Abstract
Adaptation is essential for maximizing cell survival and for cell fitness in response to sudden changes in the environment. Several aspects of cell physiology change during adaptation. Major changes in gene expression are associated with cell exposure to environmental changes, and several aspects of mRNA biogenesis appear to be targeted by signaling pathways upon stress. Exhaustive reviews have been written regarding adaptation to stress and regulation of gene expression. In this review, using osmostress in yeast as a prototypical case study, we highlight those aspects of regulation of gene induction that are general to various environmental stresses as well as mechanistic aspects that are potentially conserved from yeast to mammals.
Keywords: gene expression, Hog1, mammals, osmostress, p38, SAPK, stress responses, yeast
Abbreviations
- lncRNA
long non‐coding RNA
- MAPK
mitogen‐activated protein kinase
- RSC
chromatin structure remodeling complex
- SAPK
stress‐activated protein kinase
Stress responses
Environmental stresses include alterations in temperature, pH or oxygen concentration as well as nutrient deprivation, radiation or increases in the extracellular osmolarity. A critical property of living cells is their ability to sense and robustly respond to these fluctuations in their environment in order to maintain cellular functions. Thus, the immediate response to these stresses is crucial for cell survival, and is tightly controlled by multiple mechanisms. A common feature of many stress responses is the regulation of gene expression. Although the immediate role of gene expression in rapid adaptation is still being debated, it is generally held that gene expression is important for long‐term adaptation to stress and for protection against future stress 1, 2, 3, 4, 5.
Changes in the environment trigger a large common transcriptional response, called the environmental stress response, in budding yeast (Saccharomyces cerevisiae), which is characterized by re‐direction of resources from rapid proliferation to stress protection. In the environmental stress response, stress‐induced genes include genes for defense against reactive oxygen species and DNA damage, carbohydrate metabolism and energy generation functions, whereas most stress‐repressed genes have growth‐related functions, such as translation and ribosome biogenesis 6, 7, 8. Although environmental stress responses have common features, some aspects of the responses are unique to each individual stress. For instance, osmolyte‐synthesizing enzymes are specifically induced upon osmostress 9, 10. Transcriptional stress responses are sophisticated and fine‐tuned; the magnitude and duration of the response is proportional to the severity of the perturbation, and different perturbations result in distinct expression signatures 7. Stress responses are usually repressed or under tight control under non‐induced conditions. Indeed, cells that display an increased basal transcription in response to environmental stress in the absence of stress show reduced fitness and slow growth phenotypes 11. Correspondingly, sustained activation of stress responses leads to detrimental cellular growth or apoptosis 12. This review highlights some of the emerging principles underlying the molecular events that trigger gene expression in response to osmostress, which is used as an example of a transcriptionally regulated response.
Signal transduction and the regulation of gene expression
Stressors activate a complex network of sensors and effectors from multiple signaling pathways that coordinate the required adaptive responses. An enormous effort has been made to understand the properties of these signaling pathways. One of the most intensely studied pathways is the high osmolarity glycerol (HOG) pathway, which was discovered 20 years ago, and is composed of membrane‐associated osmosensors, an intracellular signaling pathway whose core is the Hog1 stress‐activated protein kinase (SAPK), and cytoplasmic and nuclear effectors. Several studies have greatly advanced our understanding of the details of the architecture of the HOG pathway, and its specific regulatory properties have been reviewed elsewhere 10, 13, 14, 15, 16. The emphasis here is on transcriptional mechanisms regulated by the Hog1 SAPK, for which emerging data are providing an understanding of how a signaling pathway rapidly, accurately and efficiently fine‐tunes the full balance of a transcriptional program in response to extracellular stimuli 3, 6, 17, 18. As outlined below, this research in yeast is uncovering conserved principles in multicellular organisms that underlie regulatory strategies in response to changing environments.
The systems dynamics of the HOG signaling pathway have been recently studied by monitoring the behavior of single cells under controlled environmental conditions 19, 20, 21. Signaling increases linearly, while transcription appears to be bimodal at low osmolarities. The transcriptional outcome measured by the expression of a fluorescent reporter under weak osmostress conditions is not continuously distributed, as seen by the presence of two distinct cell sub‐populations: cells that are non‐responsive and cells that fully express an osmoresponsive gene in response to identical Hog1 activation. In contrast, under stronger stress conditions, populations respond homogeneously (Fig. 1A) 22. Chromatin structure appears to be one of the determinants of this stochastic expression, as a number of mutants in chromatin remodelers display altered bimodal responses (e.g. gcn5 or rsc9ts) even at concentrations at which the transcriptional output of wild‐type cells is uniform 22. In addition, histone eviction (measured at the population level) at osmoresponsive genes is partial upon weak stress, suggesting that only a fraction of the population of cells remodels chromatin to allow efficient transcription. Interestingly, bimodal expression is not specific to osmostress, as oxidative and heat stress also result in bimodal expression, suggesting that bimodal behavior may be a general feature of stress‐induced genes 22. Based on these observations, several models have been developed to identify and predict the transcriptional output of osmostress stochastic gene regulation 23, 24, 25. Thus, single‐cell approaches may provide unbiased quantification of signaling/transcriptional outcome processes, and such approaches will be a powerful tool for defining new signaling features in response to stress in any type of cell and signaling pathway.
Figure 1.
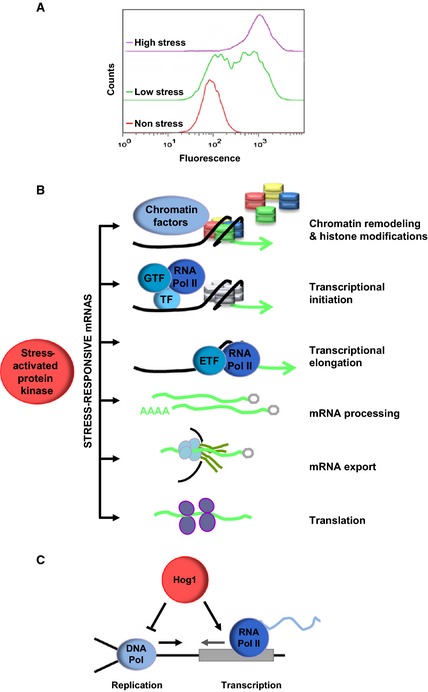
Stress‐induced gene expression by Hog1. (A) Schematic graph of bimodal expression of a fluorescent osmoresponsive reporter under low (0.1 m NaCl), high (0.4 m NaCl) and non‐osmostress conditions, measured by single‐cell flow cytometry. (B) Control of stress‐responsive mRNA biogenesis by the Hog1 SAPK. Once activated, Hog1 associates with stress‐responsive loci to modulate chromatin remodeling and histone modifications, transcriptional initiation and elongation. Hog1 also participates in mRNA processing, mRNA export and translation. (C) The Hog1 SAPK coordinates transcription with DNA replication. Upon osmostress, the activated Hog1 activates transcription and delays DNA replication by phosphorylating Mrc1. This coordinated regulation of the two processes prevents collisions between transcription and replication machineries and protects genomic integrity.
