Summary
Choline is ubiquitous in marine eukaryotes and appears to be widely distributed in surface marine waters; however, its metabolism by marine bacteria is poorly understood. Here, using comparative genomics and molecular genetic approaches, we reveal that the capacity for choline catabolism is widespread in marine heterotrophs of the marine Roseobacter clade (MRC). Using the model bacterium R uegeria pomeroyi, we confirm that the bet A, bet B and bet C genes, encoding choline dehydrogenase, betaine aldehyde dehydrogenase and choline sulfatase, respectively, are involved in choline metabolism. The bet T gene, encoding an organic solute transporter, was essential for the rapid uptake of choline but not glycine betaine (GBT). Growth of choline and GBT as a sole carbon source resulted in the re‐mineralization of these nitrogen‐rich compounds into ammonium. Oxidation of the methyl groups from choline requires formyltetrahydrofolate synthetase encoded by fhs in R . pomeroyi, deletion of which resulted in incomplete degradation of GBT. We demonstrate that this was due to an imbalance in the supply of reducing equivalents required for choline catabolism, which can be alleviated by the addition of formate. Together, our results demonstrate that choline metabolism is ubiquitous in the MRC and reveal the role of Fhs in methyl group oxidation in R . pomeroyi.
Introduction
Choline is an essential constituent of eukaryotic cells where it can either be incorporated into the polar head group of the phospholipid phosphatidylcholine or sphingolipids (Ohvo‐Rekilä et al., 2002; Li and Vance, 2008). In mammals, choline plays an essential role in the transfer of methyl groups between cellular compounds and can be transformed into the neurotransmitter, acetylcholine (Ikawa and Taylor, 1973; Ueland, 2011). Choline also occurs in marine microalgae, e.g. in diatoms (Ikawa and Taylor, 1973), and a variety of coastal plants in the form of choline O‐sulfate (COS) (Catalfomo et al., 1972; Hanson and Gage, 1991; Hanson et al., 1991; 1994), and is a known osmoprotectant used by bacteria (Cánovas et al., 1996; Nau‐Wagner et al., 1999) and plants (Hanson and Gage, 1991; Hanson et al., 1991). Choline can be liberated from phosphatidylcholine through the action of phosphodiesterases which are present in the majority of plants, as well as viruses, bacteria, fungi and animals (Jenkins and Frohman, 2005). Due to its widespread occurrence in marine eukaryotes, choline appears to be ubiquitous in the marine water column, being detected in regions ranging from productive coastal waters of the English Channel to the oligotrophic North Atlantic gyre (Roulier et al., 1990; Airs and Archer, 2010).
It is known that marine bacteria can rapidly acquire choline from seawater (Kiene, 1998; Kiene et al., 1998) with the standing concentrations of choline being in the low nM range (Roulier et al., 1990; Airs and Archer, 2010). Choline, through its conversion to glycine betaine (GBT), serves as a potent osmoprotectant (Landfald and Strøm, 1986; Styrvold et al., 1986; Graham and Wilkinson, 1992; Boch et al., 1994). It is known, for example, that certain Vibrio spp. can convert choline to GBT to facilitate their survival in saline environments when they are not in association with their chosen hosts (Kapfthammer et al., 2005). In addition to being the precursor for the osmoprotectant GBT, choline is also a nutrient for bacteria. However, its catabolism in marine surface waters is not well understood (Kiene, 1998). In many bacteria, such as Sinorhizobium meliloti and Pseudomonas aeruginosa, catabolism of choline provides a growth advantage when forming close associations with their eukaryotic hosts (Smith et al., 1988; Barra et al., 2006; Sun et al., 2014).
The marine Roseobacter clade (MRC) are a monophyletic group of Alphaproteobacteria that are frequently detected during eukaryotic phytoplankton blooms and are often found in close association with a range of eukaryotic biota (González et al., 2000; Buchan et al., 2005; Porsby et al., 2008; Hahnke et al., 2013). These associations can change from a beneficial to an antagonistic relationship depending on the physiological state of either the host or the bacterium (Seyedsayamdost et al., 2011). Due to their high level of metabolic diversity and high in situ metabolic activity, the MRC plays a major role in carbon, sulfur and nitrogen cycling within dynamic coastal surface waters (González et al., 2000; Buchan et al., 2005; Moran and Miller, 2007; Chen, 2012; Lidbury et al., 2014b). MRC bacteria are also known for their competitive success (probiotic effect) against a number of marine‐associated pathogens through the production of antagonistic secondary metabolites (Porsby et al., 2008; Prado et al., 2009). It is, therefore, likely that MRC bacteria are capable of utilizing choline as an essential nutrient. Indeed, it has been reported that Phaeobacter gallaeciensis 2.10 and Phaeobacter gallaeciensis BS107, two isolates from the MRC, show weak growth on choline and its downstream metabolite, GBT (Thole et al., 2012). However, a comprehensive study of choline metabolism by members of the MRC has not been conducted.
Recent studies have revealed that methylated compounds, such as GBT (a metabolite of choline metabolism), methanol, dimethylsulfoniopropionate (DMSP) and trimethylamine (TMA), can be oxidized by members of the MRC and SAR11 clade to augment their growth on other organic substrates, and to maintain cell viability during times of carbon starvation through the generation of reducing equivalents and ATP (Sun et al., 2011; Lidbury et al., 2014b). SAR11 clade bacteria can also grow on GBT as a sole carbon source through its sequential demethylation to glycine and then pyruvate (Sun et al., 2011; Carini et al., 2013). Marine bacteria, including representatives from the MRC and SAR11 clade, lack the genes required for the oxidation of C1 groups to CO2 via the cofactor, tetrahydromethanopterin (H4MPT). It has, therefore, been proposed that in these marine heterotrophic bacteria, oxidation of the methyl groups from these compounds requires tetrahydrofolate as the cofactor (Chistoserdova, 2011; Sun et al., 2011; Chen, 2012; Lidbury et al., 2014b), involving several key enzymes, including formyltetrahydrofolate synthetase (Fhs). However, this has yet to be experimentally validated. Using comparative genomics and mutagenesis approaches, here, we studied choline metabolism in the MRC clade and investigated the role of Fhs in methyl group oxidation during choline metabolism using the model MRC bacterium Ruegeria pomeroyi.
Results
Choline and COS catabolism to GBT in R . pomeroyi requires three genes encoded by betABC
Figure 1 shows the proposed pathway for the catabolism of choline and its metabolites in the model marine bacterium, R. pomeroyi. Enzymes required for the conversion of choline and COS to GBT (encoded by the betA, betB and betC genes) were identified in R. pomeroyi using blastp analysis, and they had 69%, 70% and 75% amino acid identity, respectively, to the characterized enzymes from the terrestrial bacterium S. meliloti (Smith et al., 1988; Østerås et al., 1998; Barra et al., 2006).
Figure 1.
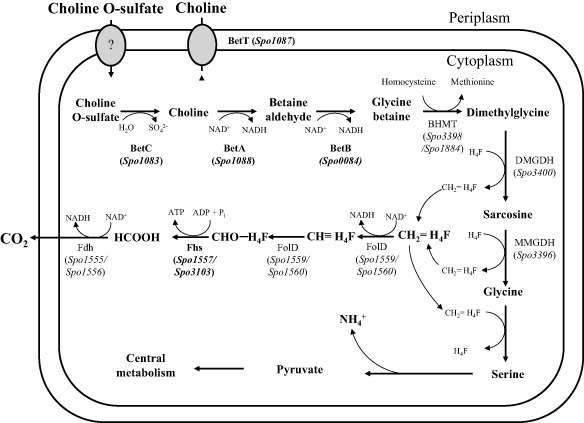
Proposed pathway of choline catabolism in R uegeria pomeroyi DSS‐3 and other related marine R oseobacter clade bacteria. HCOOH, formate; CHO‐H4F, formyl‐tetrahydrofolate; CHO = H4F, 5, 10‐methylene‐tetrahydrofolate; CHOΞH4F, 5, 10‐methenyl‐tetrahydrofolate; H4F, tetrahydrofolate; BetA, choline dehydrogenase; BetB, betaine aldehyde dehydrogenase; BetC, choline sulfatase; BetT, choline transporter; BHMT, glycine betaine: homocysteine methyltransferase; DMGDH, dimethylglycine dehydrogenase; MMGDH, sarcosine dehydrogenase; Fdh, formate dehydrogenase; FolD, 5,10‐methylene‐H4F dehydrogenase/ methenyl‐H4F cyclohydrolase; Fhs, formyl‐H4F synthetase; CO2, carbon dioxide.
