [Google Scholar]
- 28.Shiang R, Thompson LM, Zhu YZ, Church DM, Fielder TJ, Bocian M, Winokur ST, Wasmuth JJ. Mutations in the transmembrane domain of FGFR3 cause the most common genetic form of dwarfism, achondroplasia. Cell. 1994;78:335–342. doi: 10.1016/0092-8674(94)90302-6. [DOI] [PubMed] [Google Scholar]
- 29.Rousseau F, Bonaventure J, Legeai-Mallet L, Pelet A, Rozet JM, Maroteaux P, Le Merrer M, Munnich A. Mutations in the gene encoding fibroblast growth factor receptor-3 in achondroplasia. Nature. 1994;371:252–254. doi: 10.1038/371252a0. [DOI] [PubMed] [Google Scholar]
- 30.Campbell IM, Yuan B, Robberecht C, Pfundt R, Szafranski P, McEntagart ME, Nagamani SC, Erez A, Bartnik M, Wisniowiecka-Kowalnik B, et al. Parental somatic mosaicism is underrecognized and influences recurrence risk of genomic disorders. Am J Hum Genet. 2014;95:173–182. doi: 10.1016/j.ajhg.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebberink MS, Csanyi B, Chong WK, Denis S, Sharp P, Mooijer PA, Dekker CJ, Spooner C, Ngu LH, De Sousa C, et al. Identification of an unusual variant peroxisome biogenesis disorder caused by mutations in the PEX16 gene. J Med Genet. 2010;47:608–615. doi: 10.1136/jmg.2009.074302. [DOI] [PubMed] [Google Scholar]
- 32.Levenson D. Whole-exome sequencing emerges as clinical diagnostic tool: testing method proves useful for diagnosing wide range of genetic disorders. Am J Med Genet A. 2014;164A:ix–x. doi: 10.1002/ajmg.a.36385. [DOI] [PubMed] [Google Scholar]
- 33.van Zelst-Stams WA, Scheffer H, Veltman JA. Clinical exome sequencing in daily practice: 1,000 patients and beyond. Genome Med. 2014;6:2. doi: 10.1186/gm521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krumm N, O'Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arboleda VA, Lee H, Dorrani N, Zadeh N, Willis M, Macmurdo CF, Manning MA, Kwan A, Hudgins L, Barthelemy F, et al. De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay. Am J Hum Genet. 2015;96:498–506. doi: 10.1016/j.ajhg.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O'Daniel JM, Ormond KE, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med. 2015;17:68–69. doi: 10.1038/gim.2014.151. [DOI] [PubMed] [Google Scholar]
- 38.Fogel BL. Interpretation of genetic testing: variants of unknown significance. Continuum. 2011;17:347–352. doi: 10.1212/01.CON.0000396975.87637.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 40.Gray VE, Kukurba KR, Kumar S. Performance of computational tools in evaluating the functional impact of laboratory-induced amino acid mutations. Bioinformatics. 2012;28:2093–2096. doi: 10.1093/bioinformatics/bts336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castellana S, Mazza T. Congruency in the prediction of pathogenic missense mutations: state-of-the-art web-based tools. Brief Bioinform. 2013;14:448–459. doi: 10.1093/bib/bbt013. [DOI] [PubMed] [Google Scholar]
- 42.Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 43.Thomas JB, Brier MR, Bateman RJ, Snyder AZ, Benzinger TL, Xiong C, Raichle M, Holtzman DM, Sperling RA, Mayeux R, et al. Functional connectivity in autosomal dominant and late-onset Alzheimer disease. JAMA Neurol. 2014;71:1111–1122. doi: 10.1001/jamaneurol.2014.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naj AC, Jun G, Reitz C, Kunkle BW, Perry W, Park YS, Beecham GW, Rajbhandary RA, Hamilton-Nelson KL, Wang LS, et al. Effects of multiple genetic loci on age at onset in late-onset Alzheimer disease: a genome-wide association study. JAMA Neurol. 2014;71:1394–1404. doi: 10.1001/jamaneurol.2014.