Global impact of stress on gene expression
Genome‐wide studies have provided a global view of gene expression in response to osmostress. By following the kinetics of transcription over time in cells subjected to osmostress, a broader role of Hog1 as a master regulator of the massive transcriptional reprogramming that occurs was confirmed, and a relationship between the level of gene induction and dependence on the HOG pathway was suggested 7, 26, 27, 28.
Recently, high resolution genome‐wide approaches such ChIP‐seq and tiling arrays have been instrumental in revealing the localization of the Hog1 SAPK, and of key components that drive osmoresponsive transcription, as well as in detection of new transcripts and complex transcriptional architectures 29, 30. In parallel with the induction of stress‐responsive genes, a general phenomenon that occurs upon stress is a major down‐regulation of general transcription that is possibly caused by a high rate of eviction of general transcription factors from chromatin caused by the osmotic imbalance 31. In this scenario, Hog1 specifically targets RNA polymerase II to stress‐responsive genes in order to bypass this general down‐regulation of gene expression. This activity of Hog1 results in redistribution of RNA polymerase II away from other genes and towards those stress‐responsive genes. Studies of Hog1 and RNA polymerase II co‐localization showed that stronger binding of these proteins to chromatin positively correlated with maximal gene expression. Thus, it appears that there is a dedicated mechanism controlled by Hog1 that specifically targets gene induction under globally repressive conditions.
Association of signaling kinases with chromatin
Regulation of gene expression is a consequence of very fast signal propagation in response to extracellular stimuli. As mentioned above, a key observation that led to an understanding of how signaling translates into gene regulation was the observation that Hog1 associates with chromatin upon stress 32. Additional mitogen‐activated protein kinases in yeast such as Fus3, Kss1 and Mpk1 have also been found to associate with chromatin 33, 34. Similarly, structurally and functionally unrelated signaling kinases, including Snf1 35, Tor1 36 and protein kinase A 33, have been reported to be recruited to chromatin. These findings suggest that kinases function as integral components of the transcriptional regulatory machinery.
In mammals, the Hog1 ortholog p38 also associates with chromatin 37, 38, as do other mitogen‐activated protein kinases (MAPKs) such as extracellular signal‐regulated kinase and c‐Jun N‐terminal kinase 39, 40, 41, 42. Remarkably, the chromatin association of signaling components is not restricted to such kinases, but in some cases also includes their upstream activating kinases, such as mitogen‐activated protein kinase 6 and meiotic kinase 1, or their downstream target kinases such as Msk1, or even their regulatory protein phosphatases (e.g. mitogen‐activated protein (MAP) kinase phosphatase 1 phosphatase or calcineurin) 43, 44. These findings suggest a general model by which signaling complexes and their regulatory elements associate with chromatin as an additional layer of gene expression regulation.
Role of chromatin in gene regulation
Increased association of Hog1 with stress‐responsive genes strongly correlated with chromatin remodeling and increased gene expression. Genome‐wide nucleosome positioning by MNase‐Seq analysis showed that, whereas genome‐wide chromatin structure is not significantly altered upon stress, there is strong chromatin remodeling at stress‐responsive genes that display Hog1 association 29. The possibility that chromatin remodeling is an essential step for proper adaptation to extracellular stimuli is suggested by the fact that a number of mutants in the chromatin structure remodeling complex (RSC) are osmosensitive. Hog1 targets the RSC chromatin remodeling complex to stress‐responsive genes, and RSC‐deficient cells display reduced osmostress induction of gene expression 45. External stimuli induce changes in genome‐wide RSC occupancy that correlate with the induction or repression of specific families of genes that are regulated by stress 45, 46, 47. In contrast, the INO80 chromatin remodeling complex appears to be important for reassembly of chromatin during adaptation 48. The dynamic patterns of nucleosome positioning are central to repression of gene expression in the absence of stress and to establish a threshold for gene induction upon Hog1 activation 22.
In addition to chromatin remodeling, chromatin at stress‐responsive genes is subjected to a number of modifications that establish a new landscape of histone marks 17. A few studies have shown that, upon stress, several chromatin‐modifying enzymes such as Spt‐Ada‐Gcn5‐acetyl transferase complex (SAGA) histone deacetylases and methylases have an impact on stress‐responsive gene expression 49, 50, 51. For instance, the Rpd3 histone deacetylase is important for gene induction not only in response to osmostress but also other stresses 52, 53. Also, methylation of the histone H3K4 by the histone methyltransferase Set1 determines whether RSC or Swr1 complexes remodel nucleosomes at stress‐responsive loci upon osmostress 54. This observation suggests a new function for H3K4 monomethylation in governing the selectivity of chromatin remodelers for stress‐responsive genes. Overall, based on examples of histone‐modifying enzymes required for stress‐responsive genes, it may be concluded that stress genes may be controlled in a manner that is distinct from the control of other housekeeping genes 3, 41, 55, 56. Such control may be necessary due to the specific dynamics of induction and repression of these genes.