To confirm that betABC is essential for growth on choline and COS in R. pomeroyi, mutants of betA, betB and betC were constructed. Wild‐type R. pomeroyi grew well on choline (μ = 0.073 ± 0.003 h−1) and GBT (μ = 0.0860 ± 0.001 h−1), and a total of 5 mM GBT or choline was completely depleted from the culture medium after 72 and 95 h respectively (Fig. 2A). Growth on COS was slightly slower (μ = 0.043 ± 0.000 h−1). However, the final OD540 was comparable to that of choline and GBT (Fig. 2A). For the mutant strains, ΔbetA::Gm, ΔbetB::Gm, growth on GBT was not affected, while growth on choline was either completely or partially inhibited. Thus, the ΔbetA::Gm mutant failed to grow on choline as a sole carbon source, and no depletion of choline in the medium was observed (Fig. 2C). The ΔbetB::Gm (Fig. 2D) mutant could grow on choline as a sole carbon source; however, the growth rate was reduced (μ = 0.047 ± 0.008 h−1) compared with that of the wild‐type. During growth experiments on choline with ΔbetB::Gm, a transient build‐up of betaine aldehyde was detected in the culture medium, which was not evident in wild‐type cultures (data not shown). R. pomeroyi has a number of genes that may encode an aldehyde dehydrogenase similar to BetB, and it is likely that one of these enzymes was able to perform the same function as BetB, albeit at a reduced efficiency. As expected, the ΔbetC::Gm mutant could still utilize choline and GBT as a sole carbon source (Fig. 2B). However, this mutant strain failed to utilize COS as a carbon source, confirming that betC is essential for growth on COS.
Figure 2.
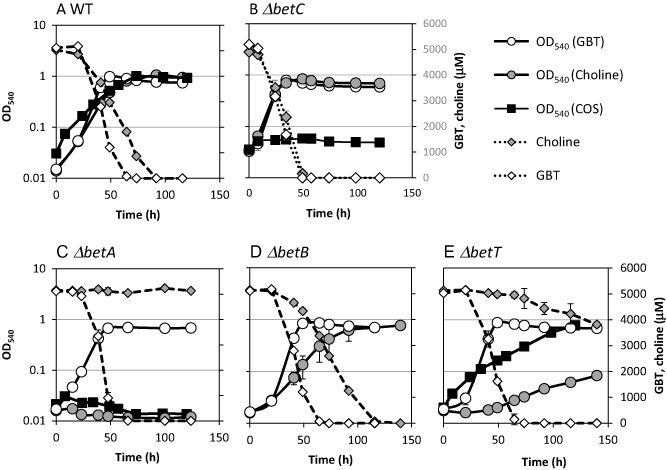
Growth of R uegeria pomeroyi (A) wild‐type (WT), (B) bet C mutant, (C) bet A mutant, (D) bet B mutant and (E) bet T mutant on either choline (grey circles), GBT (white circles) or COS (black squares) as the sole carbon source. Concentrations of choline (grey diamonds) and GBT (white diamonds) were quantified throughout the experiment. Cultures were grown in triplicate. Error bars denote standard deviations. GBT, glycine betaine; COS, choline O‐sulfate.
BetT is required for the uptake of, and growth on, choline in R . pomeroyi
In R. pomeroyi, directly upstream of betA is a putative betT gene (SPO1087), encoding a betaine‐carnitine‐choline transporter (BCCT), which is known to be responsible for the uptake of extracellular choline in Escherichia coli (Lamark et al., 1996). To investigate the role of betT in choline metabolism in R. pomeroyi, a ΔbetT::Gm mutant was constructed. The mutant could grow on choline as a sole carbon and energy source; however, the growth rate (μ = 0.012 ± 0.002 h−1) was severely reduced compared with that of the wild‐type (μ = 0.073 ± 0.003 h−1). Consequentially, the rate of choline depletion was also severely reduced (84%) compared with that of the wild‐type (Fig. 2E). Growth of the ΔbetT::Gm mutant on COS (μ = 0.027 ± 0.001 h−1) was also affected (wild‐type μ = 0.043 ± 0.000 h−1), showing a 38% reduction in growth rate.
Degradation of choline and GBT by R . pomeroyi releases ammonium
We showed previously that turnover of nitrogen‐rich methylated amines by marine bacteria, primarily as a source of supplementary energy, resulted in the remineralization of organic nitrogen in the form of ammonium (Lidbury et al., 2014b). Similarly, when either choline or GBT was used as the sole carbon and nitrogen source for R. pomeroyi, ammonium accumulation in the culture medium was observed (Fig. 3A). Moreover, the addition of glucose to the medium (increasing the carbon : nitrogen ratio above cell stoichiometry and thus making nitrogen the limiting nutrient) resulted in no accumulation of ammonium in the culture medium, despite the degradation of choline or GBT (Fig. 3B).
Figure 3.
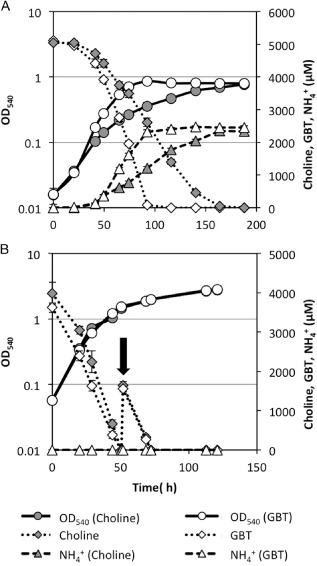
Growth of R . pomeroyi on choline (grey circles) or GBT (white circles) as a sole carbon and nitrogen source (5 mM) (A) or on choline or GBT (4 mM) as the sole nitrogen source with glucose (10 mM) added to the medium (B). Concentrations of choline (grey diamonds) and GBT (white diamonds) were quantified throughout the experiment. NH4 + was also quantified during the experiment in either GBT‐grown (white triangles) or choline‐grown cultures (grey triangles). Arrow indicates a second addition of either choline or GBT (∼ 2 mM). Cultures were grown in triplicate and error bars denote standard deviation. GBT, glycine betaine.
betABC and choline transporters are widely distributed in marine bacteria of the A lphaproteobacteria, the MRC clade and some G ammaproteobacteria
To better understand the potential importance of choline metabolism in the MRC, the genome sequences of isolates from the MRC were screened for the presence of the betABC genes required for choline metabolism. Out of 52 MRC genomes, 51 have the betA gene in their genomes, while 48 and 37 contain betC and betB respectively (Table 1). It is interesting that some strains lack betB as this gene was clearly involved but not essential for growth on choline in R. pomeroyi (Fig. 2). In the majority of isolates from the MRC, the betABC genes, together with the regulator betI, are found in a regulon, for example in isolates Roseobacter sp. Azwk‐3b and Sagittula stellata E‐37 (Fig. 4). Fourteen MRC isolates were screened for their ability to grow on choline and GBT as a sole carbon and energy source (Table 1). Growth on choline and COS as a sole carbon and energy source directly correlated with the presence of the betABC genes in their genomes. The genetic potential to use choline appears to be more widespread within the MRC than their ability to utilize methylamines, such as TMA (Chen et al., 2011) or monomethylamine (MMA) (Chen, 2012).
Table 1.