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Al-Chalabi A, Fang F, Hanby MF, Leigh PN, Shaw CE, Ye W, Rijsdijk F. An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry. 2010;81:1324–1326. doi: 10.1136/jnnp.2010.207464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keller MF, Ferrucci L, Singleton AB, Tienari PJ, Laaksovirta H, Restagno G, Chio A, Traynor BJ, Nalls MA. Genome-wide analysis of the heritability of amyotrophic lateral sclerosis. JAMA Neurol. 2014;71:1123–1134. doi: 10.1001/jamaneurol.2014.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fogh I, Ratti A, Gellera C, Lin K, Tiloca C, Moskvina V, Corrado L, Soraru G, Cereda C, Corti S, et al. A genome-wide association meta-analysis identifies a novel locus at 17q11.2 associated with sporadic amyotrophic lateral sclerosis. Hum Mol Genet. 2014;23:2220–2231. doi: 10.1093/hmg/ddt587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andreassen OA, Harbo HF, Wang Y, Thompson WK, Schork AJ, Mattingsdal M, Zuber V, Bettella F, Ripke S, Kelsoe JR, et al. Genetic pleiotropy between multiple sclerosis and schizophrenia but not bipolar disorder: differential involvement of immune-related gene loci. Mol Psychiatry. 2014;20:207–14. doi: 10.1038/mp.2013.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guipponi M, Santoni FA, Setola V, Gehrig C, Rotharmel M, Cuenca M, Guillin O, Dikeos D, Georgantopoulos G, Papadimitriou G, et al. Exome sequencing in 53 sporadic cases of schizophrenia identifies 18 putative candidate genes. PLoS One. 2014;9:e112745. doi: 10.1371/journal.pone.0112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruderfer DM, Lim ET, Genovese G, Moran JL, Hultman CM, Sullivan PF, McCarroll SA, Holmans P, Sklar P, Purcell SM. No evidence for rare recessive and compound heterozygous disruptive variants in schizophrenia. Eur J Hum Genet. 2014;23:555–557. doi: 10.1038/ejhg.2014.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rudd DS, Axelsen M, Epping EA, Andreasen NC, Wassink TH. A genome-wide CNV analysis of schizophrenia reveals a potential role for a multiple-hit model. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:619–626. doi: 10.1002/ajmg.b.32266. [DOI] [PubMed] [Google Scholar]
- 52.Kauppi K, Westlye LT, Tesli M, Bettella F, Brandt CL, Mattingsdal M, Ueland T, Espeseth T, Agartz I, Melle I, et al. Polygenic Risk for Schizophrenia Associated With Working Memory-related Prefrontal Brain Activation in Patients With Schizophrenia and Healthy Controls. Schizophr Bull. 2014;41:736–743. doi: 10.1093/schbul/sbu152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shashi V, McConkie-Rosell A, Rosell B, Schoch K, Vellore K, McDonald M, Jiang YH, Xie P, Need A, Goldstein DB. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet Med. 2014;16:176–182. doi: 10.1038/gim.2013.99. [DOI] [PubMed] [Google Scholar]
- 54.Xue Y, Ankala A, Wilcox WR, Hegde MR. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med. 2015;17:444–451. doi: 10.1038/gim.2014.122. [DOI] [PubMed] [Google Scholar]
- 55.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics. 2013;14(Suppl 11):S1. doi: 10.1186/1471-2105-14-S11-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Retterer K, Scuffins J, Schmidt D, Lewis R, Pineda-Alvarez D, Stafford A, Schmidt L, Warren S, Gibellini F, Kondakova A, et al. Assessing copy number from exome sequencing and exome array CGH based on CNV spectrum in a large clinical cohort. Genet Med. 2014 doi: 10.1038/gim.2014.160. [DOI] [PubMed] [Google Scholar]
- 61.Fogel BL, Vickrey BG, Walton-Wetzel J, Lieber E, Browner CH. Utilization of genetic testing prior to subspecialist referral for cerebellar ataxia. Genet Test Mol Biomarkers. 2013;17:588–594. doi: 10.1089/gtmb.2013.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farwell KD, Shahmirzadi L, El-Khechen D, Powis Z, Chao EC, Davis BT, Baxter RM, Zeng W, Mroske C, Parra MC, et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med. 2014 doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]