Multiple controls in mRNA biogenesis
Control of mRNA biogenesis may be exerted at various levels. Hog1 association with stress‐responsive loci indicates that it has an intimate relationship with the transcriptional machinery. Indeed, Hog1 modulates several key steps during mRNA biogenesis (Fig. 1B and Table 1). Thus, initially Hog1 regulates initiation of transcription by (a) direct phosphorylation of specific osmostress transcription factors 57, 58, (b) recruitment of RNA polymerase II and coactivators to osmoresponsive promoters 50, 59, and (c) recruitment of chromatin modification and remodeling activities 29, 45, 51, 60. Hog1 also binds to the coding regions of osmoresponsive genes, where it acts as a transcription elongation factor specific for stress 61. Remarkably, as described below, the role of Hog1 is not restricted to mRNA synthesis but is also important in modulation of additional aspects of mRNA regulation such as mRNA stability 62, 63, mRNA export 64 and translation 65 in response to stress.
Table 1.
Factors involved in mRNA biogenesis and cell‐cycle progression regulated by the Hog1 SAPK
| Factor | Reference |
|---|---|
| mRNA biogenesis factors | |
| RSC | 45 |
| Swr1 chromatin remodeling complex | 54 |
| SWI/SNF chromatin remodeling complex | 49 |
| SAGA | 49, 50 |
| Rpd3 histone deacetylases | 51 |
| Set1 histone methylases | 54 |
| Smp1 transcription factor | 57 |
| Sko1 transcription factor | 58 |
| Hot1 transcription factor | 59 |
| Ubp3 ubiquitin protease | 60 |
| Nup1, Nup2 and Nup60 nuclear pore proteins | 64 |
| Rck2 kinase | 65, 73 |
| Cell‐cycle regulators | |
| Sic1 CDK inhibitor | 80 |
| Whi5 and Msa1 transcription factors | 82 |
| Hsl1 kinase | 85 |
| Mediator of replication checkpoint protein Mrc1 | 88 |
Changes in mRNA levels in response to environmental fluctuations are influenced not only by mRNA synthesis but also by mRNA degradation rates. The use of genomic run‐on and dynamic transcription analysis assays has revealed that mRNA stability contributes specifically and dynamically to regulation of the gene expression program in response to osmostress 62, 63, 66. Several recent reports have pointed out that the gene expression process may be ‘circular’, with the initial and final stages of the process being connected and mRNA levels being buffered by compensatory changes in mRNA synthesis and decay rates 67, 68. If this is the case in response to osmostress, there should be a transient link between transcription and mRNA degradation, and such a link may be controlled by the Hog1 SAPK.
Export of mRNAs from the nucleus is critical for efficient mRNA translation. In the initial minutes after stress application, general mRNA export appears to be slightly impaired, but stress‐responsive mRNAs are still exported to the cytoplasm. Hog1 associates with components of the nuclear pore complex, and directly phosphorylates the Nup1, Nup2 and Nup60 components of the inner nuclear basket 64, suggesting that proper mRNA biogenesis of stress‐responsive genes requires coordination of the action of synthesis and export machineries by the Hog1 SAPK.
Upon stress, there is a transient decrease in protein synthesis that is caused by a decrease in amino acid uptake, repression of ribosomal protein gene expression, and a decrease in translation efficiency 69, 70, 71. The Hog1 SAPK does not appear to be involved in the initial inhibition of translation, but rather in reactivation of translation under stress, which functions as an adaptation mechanism 63, 70, 72. The cytoplasmic Rck2 kinase, which is structurally homologous to mammalian calmodulin kinases, is directly phosphorylated and regulated by Hog1, and appears to be important for the regulation of translation 65, 73.
Overall, a global picture is emerging in which, upon stress, RNA polymerase II accumulates at stress‐responsive loci but its global accumulation at housekeeping gene loci is reduced. At the same time, Hog1 controls several aspects of mRNA biogenesis and protein translation of stress‐responsive genes. This suggests the presence of a dedicated machinery for response to stress that is rapidly assembled to maximize the ability of cells to survive a sudden change in extracellular osmolarity. Similar dedicated processes may be assembled by other signaling kinases when cells are challenged with environmental changes that pose a risk to their survival.
Beyond mRNAs: lncRNAs
As described above, mRNA biogenesis is strongly regulated upon stress. However, the increased sensitivity and coverage of recent analyses have greatly expanded our understanding of the role of Hog1 in additional processes related to mRNA expression. For instance, new transcriptional roles have been identified for the Hog1 SAPK, such as modulation of RNA polymerase III‐dependent genes 29, and the regulation of a novel class of functional long non‐coding RNAs in response to osmostress 74.
Long non‐coding RNAs (lncRNAs, > 200 nt long) have been identified in virtually all studied organisms regardless of genome size or complexity. However, our understanding of their properties is frequently only descriptive (i.e. size, stability and genomic location), and in most cases their function has not been assessed 75. Expression of hundreds of lncRNAs is induced by the Hog1 SAPK upon osmostress 74. The induction of a large number of lncRNAs upon stress is an additional layer of regulatory control that may affect gene expression and translation as well as enzyme function. For example, one gene that expresses a Hog1‐dependent lncRNA in an antisense orientation is CDC28, which encodes the cyclin‐dependent kinase 1 that controls the cell cycle in yeast. Cdc28 lncRNA mediates the induction of CDC28 expression, and this increase in the level of Cdc28 results in more efficient re‐entry of the cells into the cell cycle after stress. Thus, control of lncRNA expression is a mechanism for the regulation of cell‐cycle progression in response to environmental stress 76. Therefore, genome‐wide expression and association analysis is expected to have a profound impact on the identification of new roles for SAPKs and signaling kinases in general in the regulation of gene expression.
Coordinated control of replication and transcription
The observed dramatic changes in gene expression that are mediated by Hog1 upon stress are coincident with a delay in cell‐cycle progression. Hog1 regulates multiple stages of the cell cycle by acting on core components of the cell‐cycle machinery 77, 78, 79 (Table 1). For instance, Hog1 controls the G1/S transition by down‐regulating cyclin expression and stabilizing expression of the Sic1 cyclin‐dependent kinase inhibitor 80, 81, 82. Similarly in mammals, p38 down‐regulates cyclin expression and phosphorylates the p57 cyclin‐dependent kinase inhibitor during G1 in response to osmostress 83, 84. Cells that are unable to delay cell‐cycle progression upon osmostress display reduced viability under those conditions, both in yeast and mammals 80, 83. Thus, the regulation of cell‐cycle progression is critical for maximization of cell survival upon stress. The Hog1 and p38 SAPKs are not only important for regulation of the G1/S transition but also regulate other phases of the cell cycle such as the G2/M transition in response to stress 77, 85, suggesting that, in the presence of stress, cells need to delay the cell cycle to permit generation of adaptive responses before progressing into the next phase of the cell cycle.