Comparative genomic analysis of genes involved in the catabolism of choline (CHO) and choline O‐sulfate (COS)
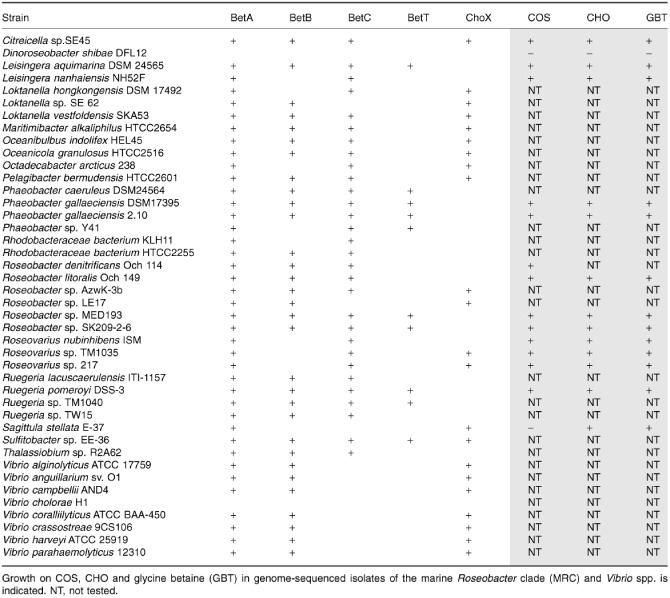
Figure 4.
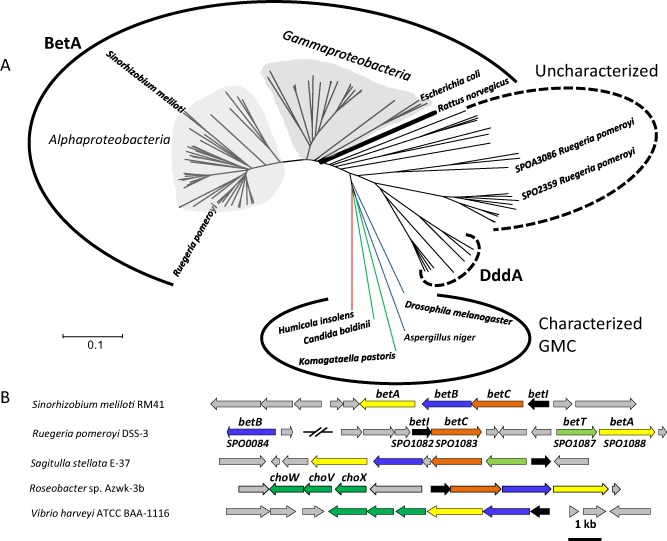
Phylogenetic analysis of choline dehydrogenase (BetA) in relation to other oxidoreductases of the glucose‐methanol‐choline (GMC) oxidoreductases family using the neighbour‐joining method (A). Bootstrap values from 500 replications were omitted for clarity. The tree was constructed using mega 5.2 (Tamura et al., 2011). A number of putative GMC oxidoreductases that are closely related to BetA were identified in marine heterotrophs, which have no assigned function, including two from R . pomeroyi. The blue lines denote glucose dehydrogenases, the red line denotes a cellobiose dehydrogenase, and the green lines represent methanol/alcohol dehydrogenases. (B) The genetic neighbourhood of the bet genes in representative marine bacterial isolates. DddA, 3‐hydroxypropionate dehydrogenase; BetA, choline dehydrogenase; cho X, periplasmic binding protein of the choline ABC transporter; cho V, ATP‐binding domain of the choline ABC transporter; cho W, transmembrane permease of the choline ABC transporter; bet I, regulator of bet operon; bet T, choline permease; bet A, choline dehydrogenase; bet B, betaine aldehyde dehydrogenase; bet C, choline sulfatase.
To better understand the distribution of choline metabolism genes among marine heterotrophs, we used BetA from R. pomeroyi as the query sequence to perform a blastp alignment scrutinizing the genomes of marine heterotrophs deposited in the Integrated Microbial Genomes database (http://img.jgi.doe.gov/). BetA belongs to the glucose‐methanol‐choline (GMC) oxidoreductase family (Cavener, 1992), including the characterized 3‐hydroxypropionate dehydrogenase (DddA) which is involved in DMSP catabolism (Curson et al., 2011). In addition to the MRC clade, BetA is also found in many isolates from the Gammaproteobacteria, including Vibrio spp. and Alteromonas spp. (Fig. 4). BetA homologues were also present in a number of single‐cell amplified genomes from abundant bacteria of the Alphaproteobacteria and Gammaproteobacteria, which were retrieved from marine surface waters (Swan et al., 2013) (Fig. 4, Fig. S3). While representatives from the MRC possessed the betC gene required for COS degradation to choline, as well as genes required for the further catabolism of GBT to glycine (Table 1), representatives of Vibrio spp. did not. Furthermore, no BetA homologues were retrieved from the genomes of SAR11 clade bacteria.
The BCCT‐type choline transporter BetT is not present in all MRC bacteria (Fig. 4, Table 1). Instead, some MRC bacteria (e.g. Roseovarius sp. 217, Octadecabacter arcticus 238) have three open reading frames (ORFs) immediately upstream of the betIABC genes, which are annotated as genes encoding three subunits of an ABC‐type choline transporter (ChoXWV) (Chen et al., 2010). Phylogenetic analysis of the substrate‐binding protein, ChoX, from MRC bacteria reveals a close relationship with the ChoX from S. meliloti (Fig. S2; Chen et al., 2010), suggesting that this gene is likely to be involved in choline metabolism. The BetT‐type and the ChoX‐type choline transporters seem mutually exclusive in almost all MRC bacteria isolates (Table 1). The presence of the BetT‐type transporter is associated with MRC subclades one and two as defined by Newton and colleagues (2010), while the ABC‐type choline transporter, ChoXWV, is associated with MRC subclades three and four and Vibrio spp.
The role of formyl tetrahydrofolate synthetase (Fhs) during choline metabolism in R . pomeroyi
In R. pomeroyi, complete degradation of choline to pyruvate results in the release of ammonium (Figs 1 and 3), while two of the three methyl groups arising from choline degradation are hypothesized to be conjugated to the carrier tetrahydrofolate (H4F) and further oxidized (Fig. 1). The other methyl group is predicted to be oxidized and conjugated to homocysteine, producing methionine (Fig. 1). Indeed, H4F‐binding domains were found in several key enzymes involved in choline catabolism, including dimethylglycine (DMG) dehydrogenase (SPO3400) and sarcosine dehydrogenase (SPO3396). It was hypothesized that complete oxidation of 5, 10‐methylenetetrahydrofolate (CH2 = H4F) through formyltetrahydrofolate synthetase (Fhs) provides reducing power in the form of NADH and ATP (Sun et al., 2011; Lidbury et al., 2014b). R. pomeroyi has two nearly identical copies (99.3% identity in nucleotide sequence, 100% identical in amino acid sequence) of the fhs gene (Fig. S4).
To determine the role of fhs in the oxidation of methyl groups in R. pomeroyi, both copies of fhs in this bacterium were deleted, generating the double mutant, Δfhs‐1::Gm/Δfhs‐2::Spc (hereafter refer to as the fhs null mutant). Compared with the growth rate of the wild‐type on choline (μ = 0.073 ± 0.003 h−1), the fhs null mutant had a significantly reduced growth rate (μ = 0.033 ± 0.008 h−1) as well as a reduced final growth yield (fhs null mutant OD540 = 0.27, wild‐type OD540 = 1.17) (Fig. 5A). In cultures of the fhs null mutant, the initial rate of choline depletion was slower than that of the wild‐type; however, complete degradation of choline still occurred. During the experiment, there was a gradual build‐up of the metabolite GBT in cultures of the fhs null mutant (Fig. 5B). However, in wild‐type cultures, only a transient spike in GBT was observed. The complemented fhs mutant had a partially restored growth rate (μ = 0.050 ± 0.002 h−1) and final growth yield (OD540 = 0.84) (Fig. 5A) due to the restored ability to utilize GBT (Fig. 5B).
Figure 5.
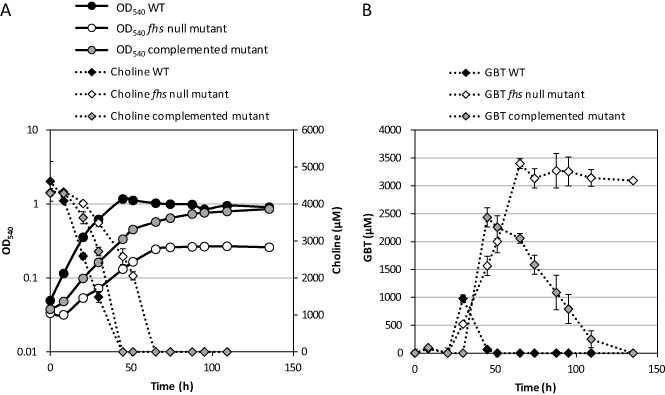
Growth (circles) of R . pomeroyi wild‐type, fhs null mutant and the complemented mutant on choline (5 mM) (diamonds) as a sole carbon, nitrogen and energy source (A). GBT (diamonds) in the culture medium was quantified throughout growth (B). Cultures were grown in triplicate. Error bars denote standard deviations.
To determine whether the fhs mutation also affected growth on other downstream metabolites of choline metabolism, the wild‐type, the fhs null mutant and the complemented mutant were all grown on either glycine, sarcosine, DMG or GBT as a sole carbon and energy source. For the wild‐type, the final OD540 of the cultures showed a positive correlation with the increasing number of methyl groups, with growth on glycine resulting in the lowest OD540 and growth on GBT resulting in the highest yield (Fig. 6A). In the fhs null mutant, a similar phenotype was observed, but the null mutant failed to grow on GBT (Fig. 6B). Growth on GBT was, however, restored in the complemented mutant (Fig. 6C).
Figure 6.
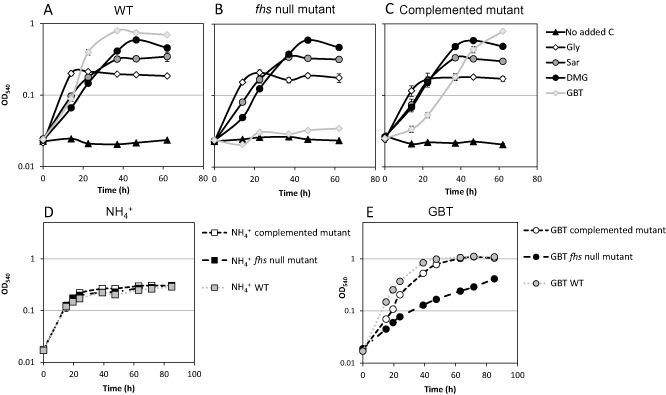
Growth of R . pomeroyi wild‐type (A), the fhs null mutant (B) and the complemented mutant (C) on 4 mM glycine (Gly), sarcosine (Sar), dimethylglycine (DMG) or glycine betaine (GBT) as the sole carbon source and ammonium (5 mM) as the nitrogen source. Negative growth control was set up without added carbon source. These three strains were also grown on 0.5 mM ammonium (NH4 +) (D) or GBT (E) as a sole nitrogen source with glucose as the carbon source (10 mM). Cultures were grown in triplicate. Error bars denote standard deviations.
Although the null mutant cannot grow on GBT as a sole carbon source, we noticed that it could still grow on GBT as a nitrogen source, suggesting that sequential demethylation of GBT was still occurring in the null mutant, liberating ammonium as the source of nitrogen (Figs 1 and 6E). This suggests that the inability to demethylate GBT as the sole carbon source in the fhs null mutant was not due to a lack of recycled H4F. Indeed, experiments supplementing the fhs null mutant with either homocysteine or H4F failed to restore growth on GBT as the sole carbon source (Figs S5 and S6). Therefore, we hypothesized that GBT catabolism cannot function without Fhs due to an imbalance in the reducing state of the cell, i.e. the cell is limited by either the production of reducing equivalents and/or ATP. To test this hypothesis, the fhs null mutant was grown on choline and supplemented with formate to provide NADH through formate dehydrogenase (Fig. 1). The fhs null mutant grown in the presence of choline‐only reached a final OD540 ∼ 0.27, whereas supplementation with formate (13 mM total) resulted in almost double the amount of growth (OD540 ∼ 0.52) (Fig. 7A). Consequently, the concentration of GBT in cultures supplemented with formate was significantly reduced (Fig. 7B). The addition of more choline (∼ 8 mM) to half the choline‐only cultures resulted in the continuation of growth after cultures had reached stationary phase; meanwhile, a stepwise build‐up of GBT was also observed (Fig. 7B). Together, these data show that in R. pomeroyi, the reducing equivalents, as well as the ATP generated through oxidation of the C1 groups, are essential for maintaining adequate reducing power during choline, and specifically GBT catabolism.
Figure 7.
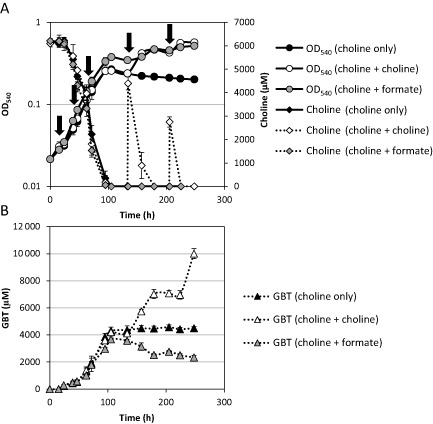
(A) Growth of the fhs null mutant (circles) on choline supplemented with either formate (grey circles) or additional choline (white circles). Choline was quantified throughout the experiment. Arrows denote additions of formate (13 mM total). The first three additions were with 1 mM formate, the latter two with 5 mM. (B) Quantification of GBT in the culture medium during the experiment. All cultures were grown in triplicate, and error bars denote standard deviations.
Discussion
In this study, the key genes responsible for the metabolism and subsequent growth on choline by members of the MRC have been experimentally confirmed. Marine eukaryotic flora accumulate COS as an osmolyte (Hanson and Gage, 1991; Hanson et al., 1991; Murakeözy et al., 2003), and the work in this study has confirmed that isolates from the MRC can utilize COS as a nutrient, using BetC. Phosphatidylcholine often accounts for > 50% of the phospholipid pool in certain eukaryotic fauna and flora (van Meer et al., 2008); therefore, phosphatidylcholine may provide a significant source of choline in niches associated with eukaryotic biota. The ability to metabolize choline and COS is ubiquitous in the MRC, and we speculate that these compounds may be an important nutrient source for these bacteria that are known to form close associations with eukaryotic biota (Ikawa and Taylor, 1973; González et al., 2000; Hjelm et al., 2004; Buchan et al., 2005; Porsby et al., 2008; Lema et al., 2014).
Bacteria related to the SAR11 clade can catabolize GBT, and homologues of the key enzymes involved in GBT metabolism, betaine homocysteine methyltransferase, DMG dehydrogenase and sarcosine dehydrogenase, have been identified in their genomes (Sun et al., 2011). However, to date, there is no physiological evidence to suggest that choline is a nutrient source for this clade. Furthermore, the genes involved in the uptake of choline (betT, choX) and subsequent catabolism (betABC) are absent from their genomes. It was previously shown that choline can be rapidly taken up by coastal marine bacteria and transformed to GBT (Gauthier and Le Rudulier, 1990; Ghoul et al., 1990; Kiene, 1998), and different phytoplankton species can also synthesize and/or acquire extracellular GBT to aid in osmoregulation (Keller et al., 1999; 2004). Consequentially, the concentration of particulate GBT is significantly higher than that of particulate choline in marine surface waters (Airs and Archer, 2010). In addition, a proportion of intracellular GBT can also be released back into the marine environment, through both passive and active mechanisms (Kapfthammer et al., 2005). Together, these data suggest that GBT is likely to be more widespread within the water column compared with choline, which may be a nutrient more commonly associated with niches surrounding eukaryotic biota. In support of this hypothesis, all characterized choline‐specific BCCT‐type transporters have only been identified in bacteria that form close associations with either a plant or animal host (Andresen et al., 1988; Fan et al., 2003; Chen and Beattie, 2008). Unlike many MRC bacteria, SAR11 bacteria are free‐living, oligotrophic cells that are not typically associated with eukaryotic flora or fauna (Morris et al., 2002; Giovannoni et al., 2005; Luo et al., 2013). The contrasting ability of choline and GBT catabolism in the MRC and the SAR11 clade bacteria may, therefore, reflect their different lifestyles and thus ecological niche separation.