Coordination between the cell cycle and transcription is even more necessary during S phase, where transcription needs to be spatially and temporally coordinated with DNA replication to prevent collisions between the transcription and replication machineries. Cells have evolved a number of mechanisms to ensure that both processes are compatible under normal growth conditions 86. When yeast cells are stressed during S phase, Hog1 promotes gene induction, and, remarkably, also delays replication 87. Hog1 affects early origin firing and fork progression by directly targeting Mrc1, a protein that links the Cdc45 helicase with DNA polymerase 88. By delaying replication, Hog1 plays a key role in preventing conflicts between RNA and DNA polymerases (Fig. 1C). The phosphorylation of Mrc1 may be relevant not only for responding to osmostress, but also for coordination of DNA replication with any induced outburst of gene expression that occurs during S phase 77. Thus, cells activate checkpoint surveillance mechanisms in response to extracellular stimuli to modulate cell‐cycle progression and to permit adaptation to changing environmental conditions.
Conservation of the regulation of gene expression between yeast and mammals
The entire HOG SAPK pathway is conserved in higher eukaryotes, including humans, with the mammalian p38 SAPK being the structural and functional homolog of the yeast Hog1 SAPK 89, 90. p38 plays a key role not only in regulation of cellular responses to many types of stresses, but also in the regulation of proliferation, differentiation, survival and development of specific cell types 91. A large body of evidence over recent years has highlighted that abnormalities in this pathway trigger pathological conditions, such as cancer, inflammation‐related diseases and metabolic dysregulation 92, 93. Thus, we anticipate that the regulatory functions as well as the mechanisms of action of the HOG SAPK that have been identified in yeast may be relevant to understanding diseases related to SAPKs in humans.
It is clear that the p38 pathway has a pivotal role in stress‐induced transcriptional responses. For instance, expression of between 60 and 88% of the genes induced in response to three distinct stress stimuli (tumor necrosis factor α, high osmolarity, and the protein synthesis inhibitor anisomycin) was dependent on the p38 SAPK 94. Gene expression profiles changed, and the dependence on p38 decreased over time with stress 94, 95, 96, underlying the importance of p38 for the early transcriptional responses to stress. Indeed, the stress‐induced genes were clearly enriched in transcription factors, suggesting that cells require an extensive program of gene expression for long‐term adaptation to stress. Remarkably, only a small core set of key genes are commonly activated by all stresses. In addition, there are also differences in gene expression between cell types. Thus, it will be essential to elucidate gene expression patterns in specific cell types and in response to various stresses in order to fully understand the regulatory role of p38.
How does p38 regulate a gene expression program to allow cells to respond to specific stimuli? There has been intense research effort to answer this question 42, 97, 98. Multiple mechanisms of transcriptional regulation have been described, focusing specifically on p38 and based on its homology with Hog1. As mentioned above, p38 associates with chromatin. p38 is recruited to chromatin via its interaction with specific transcription factors to ensure its specific localization in response to a stimulus 99, 100. Such binding of p38 allows recruitment of the RNA polymerase II machinery to trigger gene expression initiation 38. Active p38 is also found within the coding region of the target genes and beyond, suggesting that p38 (similarly to Hog1 in yeast) travels with the RNA polymerase II machinery. How the p38 kinase is recruited to the stress‐activated gene bodies, and what its mechanism of action is in transcriptional elongation, remain unclear in mammals. p38 also directly phosphorylates several transcription factors as well as chromatin modification and remodeling factors, resulting in alteration of their protein stability or localization, or their affinity for DNA and protein partners. For example, p38 targets the myocyte enhancer factor 2D transcription factor by phosphorylation to allow recruitment of the myeloid/lymphoid or mixed‐lineage leukemia‐like protein methyltransferase complex, which trimethylates K4 of histone 3, to muscle‐specific promoters during myoblast differentiation 101. Another example is BAF60c, a subunit of the SWI/SNF complex, which is directly phosphorylated by p38 to allow chromatin remodeling and transcriptional activation of muscle‐specific promoters 102. Although our knowledge regarding the repertoire of p38 substrates related to transcription has increased over the years 98, it is clear that many unidentified p38 substrates remain.
Beyond mRNA biogenesis, p38 has also been found to regulate mRNA stability and translation 103, 104. p38 regulates the stability of various cytokine mRNAs, which involves mRNA elements such as AU‐rich motifs, as well as mRNA‐binding proteins such as tristetraprolin, HuR and hnRNPK homology‐type splicing regulatory protein. For instance, kinases downstream of p38 regulate the stability of proinflammatory cytokine transcripts by phosphorylation of AU‐rich element‐binding proteins that interact with AU‐rich elements in cytokine mRNA 3’ UTRs 105. Also, p38 acts on some mRNAs through regulation of the RNA binding protein HuR 106, 107, 108. In addition to control of mRNA stability, p38 activation leads to rapid adjustment of protein synthesis. A known direct link of p38 to the translation machinery is the MAPK signal‐integrating kinase Mnk. Upon activation by p38 MAPK, Mnk1 binds to eukaryotic initiation factor 4G and catalyzes the phosphorylation of eukaryotic initiation factor 4E 109, 110. All of these data clearly suggest that p38, similar to Hog1, is able to modulate several steps in mRNA biogenesis to modulate gene expression.
In summary, there is no doubt that SAPKs plays a central role in the regulation of chromatin and transcription in response to a stimulus. Using both genome‐wide scale and context‐specific approaches, together with mathematical modeling and single‐cell analysis, it is expected that in the coming years we will learn more about how these SAPK pathways regulate gene expression, as well as the special characteristics of stress‐responsive genes.
Author contributions
EdN and FP wrote the manuscript.