The BCCT‐type transporter, BetT, found in the genomes of R. pomeroyi and other MRC bacteria appears different from the previous BetT choline transporters characterized from either E. coli or P. syringae in that those BetT proteins are over 100 amino acids longer (Chen and Beattie, 2008). In the same study, it was experimentally confirmed that the presence of an elongated C‐terminus is required for the uptake of choline under hyperosmotic stress. The authors proposed that the addition of an elongated C‐terminus could be used to predict the function of BetT, where the presence of an elongated C‐terminus denotes a role in osmoregulation and its absence denotes a role in the uptake of choline as a nutrient source (Chen and Beattie, 2008). Therefore, BetT in the MRC may primarily have a role in the uptake of choline as a nutrient. The ΔbetT::Gm mutant of R. pomeroyi showed no change in growth on GBT, suggesting that it is not a GBT transporter. This is in line with the fact that the majority of BCCT‐type transporters are characterized by having a narrow substrate range (Choquet et al., 2005; Chen and Beattie, 2008). The GBT transporter in R. pomeroyi awaits further experimental validation, and there are a number of potential candidates within its genome. We observed that the ΔbetT::Gm mutant showed reduced growth rates on choline and COS. This suggests that there is another transport system in place for the uptake of these compounds, similar to that of Bacillus subtilis (Nau‐Wagner et al., 2012).
The catabolism of nitrogen‐rich compounds provides a route for the remineralization of organic nitrogen back into ammonium, which can stimulate the growth of another bacterium in co‐culture (Lidbury et al., 2014b). Here, we provide further evidence that growth of marine heterotrophs on choline or GBT as a sole carbon source can also result in the remineralization of ammonium. Bacterioplankton in the Sargasso Sea, a region typified by prolonged periods of phosphate limitation and not nitrogen limitation (Wu et al., 2000), may well release ammonium during the oxidation of different nitrogen‐rich methylated compounds (Sun et al., 2011). The notion that nitrogen‐rich methylated compounds are primarily oxidized for carbon or energy is in line with the observation that phytoplankton seston is rapidly degraded by the bacterioplankton resulting in an increase of inorganic nitrogen, in the form of ammonium (Garber, 1984).
In R. pomeroyi and other marine heterotrophs, Fhs (encoded by fhs), which is involved in H4F‐mediated oxidation of methyl groups, is predicted to convert formyl‐H4F to formate, which can then be oxidized to CO2 (Chen, 2012). Fhs, therefore, plays an essential role not only in the recycling of H4F, making it available for further methyl group acceptance, but also providing ATP and reducing equivalents in the form of NADH resulting from the further catabolism of formate (Fig. 1) (Chistoserdova et al., 2004; Sun et al., 2011; Chen, 2012). Our experiments show that the turnover of GBT is affected in the fhs null mutant during growth, particularly when this compound represents the only source of ATP and reducing equivalents for the cell. However, our experiments do not clarify whether or not complete oxidation of the methyl groups to CO2 has been terminated. R. pomeroyi does possess the genes required for C1 oxidation through the glutathione‐linked (GSH) C1 oxidation pathway, which has previously been shown to alleviate stress caused by formaldehyde toxicity (Harms et al., 1996; Marx et al., 2003; Martinez‐Gomez et al., 2013). The GSH‐linked pathway usually requires the enzyme formaldehyde‐activating enzyme (Fae) to facilitate the conjugation of formaldehyde to GSH or to the alternative cofactor H4MPT (increasing the rate of conjugation by up to 10‐fold) (Goenrich et al., 2002; Chen, 2012). Unlike the majority of non‐marine representative methylotrophs, isolates from the MRC, including R. pomeroyi, lack fae. Therefore, it is unclear whether or not the GSH‐linked pathway can deal with any potential build‐up of formaldehyde during the catabolism of choline and GBT in the fhs null mutant. In Methylobacterium extorquens PA1, formaldehyde leakage via the gamma‐glutamylmethylamide/N‐methylglutamate pathway was observed during growth on MMA (Nayak and Marx, 2014). Therefore, in the fhs null mutant, formaldehyde leakage, due to a potential lack of free H4F, may present a problem for the cell and may explain the slower growth rates observed for the fhs null mutant when growing on GBT as a sole nitrogen source. In reality, a combination of impaired reducing power generation and free H4F is the likely explanation behind the phenotypes observed in the fhs null mutant. This was supported by the fact that the fhs null mutant failed to grow on GBT as a sole carbon source without the addition of another source of reducing power, such as formate.
In summary, we demonstrate that the ability to utilize choline is a universal trait of MRC bacteria which requires the enzymes BetABC. Based on comparative genomic analyses, choline metabolism appears to be absent in SAR11 clade bacteria. In addition, our study has also confirmed the hypothesis that the H4F‐linked C1 oxidation pathway has a role in the oxidation of the methyl groups released during the degradation of methylated compounds, which is required to maintain normal cell physiology.
Experimental procedures
Cultivation of bacteria
The MRC isolates were maintained on marine agar 2216 (Difco, UK) or ½ YPSS: yeast extract (2 g l−1), peptone (1.25 g l−1) and sea salts (30 g l−1, Sigma). Gentamicin (10 μg ml−1), kanamycin (80 μg ml−1) or spectinomycin (175 μg ml−1) was added to maintain R. pomeroyi mutant strains, ΔbetA::Gm, ΔbetB::Gm, ΔbetC::Gm, ΔbetT::Gm, Δfhs‐1::Gm/Δfhs‐2::Spc, and the complemented mutant strain, Δfhs‐1::GmΔ/fhs‐2::Spc + fhs‐1:DSS‐3. Choline O‐sulfate was purchased from the Cambridge Isotope Laboratories. For all growth experiments, R. pomeroyi (wild‐type and mutants) as well as other strains from the MRC were grown in a marine ammonium mineral salts (MAMS) medium with the addition of relevant carbon sources. The MAMS medium as modified by Schäfer (2007) contained the following (per litre): NaCl, 20 g; (NH4)2SO4, 1 g; MgSO4·7H2O, 1 g; CaCl2·2H2O, 0.2 g; FeSO4·7H2O, 2 mg; Na2MoO4·2H2O, 20 mg; KH2PO4, 0.36 g; K2HPO4, 2.34 g; plus 1 ml of SL‐10 trace metals solution (Schäfer, 2007). Vitamins were prepared as described previously (Chen, 2012). To determine if choline and GBT were used as a nitrogen source and whether growth on choline or GBT led to the release of ammonium, (NH4)2SO4 was removed from the standard MAMS recipe, and either GBT or choline was added to the medium (at concentrations of 4 or 5 mM) with 10 mM glucose.
Genetic manipulation of R . pomeroyi
A full list of strain and plasmids used in this study is outlined in Table 2. To construct mutants in R. pomeroyi, two regions of genomic DNA were amplified, one towards the 5′ end (with restriction sites engineered at either end) and the other towards the 3′ end (with restriction sites engineered at each end) of the target genes. Sequence integrity after subcloning into the pGEM‐T vector was confirmed via DNA sequencing. The complete list of primers used, and restriction sites introduced to generate the mutants used in this study, is shown in Table S1. An upstream and downstream fragment of the target gene, along with either the gentamicin gene cassette, amplified from p34S‐Gm (Dennis and Zylstra, 1998), or the spectinomycin cassette, amplified from pHP45Ω (Prentki and Krisch, 1984), were subcloned into the cloning vector pGEM‐T (Promega). The entire construct was then excised from pGEM‐T and ligated into the suicide vector pK18mobsacB (Schäfer et al., 1994). The resulting plasmid was transformed into Escherichia coli S17.1 via electroporation and mobilized into R. pomeroyi via conjugation onto a 0.22 μm pore‐size, 47 mm sterile filter (Millipore, UK), using 1/2 YTSS (Deutsche Sammlung von Mikroorganismen und Zellkulturen, DSMZ) as the medium. Transconjugants were selected for on the sea salts minimal medium as described previously with gentamicin (10 μg ml−1) and MMA (3 mM) as a sole nitrogen source (Lidbury et al., 2014a). Double cross‐over mutants were selected by their sensitivity to kanamycin, and homologous recombination was confirmed by polymerase chain reaction (PCR) and DNA sequencing.
Table 2.