Acknowledgements
The laboratory of F.P. and E.N. is supported by grants from the Spanish Ministry of Economy and Competitiveness (BFU2012‐33503 and FEDER, BFU2014‐52125‐REDT and BFU2014‐51672‐REDC to F.P. and BFU2014‐52333‐P to E.N.), the Catalan Government (2014 SGR 599) and an ERC Advanced Grant (number 294294) from the EU seventh framework program (SYNCOM) to F.P. This project is supported by the Fundación Botín and by Banco Santander through its Santander Universities Global Division to F.P. F.P. and E.N. are recipients of an ICREA Acadèmia (Generalitat de Catalunya).
References
- 1. Berry DB & Gasch AP (2008) Stress‐activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell 19, 4580–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Westfall PJ, Patterson JC, Chen RE & Thorner J (2008) Stress resistance and signal fidelity independent of nuclear MAPK function. Proc Natl Acad Sci USA 105, 12212–12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Nadal E, Ammerer G & Posas F (2011) Controlling gene expression in response to stress. Nat Rev Genet 12, 833–845. [DOI] [PubMed] [Google Scholar]
- 4. Bouwman J, Kiewiet J, Lindenbergh A, van EK, Siderius M & Bakker BM (2011) Metabolic regulation rather than de novo enzyme synthesis dominates the osmo‐adaptation of yeast. Yeast, 28, 43–53. [DOI] [PubMed] [Google Scholar]
- 5. Babazadeh R, Adiels CB, Smedh M, Petelenz‐Kurdziel E, Goksor M & Hohmann S (2013) Osmostress‐induced cell volume loss delays yeast Hog1 signaling by limiting diffusion processes and by Hog1‐specific effects. PLoS One 8, e80901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martinez‐Montanes F, Pascual‐Ahuir A & Proft M (2010) Toward a genomic view of the gene expression program regulated by osmostress in yeast. OMICS 14, 619–627. [DOI] [PubMed] [Google Scholar]
- 7. Gasch AP, Spellman PT, Kao CM, Carmel‐Harel O, Eisen MB, Storz G, Botstein D & Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11, 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lopez‐Maury L, Marguerat S & Bahler J (2008) Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet 9, 583–593. [DOI] [PubMed] [Google Scholar]
- 9. Hohmann S, Krantz M & Nordlander B (2007) Yeast osmoregulation. Methods Enzymol 428, 29–45. [DOI] [PubMed] [Google Scholar]
- 10. Saito H & Posas F (2012) Response to hyperosmotic stress. Genetics 192, 289–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Duibhir E, Lijnzaad P, Benschop JJ, Lenstra TL, van LD, Groot Koerkamp MJ, Margaritis T, Brok MO, Kemmeren P & Holstege FC (2014) Cell cycle population effects in perturbation studies. Mol Syst Biol, 10, 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vendrell A, Martinez‐Pastor M, Gonzalez‐Novo A, Pascual‐Ahuir A, Sinclair DA, Proft M & Posas F (2011) Sir2 histone deacetylase prevents programmed cell death caused by sustained activation of the Hog1 stress‐activated protein kinase. EMBO Rep 12, 1062–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gehart H, Kumpf S, Ittner A & Ricci R (2010) MAPK signalling in cellular metabolism: stress or wellness? EMBO Rep 11, 834–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Engelberg D, Perlman R & Levitzki A (2014) Transmembrane signaling in Saccharomyces cerevisiae as a model for signaling in metazoans: state of the art after 25 years. Cell Signal 26, 2865–2878. [DOI] [PubMed] [Google Scholar]
- 15. Brewster JL & Gustin MC (2014) Hog1: 20 years of discovery and impact. Sci Signal, 7, re7. [DOI] [PubMed] [Google Scholar]
- 16. Chasman D, Ho YH, Berry DB, Nemec CM, MacGilvray ME, Hose J, Merrill AE, Lee MV, Will JL, Coon JJ et al (2014) Pathway connectivity and signaling coordination in the yeast stress‐activated signaling network. Mol Syst Biol 10, 759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Nadal E & Posas F (2010) Multilayered control of gene expression by stress‐activated protein kinases. EMBO J 29, 4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weake VM & Workman JL (2010) Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet 11, 426–437. [DOI] [PubMed] [Google Scholar]
- 19. Mettetal JT, Muzzey D, Gomez‐Uribe C & van Oudenaarden A (2008) The frequency dependence of osmo‐adaptation in Saccharomyces cerevisiae. Science 319, 482–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muzzey D, Gomez‐Uribe CA, Mettetal JT & van Oudenaarden A (2009) A systems‐level analysis of perfect adaptation in yeast osmoregulation. Cell 138, 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Patterson JC, Klimenko ES & Thorner J (2010) Single‐cell analysis reveals that insulation maintains signaling specificity between two yeast MAPK pathways with common components. Sci Signal, 3, ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pelet S, Rudolf F, Nadal‐Ribelles M, de Nadal E, Posas F & Peter M (2011) Transient activation of the HOG MAPK pathway regulates bimodal gene expression. Science 332, 732–735. [DOI] [PubMed] [Google Scholar]
- 23. Neuert G, Munsky B, Tan RZ, Teytelman L, Khammash M & van Oudenaarden A (2013) Systematic identification of signal‐activated stochastic gene regulation. Science 339, 584–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zechner C, Ruess J, Krenn P, Pelet S, Peter M, Lygeros J & Koeppl H (2012) Moment‐based inference predicts bimodality in transient gene expression. Proc Natl Acad Sci USA 109, 8340–8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hao N, Budnik BA, Gunawardena J & O'Shea EK (2013) Tunable signal processing through modular control of transcription factor translocation. Science 339, 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nadal E & Arino J (2000) The transcriptional response of yeast to saline stress. J Biol Chem 275, 17249–17255. [DOI] [PubMed] [Google Scholar]
- 27. Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES & Young RA (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12, 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Rourke SM & Herskowitz I (2004) Unique and redundant roles for HOG MAPK pathway components as revealed by whole‐genome expression analysis. Mol Biol Cell 15, 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nadal‐Ribelles M, Conde N, Flores O, Gonzalez‐Vallinas J, Eyras E, Orozco M, de Nadal E & Posas F (2012) Hog1 bypasses stress‐mediated down‐regulation of transcription by RNA polymerase II redistribution and chromatin remodeling. Genome Biol 13, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cook KE & O'Shea EK (2012) Hog1 controls global reallocation of RNA Pol II upon osmotic shock in Saccharomyces cerevisiae. G3: Genes ‐ Genomes ‐ Genetics, 2, 1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Proft M & Struhl K (2004) MAP kinase‐mediated stress relief that precedes and regulates the timing of transcriptional induction. Cell 118, 351–361. [DOI] [PubMed] [Google Scholar]
- 32. Alepuz PM, Jovanovic A, Reiser V & Ammerer G (2001) Stress‐induced map kinase Hog1 is part of transcription activation complexes. Mol Cell 7, 767–777. [DOI] [PubMed] [Google Scholar]
- 33. Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB & Young RA (2006) Activated signal transduction kinases frequently occupy target genes. Science 313, 533–536. [DOI] [PubMed] [Google Scholar]
- 34. Kim KY, Truman AW & Levin DE (2008) Yeast Mpk1 mitogen‐activated protein kinase activates transcription through Swi4/Swi6 by a noncatalytic mechanism that requires upstream signal. Mol Cell Biol 28, 2579–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lo WS, Gamache ER, Henry KW, Yang D, Pillus L & Berger SL (2005) Histone H3 phosphorylation can promote TBP recruitment through distinct promoter‐specific mechanisms. EMBO J 24, 997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li H, Tsang CK, Watkins M, Bertram PG & Zheng XF (2006) Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 442, 1058–1061. [DOI] [PubMed] [Google Scholar]
- 37. Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L & Puri PL (2004) p38 pathway targets SWI‐SNF chromatin‐remodeling complex to muscle‐specific loci. Nat Genet 36, 738–743. [DOI] [PubMed] [Google Scholar]
- 38. Ferreiro I, Barragan M, Gubern A, Ballestar E, Joaquin M & Posas F (2010) The p38 SAPK is recruited to chromatin via its interaction with transcription factors. J Biol Chem 285, 31819–31828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lawrence MC, McGlynn K, Shao C, Duan L, Naziruddin B, Levy MF & Cobb MH (2008) Chromatin‐bound mitogen‐activated protein kinases transmit dynamic signals in transcription complexes in beta‐cells. Proc Natl Acad Sci USA 105, 13315–13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suganuma T, Mushegian A, Swanson SK, Abmayr SM, Florens L, Washburn MP & Workman JL (2010) The ATAC acetyltransferase complex coordinates MAP kinases to regulate JNK target genes. Cell 142, 726–736. [DOI] [PubMed] [Google Scholar]
- 41. Suganuma T & Workman JL (2012) MAP kinases and histone modification. J Mol Cell Biol 4, 348–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Klein AM, Zaganjor E & Cobb MH (2013) Chromatin‐tethered MAPKs. Curr Opin Cell Biol 25, 272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nelson JD, LeBoeuf RC & Bomsztyk K (2011) Direct recruitment of insulin receptor and ERK signaling cascade to insulin‐inducible gene loci. Diabetes 60, 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lawrence MC, Shao C, McGlynn K, Naziruddin B, Levy MF & Cobb MH (2009) Multiple chromatin‐bound protein kinases assemble factors that regulate insulin gene transcription. Proc Natl Acad Sci USA 106, 22181–22186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mas G, de Nadal E, Dechant R, Rodriguez de la Concepcion ML, Logie C, Jimeno‐Gonzalez S, Chavez S, Ammerer G & Posas F (2009) Recruitment of a chromatin remodelling complex by the Hog1 MAP kinase to stress genes. EMBO J 28, 326–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ng HH, Robert F, Young RA & Struhl K (2002) Genome‐wide location and regulated recruitment of the RSC nucleosome‐remodeling complex. Genes Dev 16, 806–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Damelin M, Simon I, Moy TI, Wilson B, Komili S, Tempst P, Roth FP, Young RA, Cairns BR & Silver PA (2002) The genome‐wide localization of Rsc9, a component of the RSC chromatin‐remodeling complex, changes in response to stress. Mol Cell 9, 563–573. [DOI] [PubMed] [Google Scholar]
- 48. Klopf E, Paskova L, Sole C, Mas G, Petryshyn A, Posas F, Wintersberger U, Ammerer G & Schuller C (2009) Cooperation between the INO80 complex and histone chaperones determines adaptation of stress gene transcription in the yeast Saccharomyces cerevisiae. Mol Cell Biol 29, 4994–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Proft M & Struhl K (2002) Hog1 kinase converts the Sko1‐Cyc8‐Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell 9, 1307–1317. [DOI] [PubMed] [Google Scholar]
- 50. Zapater M, Sohrmann M, Peter M, Posas F & de Nadal E (2007) Selective requirement for SAGA in Hog1‐mediated gene expression depending on the severity of the external osmostress conditions. Mol Cell Biol 27, 3900–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de Nadal E, Zapater M, Alepuz PM, Sumoy L, Mas G & Posas F (2004) The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427, 370–374. [DOI] [PubMed] [Google Scholar]
- 52. Ruiz‐Roig C, Vieitez C, Posas F & de Nadal E (2010) The Rpd3L HDAC complex is essential for the heat stress response in yeast. Mol Microbiol 76, 1049–1062. [DOI] [PubMed] [Google Scholar]
- 53. Alejandro‐Osorio AL, Huebert DJ, Porcaro DT, Sonntag ME, Nillasithanukroh S, Will JL & Gasch AP (2009) The histone deacetylase Rpd3p is required for transient changes in genomic expression in response to stress. Genome Biol 10, R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nadal‐Ribelles M, Mas G, Millan‐Zambrano G, Sole C, Ammerer G, Chavez S, Posas F & de Nadal E (2015) H3K4 monomethylation dictates nucleosome dynamics and chromatin remodeling at stress‐responsive genes. Nucleic Acids Res doi: 10.1093/nar/gkv220. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suganuma T & Workman JL (2013) Chromatin and signaling. Curr Opin Cell Biol 25, 322–326. [DOI] [PubMed] [Google Scholar]