List of strains and plasmids used in this study
| Strains/plasmids | Description/use | Source |
|---|---|---|
| E. coli S17.1 | Electrocompetent cells used for conjugation | Lab collection |
| E. coli JM109 | Routine host for cloning | Promega |
| R. pomeroyi DSS‐3 | Wild‐type | González and colleagues (2003) |
| R. pomeroyi ΔbetA::Gm | R. pomeroyi with disrupted betA | This study |
| R. pomeroyi ΔbetB::Gm | R. pomeroyi with disrupted betB | This study |
| R. pomeroyi ΔbetC::Gm | R. pomeroyi with disrupted betC | This study |
| R. pomeroyi ΔbetT::Gm | R. pomeroyi with disrupted betT | This study |
| R. pomeroyi Δfhs‐1::Gm Δfhs‐2::Spc | R. pomeroyi with both copies of fhs disrupted | This study |
| R. pomeroyi Δfhs + fhs:DSS‐3 | fhs null mutant complemented with native fhs‐1 | This study |
| p34S‐Gm | Source of a gentamicin gene cassette | Dennis and Zylstra (1998) |
| pK18mobsacB | Suicide vector for R. pomeroyi (KanR) | Schäfer and colleagues (1994) |
| pBBR1MCS‐km | Broad‐host‐range plasmid (KanR) | Kovach and colleagues (1995) |
| pHP45Ω | Source of spectinomycin gene cassette | Prentki and Krisch (1984) |
| pKIL301 | Mutated betA and the gentamicin cassette cloned into pK18mobsacB | This study |
| pKIL302 | Mutated betB and the gentamicin cassette cloned into pK18mobsacB | This study |
| pKIL303 | Mutated betC and the gentamicin cassette cloned into pK18mobsacB | This study |
| pKIL304 | Mutated betT and the gentamicin cassette cloned into pK18mobsacB | This study |
| pKIL305 | Mutated fhs‐1 and the gentamicin cassette cloned into pK18mobsacB | This study |
| pKIL306 | Mutated fhs‐2 and the spectinomycin cassette cloned into pK18mobsacB | This study |
| pBIL301 | Native fhs and the promoter upstream on the fhs‐1 operon cloned into the vector pBBR1MCS‐km | This study |
To complement the Δfhs‐1::GmΔfhs‐2::Spc mutant, the fhs‐1 gene (encoded by SPO1557), and the native promoter for the operon were amplified and individually subcloned into pGEM‐T. The promoter and fhs‐1 were sequentially cloned into the broad‐host range plasmid, pBBR1MCS‐km (Kovach et al., 1995) using the restriction sites KpnI and SalI, and SalI and BamHI respectively. The resultant plasmid, pBIL105, was mobilized into the R. pomeroyi mutant as described above. Confirmation of the complemented mutant was carried out by PCR and DNA sequencing.
Quantification of quaternary and methylated amines
Cells were removed from culture medium by centrifugation (10 000 × g, 2 min) of spin columns (0.22 μm pore‐size, nylon, Costar, Corning, NY). All amines and ammonium, apart from COS, were quantified using a cation‐exchange ion chromatograph equipped with a Metrosep C4/250‐mm separation column and a conductivity detector (Metrohm) as described previously (Lidbury et al., 2014a).
Comparative genomic analysis of the genes involved in the metabolism of choline and related compounds
For all analyses, the Integrated Microbial Genomes database at the Joint Genome Institute (IMG/JGI) was used to identify all genes involved in the metabolism of quaternary amines. Each blast search was conducted using an E‐value of 1e‐20 with a minimum sequence identity cut‐off of 30%. Searches were conducted using all marine heterotrophic bacteria in the IMG/JGI database selecting genomes with draft/finished/permanent draft status, after which representative sequences from different bacterial clades/groups were selected. The locus tag and accession numbers (Gene ID) for the following genes used as query sequences were as follows: betA, SMc00093, 637181738; betB, SMc00094, 637181739; betC, SMc00127, 637181740; betT, 637288573; choX, 638910580. Phylogenetic analysis was conducted using the mega 5.2 package (Tamura et al., 2011). The National Centre for Biotechnology Information database was used to find a number of sequences not in the IMG/JGI database. Due to the high number of hits retrieved, a stringent E‐value < 1e‐170 was used to identify true homologues. Where appropriate, phylogenetic analysis was performed to infer function.
Supporting information
Fig. S1. Phylogenetic analysis of the substrate binding proteins (SBPs) affiliated with the putative choline ABC‐type transporter found in marine bacteria. Reference sequences from characterized SBPs were added to the alignment. Characterized SBPs, related to osmolyte SBPs, based on the structural analysis conducted by Berntsson and colleagues (2010) were used as an outgroup. The tree was aligned in mega 5.2 using the neighbour‐joining method using 500 replications for bootstrapping. The scale bar represents the number of substitutions per amino acid. ChoX, SBP specific for choline; TmoX, SBP specific for trimethylamine N‐oxide; BetX, SBP specific for glycine betaine; CaiX, SBP specific for carnitine.
Fig. S2. Detailed phylogeny of ChoX from Fig. S1 showing strain names and their corresponding accession numbers (Gene ID in IMG/JGI).
Fig. S3. Phylogenetic analysis of choline dehydrogenase (BetA). The evolutionary history was inferred using the neighbour‐joining method. For the major nodes, the percentage (> 75%) of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p‐distance method and are in the units of the number of amino acid differences per site. The analysis involved 101 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 685 positions in the final dataset. Evolutionary analyses were conducted in mega6.
Fig. S4. Gene neighbourhoods of fhs1 and fhs2. The scale bar represents the number of bases. folD, 5,10‐methylene‐H4F dehydrogenase/methenyl‐H4F cyclohydrolase; tmm, trimethylamine monooxygenase; tdm, trimethylamine N‐oxide demethylase; fhs, formyl‐H4F synthetase; tmoR, putative regulator of tmm; amt, unspecified ammonium transporter; ftsH, ATP‐dependent metalloprotease; fhdA, formate dehydrogenase alpha subunit; fhdB, formate dehydrogenase beta subunit; PBP, uncharacterized HAAT family amino acid periplasmic binding protein.
Fig. S5. Growth of the R. pomeroyi fhs null mutant on GBT (red squares), homocysteine (purple crosses) or GBT and homocysteine (green triangles) as the carbon source respectively. A positive control consisted of glucose as a carbon source and the negative control had no added carbon. Cultures were grown in triplicate. Error bars denote SD. Hcy, homocysteine.
Fig. S6. Growth of R. pomeroyi wild‐type and the fhs null mutant on glucose and GBT as the carbon and energy source and ammonium as the nitrogen source. Tetrahydrofolate (1 mM) was added to wild‐type and mutant cultures at T = 0 h and T = 21 h, and GBT consumption was recorded. Cultures were grown in triplicate. Error bars denote SD.
Table S1. List of oligonucleotides used in this study.
Acknowledgements
This work was supported by the Natural Environment Research Council (NERC) through a PhD studentship (to IL) and research grant (NE/M002233). We also thank the University of Warwick undergraduate research support scheme (URSS) for a grant to support GK.