- 56. Zanton SJ & Pugh BF (2006) Full and partial genome‐wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev 20, 2250–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. de Nadal E, Casadome L & Posas F (2003) Targeting the MEF2‐like transcription factor Smp1 by the stress‐activated Hog1 mitogen‐activated protein kinase. Mol Cell Biol 23, 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Proft M, Pascual‐Ahuir A, de Nadal E, Arino J, Serrano R & Posas F (2001) Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J 20, 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Alepuz PM, de Nadal E, Zapater M, Ammerer G & Posas F (2003) Osmostress‐induced transcription by Hot1 depends on a Hog1‐mediated recruitment of the RNA Pol II. EMBO J 22, 2433–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sole C, Nadal‐Ribelles M, Kraft C, Peter M, Posas F & de Nadal E (2011) Control of Ubp3 ubiquitin protease activity by the Hog1 SAPK modulates transcription upon osmostress. EMBO J 30, 3274–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K & Posas F (2006) The stress‐activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell 23, 241–250. [DOI] [PubMed] [Google Scholar]
- 62. Molin C, Jauhiainen A, Warringer J, Nerman O & Sunnerhagen P (2009) mRNA stability changes precede changes in steady‐state mRNA amounts during hyperosmotic stress. RNA 15, 600–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Romero‐Santacreu L, Moreno J, Perez‐Ortin JE & Alepuz P (2009) Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA 15, 1110–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Regot S, de Nadal E, Rodriguez‐Navarro S, Gonzalez‐Novo A, Perez‐Fernandez J, Gadal O, Seisenbacher G, Ammerer G & Posas F (2013) The Hog1 stress‐activated protein kinase targets nucleoporins to control mRNA export upon stress. J Biol Chem 288, 17384–17398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bilsland‐Marchesan E, Arino J, Saito H, Sunnerhagen P & Posas F (2000) Rck2 kinase is a substrate for the osmotic stress‐activated mitogen‐activated protein kinase Hog1. Mol Cell Biol 20, 3887–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Miller C, Schwalb B, Maier K, Schulz D, Dumcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dolken L et al (2011) Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol 7, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Haimovich G, Medina DA, Causse SZ, Garber M, Millan‐Zambrano G, Barkai O, Chavez S, Perez‐Ortin JE, Darzacq X & Choder M (2013) Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell 153, 1000–1011. [DOI] [PubMed] [Google Scholar]
- 68. Sun M, Schwalb B, Pirkl N, Maier KC, Schenk A, Failmezger H, Tresch A & Cramer P (2013) Global analysis of eukaryotic mRNA degradation reveals Xrn1‐dependent buffering of transcript levels. Mol Cell 52, 52–62. [DOI] [PubMed] [Google Scholar]
- 69. Norbeck J & Blomberg A (1998) Amino acid uptake is strongly affected during exponential growth of Saccharomyces cerevisiae in 0.7 M NaCl medium. FEMS Microbiol Lett 158, 121–126. [DOI] [PubMed] [Google Scholar]
- 70. Uesono Y & Toh E (2002) Transient inhibition of translation initiation by osmotic stress. J Biol Chem 277, 13848–13855. [DOI] [PubMed] [Google Scholar]
- 71. Garre E, Romero‐Santacreu L, De CN, Blasco‐Angulo N, Sunnerhagen P & Alepuz P (2012) Yeast mRNA cap‐binding protein Cbc1/Sto1 is necessary for the rapid reprogramming of translation after hyperosmotic shock. Mol Biol Cell 23, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Warringer J, Hult M, Regot S, Posas F & Sunnerhagen P (2010) The HOG pathway dictates the short‐term translational response after hyperosmotic shock. Mol Biol Cell 21, 3080–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Teige M, Scheikl E, Reiser V, Ruis H & Ammerer G (2001) Rck2, a member of the calmodulin‐protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc Natl Acad Sci USA 98, 5625–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nadal‐Ribelles M, Sole C, Xu Z, Steinmetz LM, de Nadal E & Posas F (2014) Control of Cdc28 CDK1 by a stress‐induced lncRNA. Mol Cell 53, 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pelechano V, Wei W, Jakob P & Steinmetz LM (2014) Genome‐wide identification of transcript start and end sites by transcript isoform sequencing. Nat Protoc 9, 1740–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sole C, Nadal‐Ribelles M, de Nadal E & Posas F (2014) A novel role for lncRNAs in cell cycle control during stress adaptation. Curr Genet. doi: 10.1007/s00294‐014‐0453‐y. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Duch A, de Nadal E & Posas F (2012) The p38 and Hog1 SAPKs control cell cycle progression in response to environmental stresses. FEBS Lett 586, 2925–2931. [DOI] [PubMed] [Google Scholar]
- 78. Cuadrado A & Nebreda AR (2010) Mechanisms and functions of p38 MAPK signalling. Biochem J 429, 403–417. [DOI] [PubMed] [Google Scholar]
- 79. Clotet J & Posas F (2007) Control of cell cycle in response to osmostress: lessons from yeast. Methods Enzymol 428, 63–76. [DOI] [PubMed] [Google Scholar]
- 80. Escote X, Zapater M, Clotet J & Posas F (2004) Hog1 mediates cell‐cycle arrest in G1 phase by the dual targeting of Sic1. Nat Cell Biol 6, 997–1002. [DOI] [PubMed] [Google Scholar]
- 81. Adrover MA, Zi Z, Duch A, Schaber J, Gonzalez‐Novo A, Jimenez J, Nadal‐Ribelles M, Clotet J, Klipp E & Posas F (2011) Time‐dependent quantitative multicomponent control of the G‐S network by the stress‐activated protein kinase Hog1 upon osmostress. Sci Signal, 4, ra63. [DOI] [PubMed] [Google Scholar]
- 82. Gonzalez‐Novo A, Jimenez J, Clotet J, Nadal‐Ribelles M, Cavero S, de Nadal E & Posas F (2015) Hog1 targets Whi5 and Msa1 transcription factors to down‐regulate cyclin expression upon stress. Mol Cell Biol, 35, 1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Joaquin M, Gubern A, Gonzalez‐Nunez D, Josue RE, Ferreiro I, de Nadal E, Nebreda AR & Posas F (2012) The p57 CDKi integrates stress signals into cell‐cycle progression to promote cell survival upon stress. EMBO J 31, 2952–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Joaquin M, Gubern A & Posas F (2012) A novel G1 checkpoint mediated by the p57 CDK inhibitor and p38 SAPK promotes cell survival upon stress. Cell Cycle 11, 3339–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Clotet J, Escote X, Adrover MA, Yaakov G, Gari E, Aldea M, de Nadal E & Posas F (2006) Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J 25, 2338–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Helmrich A, Ballarino M, Nudler E & Tora L (2013) Transcription‐replication encounters, consequences and genomic instability. Nat Struct Mol Biol 20, 412–418. [DOI] [PubMed] [Google Scholar]
- 87. Yaakov G, Duch A, Garcia‐Rubio M, Clotet J, Jimenez J, Aguilera A & Posas F (2009) The stress‐activated protein kinase Hog1 mediates S phase delay in response to osmostress. Mol Biol Cell 20, 3572–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Duch A, Felipe‐Abrio I, Barroso S, Yaakov G, Garcia‐Rubio M, Aguilera A, de Nadal E & Posas F (2013) Coordinated control of replication and transcription by a SAPK protects genomic integrity. Nature 493, 116–119. [DOI] [PubMed] [Google Scholar]
- 89. Galcheva‐Gargova Z, Derijard B, Wu IH & Davis RJ (1994) An osmosensing signal transduction pathway in mammalian cells. Science 265, 806–808. [DOI] [PubMed] [Google Scholar]
- 90. Han J, Lee JD, Bibbs L & Ulevitch RJ (1994) A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 265, 808–811. [DOI] [PubMed] [Google Scholar]
- 91. Kyriakis JM & Avruch J (2012) Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10‐year update. Physiol Rev 92, 689–737. [DOI] [PubMed] [Google Scholar]
- 92. Gerits N, Kostenko S & Moens U (2007) In vivo functions of mitogen‐activated protein kinases: conclusions from knock‐in and knock‐out mice. Transgenic Res 16, 281–314. [DOI] [PubMed] [Google Scholar]
- 93. Wagner EF & Nebreda AR (2009) Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 9, 537–549. [DOI] [PubMed] [Google Scholar]
- 94. Ferreiro I, Joaquin M, Islam A, Gomez‐Lopez G, Barragan M, Lombardia L, Dominguez O, Pisano DG, Lopez‐Bigas N, Nebreda AR et al (2010) Whole genome analysis of p38 SAPK‐mediated gene expression upon stress. BMC Genom 11, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Viemann D, Goebeler M, Schmid S, Klimmek K, Sorg C, Ludwig S & Roth J (2004) Transcriptional profiling of IKK2/NF‐kappaB‐ and p38 MAP kinase‐dependent gene expression in TNF‐alpha‐stimulated primary human endothelial cells. Blood 103, 3365–3373. [DOI] [PubMed] [Google Scholar]
- 96. Zer C, Sachs G & Shin JM (2007) Identification of genomic targets downstream of p38 mitogen‐activated protein kinase pathway mediating tumor necrosis factor‐alpha signaling. Physiol Genomics 31, 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Whitmarsh AJ (2007) Regulation of gene transcription by mitogen‐activated protein kinase signaling pathways. Biochim Biophys Acta 1773, 1285–1298. [DOI] [PubMed] [Google Scholar]
- 98. Yang SH, Sharrocks AD & Whitmarsh AJ (2013) MAP kinase signalling cascades and transcriptional regulation. Gene 513, 1–13. [DOI] [PubMed] [Google Scholar]
- 99. Edmunds JW & Mahadevan LC (2004) MAP kinases as structural adaptors and enzymatic activators in transcription complexes. J Cell Sci 117, 3715–3723. [DOI] [PubMed] [Google Scholar]
- 100. Chow CW & Davis RJ (2006) Proteins kinases: chromatin‐associated enzymes? Cell 127, 887–890. [DOI] [PubMed] [Google Scholar]
- 101. Rampalli S, Li L, Mak E, Ge K, Brand M, Tapscott SJ & Dilworth FJ (2007) p38 MAPK signaling regulates recruitment of Ash2L‐containing methyltransferase complexes to specific genes during differentiation. Nat Struct Mol Biol 14, 1150–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Forcales SV, Albini S, Giordani L, Malecova B, Cignolo L, Chernov A, Coutinho P, Saccone V, Consalvi S, Williams R et al (2012) Signal‐dependent incorporation of MyoD‐BAF60c into Brg1‐based SWI/SNF chromatin‐remodelling complex. EMBO J 31, 301–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tiedje C, Holtmann H & Gaestel M (2014) The role of mammalian MAPK signaling in regulation of cytokine mRNA stability and translation. J Interferon Cytokine Res 34, 220–232. [DOI] [PubMed] [Google Scholar]
- 104. Cargnello M & Roux PP (2011) Activation and function of the MAPKs and their substrates, the MAPK‐activated protein kinases. Microbiol Mol Biol Rev 75, 50–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sandler H & Stoecklin G (2008) Control of mRNA decay by phosphorylation of tristetraprolin. Biochem Soc Trans 36, 491–496. [DOI] [PubMed] [Google Scholar]
- 106. Farooq F, Balabanian S, Liu X, Holcik M & Mackenzie A (2009) p38 mitogen‐activated protein kinase stabilizes SMN mRNA through RNA binding protein HuR. Hum Mol Genet 18, 4035–4045. [DOI] [PubMed] [Google Scholar]
- 107. Lafarga V, Cuadrado A, Lopez dsI, Bengoechea R, Fernandez‐Capetillo O & Nebreda AR (2009) p38 Mitogen‐activated protein kinase‐ and HuR‐dependent stabilization of p21(Cip1) mRNA mediates the G1/S checkpoint. Mol Cell Biol, 29, 4341–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A & Gaestel M (2012) The p38/MK2‐driven exchange between tristetraprolin and HuR regulates AU‐rich element‐dependent translation. PLoS Genet 8, e1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shveygert M, Kaiser C, Bradrick SS & Gromeier M (2010) Regulation of eukaryotic initiation factor 4E (eIF4E) phosphorylation by mitogen‐activated protein kinase occurs through modulation of Mnk1–eIF4G interaction. Mol Cell Biol 30, 5160–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Lawson SK, Dobrikova EY, Shveygert M & Gromeier M (2013) p38alpha mitogen‐activated protein kinase depletion and repression of signal transduction to translation machinery by miR‐124 and ‐128 in neurons. Mol Cell Biol 33, 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]