References
- Airs, R.L. , and Archer, S.D. (2010) Analysis of glycine betaine and choline in seawater particulates by liquid chromatography/electrospray ionization/mass spectrometry. Limnol Oceanog‐Meth 8: 499–506. [Google Scholar]
- Andresen, P.A. , Kaasen, I. , Styrvold, O.B. , Boulnois, G. , and Strom, A.R. (1988) Molecular cloning, physical mapping and expression of the bet genes governing the osmoregulatory choline‐glycine betaine pathway of Escherichia coli . J Gen Microbiol 134: 1737–1746. [DOI] [PubMed] [Google Scholar]
- Barra, L. , Fontenelle, C. , Ermel, G. , Trautwetter, A. , Walker, G.C. , and Blanco, C. (2006) Interrelations between glycine betaine catabolism and methionine biosynthesis in Sinorhizobium meliloti strain 102F34. J Bacteriol 188: 7195–7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch, J. , Kempf, B. , and Bremer, E. (1994) Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J Bacteriol 176: 5364–5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan, A. , González, J.M. , and Moran, M.A. (2005) Overview of the marine Roseobacter lineage. Appl Environ Microbiol 71: 5665–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cánovas, D. , Vargas, C. , Csonka, L.N. , Ventosa, A. , and Nieto, J.J. (1996) Osmoprotectants in Halomonas elongata: high‐affinity betaine transport system and choline‐betaine pathway. J Bacteriol 178: 7221–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carini, P. , Steindler, L. , Beszteri, S. , and Giovannoni, S.J. (2013) Nutrient requirements for growth of the extreme oligotroph ‘Candidatus Pelagibacter ubique’ HTCC1062 on a defined medium. ISME J 7: 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalfomo, P. , Block, J.H. , Constantine, G.H. , and Kirk, P.W. (1972) Choline sulfate (ester) in marine higher fungi. Mar Chem 1: 157–162. [Google Scholar]
- Cavener, D.R. (1992) GMC oxidoreductases: a newly defined family of homologous proteins with diverse catalytic activities. J Mol Biol 223: 811–814. [DOI] [PubMed] [Google Scholar]
- Chen, C. , and Beattie, G.A. (2008) Pseudomonas syringae BetT is a low‐affinity choline transporter that is responsible for superior osmoprotection by choline over glycine betaine. J Bacteriol 190: 2717–2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. , Malek, A.A. , Wargo, M.J. , Hogan, D.A. , and Beattie, G.A. (2010) The ATP‐binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate‐binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol Microbiol 75: 29–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. (2012) Comparative genomics of methylated amine utilisation by marine Roseobacter clade bacteria and development of functional gene markers (tmm, gmaS). Environ Microbiol 14: 2308–2322. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Patel, N.A. , Crombie, A. , Scrivens, J.H. , and Murrell, J.C. (2011) Bacterial flavin‐containing monooxygenase is trimethylamine monooxygenase. Proc Natl Acad Sci USA 108: 17791–17796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova, L. (2011) Modularity of methylotrophy, revisited. Environ Microbiol 13: 2603–2622. [DOI] [PubMed] [Google Scholar]
- Chistoserdova, L. , Laukel, M. , Portais, J.‐C. , Vorholt, J.A. , and Lidstrom, M.E. (2004) Multiple formate dehydrogenase enzymes in the facultative methylotroph Methylobacterium extorquens AM1 are dispensable for growth on methanol. J Bacteriol 186: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet, G. , Jehan, N. , Pissavin, C. , Blanco, C. , and Jebbar, M. (2005) OusB, a broad‐specificity ABC‐type transporter from Erwinia chrysanthemi, mediates uptake of glycine betaine and choline with a high affinity. Appl Environ Microbiol 71: 3389–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curson, A.R.J. , Todd, J.D. , Sullivan, M.J. , and Johnston, A.W.B. (2011) Catabolism of dimethylsulfoniopropionate: microorganisms, enzymes and genes. Nat Rev Micro 9: 849–859. [DOI] [PubMed] [Google Scholar]
- Dennis, J.J. , and Zylstra, G.J. (1998) Plasposons: modular self‐cloning minitransposon derivatives for rapid genetic analysis of gram‐negative bacterial genomes. Appl Environ Microbiol 64: 2710–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, X. , Pericone, C.D. , Lysenko, E. , Goldfine, H. , and Weiser, J.N. (2003) Multiple mechanisms for choline transport and utilisation in Haemophilus influenzae . Mol Microbiol 50: 537–548. [DOI] [PubMed] [Google Scholar]
- Garber, J.H. (1984) Laboratory study of nitrogen and phosphorus remineralization during the decomposition of coastal plankton and seston. Estuar Coast Shelf Sci 18: 685–702. [Google Scholar]
- Gauthier, M.J. , and Le Rudulier, D. (1990) Survival in seawater of Escherichia coli cells grown in marine sediments containing glycine betaine. Appl Environ Microbiol 56: 2915–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoul, M. , Bernard, T. , and Cormier, M. (1990) Evidence that Escherichia coli accumulates glycine betaine from marine sediments. Appl Environ Microbiol 56: 551–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni, S.J. , Tripp, H.J. , Givan, S. , Podar, M. , Vergin, K.L. , Baptista, D. , et al (2005) Genome streamlining in a cosmopolitan oceanic bacterium. Science 309: 1242–1245. [DOI] [PubMed] [Google Scholar]
- Goenrich, M. , Bartoschek, S. , Hagemeier, C.H. , Griesinger, C. , and Vorholt, J.A. (2002) A glutathione‐dependent formaldehyde‐activating enzyme (Gfa) from Paracoccus denitrificans detected and purified via two‐dimensional proton exchange NMR spectroscopy. J Biol Chem 277: 3069–3072. [DOI] [PubMed] [Google Scholar]
- González, J.M. , Simó, R. , Massana, R. , Covert, J.S. , Casamayor, E.O. , Pedrós‐Alió, C. , and Moran, M.A. (2000) Bacterial community structure associated with a dimethylsulfoniopropionate‐producing North Atlantic algal bloom. Appl Environ Microbiol 66: 4237–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, J.M. , Covert, J.S. , Whitman, W.B. , Henriksen, J.R. , Mayer, F. , Scharf, B. , et al (2003) Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., dimethylsulfoniopropionate‐demethylating bacteria from marine environments. Int J Syst Evol Micro 53: 1261–1269. [DOI] [PubMed] [Google Scholar]
- Graham, J.E. , and Wilkinson, B.J. (1992) Staphylococcus aureus osmoregulation: roles for choline, glycine betaine, proline, and taurine. J Bacteriol 174: 2711–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnke, S. , Brock, N.L. , Zell, C. , Simon, M. , Dickschat, J.S. , and Brinkhoff, T. (2013) Physiological diversity of Roseobacter clade bacteria co‐occurring during a phytoplankton bloom in the North Sea. Syst Appl Microbiol 36: 39–48. [DOI] [PubMed] [Google Scholar]
- Hanson, A.D. , and Gage, D.A. (1991) Identification and determination by fast atom bombardment mass spectrometry of the compatible solute choline‐o‐sulfate in Limonium species and other halophytes. Aus J Plant Physiol 18: 317–327. [Google Scholar]
- Hanson, A.D. , Rathinasabapathi, B. , Chamberlin, B. , and Gage, D.A. (1991) Comparative physiological evidence that β‐alanine betaine and choline‐O‐sulfate act as compatible osmolytes in halophytic Limonium species. Plant Physiol 97: 1199–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, A.D. , Rathinasabapathi, B. , Rivoal, J. , Burnet, M. , Dillon, M.O. , and Gage, D.A. (1994) Osmoprotective compounds in the Plumbaginaceae: a natural experiment in metabolic engineering of stress tolerance. Proc Natl Acad Sci USA 91: 306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, N. , Ras, J. , Reijnders, W.N. , van Spanning, R.J. , and Stouthamer, A.H. (1996) S‐formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification? J Bacteriol 178: 6296–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelm, M. , Riaza, A. , Formoso, F. , Melchiorsen, J. , and Gram, L. (2004) Seasonal incidence of autochthonous antagonistic Roseobacter spp. and Vibrionaceae strains in a Turbot larva (Scophthalmus maximus) rearing system. Appl Environ Microbiol 70: 7288–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa, M. , and Taylor, R.F. (1973) Choline and related substances in Algae In Marine Pharmacognosy: Action of Marine Biotoxins at the Cellular Level. Martin D., and Padilla G.M. (eds). New York, USA: Academic Press, pp. 203–236. [Google Scholar]
- Jenkins, G.M. , and Frohman, M.A. (2005) Phospholipase D: a lipid centric review. Cell Mol Life Sci CMLS 62: 2305–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapfthammer, D. , Karatan, E. , Pflughoeft, K.J. , and Watnick, P.I. (2005) Role for glycine betaine transport in Vibrio cholerae osmoadaptation and biofilm formation within microbial communities. Appl Environ Microbiol 71: 3840–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, M.D. , Kiene, R.P. , Matrai, P.A. , and Bellows, W.K. (1999) Production of glycine betaine and dimethylsulfoniopropionate in marine phytoplankton. II. N‐limited chemostat cultures. Mar Biol 135: 249–257. [Google Scholar]
- Keller, M.D. , Matrai, P.A. , Kiene, R.P. , and Bellows, W.K. (2004) Responses of coastal phytoplankton populations to nitrogen additions: dynamics of cell‐associated dimethylsulfoniopropionate (DMSP), glycine betaine (GBT), and homarine. Can J Fish Aquat Sci 61: 685–699. [Google Scholar]
- Kiene, R.P. (1998) Uptake of choline and its conversion to glycine betaine by bacteria in estuarine waters. Appl Environ Microbiol 64: 1045–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiene, R.P. , Hoffmann Williams, L.P. , and Walker, J.E. (1998) Seawater microorganisms have a high affinity glycine betaine uptake system which also recognizes dimethylsulfoniopropionate. Aqua Microb Ecol 15: 39–51. [Google Scholar]
- Kovach, M.E. , Elzer, P.H. , Steven Hill, D. , Robertson, G.T. , Farris, M.A. , Roop Ii, R.M. , and Peterson, K.M. (1995) Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene 166: 175–176. [DOI] [PubMed] [Google Scholar]
- Lamark, T. , Røkenes, T.P. , McDougall, J. , and Strøm, A.R. (1996) The complex bet promoters of Escherichia coli: regulation by oxygen (ArcA), choline (BetI), and osmotic stress. J Bacteriol 178: 1655–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landfald, B. , and Strøm, A.R. (1986) Choline‐glycine betaine pathway confers a high level of osmotic tolerance in Escherichia coli . J Bacteriol 165: 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lema, K.A. , Bourne, D.G. , and Willis, B.L. (2014) Onset and establishment of diazotrophs and other bacterial associates in the early life history stages of the coral Acropora millepora . Mol Ecol 23: 4682–4695. [DOI] [PubMed] [Google Scholar]
- Li, Z. , and Vance, D.E. (2008) Thematic review series: Glycerolipids. Phosphatidylcholine and choline homeostasis. J Lipid Res 49: 1187–1194. [DOI] [PubMed] [Google Scholar]
- Lidbury, I. , Murrell, J.C. , and Chen, Y. (2014a) Trimethylamine N‐oxide metabolism by abundant marine heterotrophic bacteria. Proc Natl Acad Sci USA 111: 2710–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidbury, I.D.E.A. , Murrell, J.C. , and Chen, Y. (2014b) Trimethylamine and trimethylamine N‐oxide are supplementary energy sources for a marine heterotrophic bacterium: implications for marine carbon and nitrogen cycling. ISME J 9: 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, H. , Csűros, M. , Hughes, A.L. , and Moran, M.A. (2013) Evolution of divergent life history strategies in marine Alphaproteobacteria . mBio 4: e00373‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez‐Gomez, N.C. , Nguyen, S. , and Lidstrom, M.E. (2013) Elucidation of the role of the methylene‐tetrahydromethanopterin dehydrogenase MtdA in the tetrahydromethanopterin‐dependent oxidation pathway in Methylobacterium extorquens AM1. J Bacteriol 195: 2359–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx, C.J. , Chistoserdova, L. , and Lidstrom, M.E. (2003) Formaldehyde‐detoxifying role of the tetrahydromethanopterin‐linked pathway in Methylobacterium extorquens AM1. J Bacteriol 185: 7160–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer, G. , Voelker, D.R. , and Feigenson, G.W. (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, M.A. , and Miller, W.L. (2007) Resourceful heterotrophs make the most of light in the coastal ocean. Nat Rev Micro 5: 792–800. [DOI] [PubMed] [Google Scholar]
- Morris, R.M. , Rappe, M.S. , Connon, S.A. , Vergin, K.L. , Siebold, W.A. , Carlson, C.A. , and Giovannoni, S.J. (2002) SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810. [DOI] [PubMed] [Google Scholar]
- Murakeözy, É.P. , Nagy, Z. , Duhazé, C. , Bouchereau, A. , and Tuba, Z. (2003) Seasonal changes in the levels of compatible osmolytes in three halophytic species of inland saline vegetation in Hungary. J Plant Physiol 160: 395–401. [DOI] [PubMed] [Google Scholar]
- Nau‐Wagner, G. , Boch, J. , Le Good, J.A. , and Bremer, E. (1999) High affinity transport of choline‐O‐sulfate and its use as a compatible solute in Bacillus subtilis . Appl Environ Microbiol 65: 560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau‐Wagner, G. , Opper, D. , Rolbetzki, A. , Boch, J. , Kempf, B. , Hoffmann, T. , and Bremer, E. (2012) Genetic control of osmoadaptive glycine betaine synthesis in Bacillus subtilis through the choline‐sensing and glycine betaine‐responsive GbsR repressor. J Bacteriol 194: 2703–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak, D.D. , and Marx, C.J. (2014) Methylamine utilization via the N‐methylglutamate pathway in Methylobacterium extorquens PA1 involves a novel flow of carbon through c1 assimilation and dissimilation pathways. J Bacteriol 196: 4130–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, R.J. , Griffin, L.E. , Bowles, K.M. , Meile, C. , Gifford, S. , Givens, C.E. , et al (2010) Genome characteristics of a generalist marine bacterial lineage. ISME J 4: 784–798. [DOI] [PubMed] [Google Scholar]
- Ohvo‐Rekilä, H. , Ramstedt, B. , Leppimäki, P. , and Slotte, J.P. (2002) Cholesterol interactions with phospholipids in membranes. Prog Lipid Res 41: 66–97. [DOI] [PubMed] [Google Scholar]
- Østerås, M. , Boncompagni, E. , Vincent, N. , Poggi, M.‐C. , and Le Rudulier, D. (1998) Presence of a gene encoding choline sulfatase in Sinorhizobium meliloti bet operon: Choline‐O‐sulfate is metabolized into glycine betaine. Proc Natl Acad Sci USA 95: 11394–11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porsby, C.H. , Nielsen, K.F. , and Gram, L. (2008) Phaeobacter and Ruegeria species of the Roseobacter clade colonize separate niches in a Danish Turbot (Scophthalmus maximus)‐rearing farm and antagonise Vibrio anguillarum under different growth conditions. Appl Environ Microbiol 74: 7356–7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado, S. , Montes, J. , Romalde, J.L. , and Barja, J.L. (2009) Inhibitory activity of Phaeobacter strains against aquaculture pathogenic bacteria. Int Microbiol 12: 107–114. [PubMed] [Google Scholar]
- Prentki, P. , and Krisch, H.M. (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29: 303–313. [DOI] [PubMed] [Google Scholar]
- Roulier, M.A. , Palenik, B. , and Morel, F.M.M. (1990) A method for the measurement of choline and hydrogen peroxide in seawater. Mar Chem 30: 409–421. [Google Scholar]
- Schäfer, A. , Tauch, A. , Jäger, W. , Kalinowski, J. , Thierbach, G. , and Pühler, A. (1994) Small mobilizable multi‐purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum . Gene 145: 69–73. [DOI] [PubMed] [Google Scholar]
- Schäfer, H. (2007) Isolation of Methylophaga spp. from marine dimethylsulfide‐degrading enrichment cultures and identification of polypeptides induced during growth on dimethylsulfide. Appl Environ Microbiol 73: 2580–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost, M.R. , Case, R.J. , Kolter, R. , and Clardy, J. (2011) The Jekyll‐and‐Hyde chemistry of Phaeobacter gallaeciensis . Nat Chem 3: 331–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L.T. , Pocard, J.A. , Bernard, T. , and Le Rudulier, D. (1988) Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti . J Bacteriol 170: 3142–3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styrvold, O.B. , Falkenberg, P. , Landfald, B. , Eshoo, M.W. , Bjørnsen, T. , and Strøm, A.R. (1986) Selection, mapping, and characterization of osmoregulatory mutants of Escherichia coli blocked in the choline‐glycine betaine pathway. J Bacteriol 165: 856–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J. , Steindler, L. , Thrash, J.C. , Halsey, K.H. , Smith, D.P. , Carter, A.E. , et al (2011) One carbon metabolism in SAR11 pelagic marine bacteria. PLoS ONE 6: e23973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z. , Kang, Y. , Norris, M.H. , Troyer, R.M. , Son, M.S. , Schweizer, H.P. , et al (2014) Blocking phosphatidylcholine utilisation in Pseudomonas aeruginosa, via mutagenesis of fatty acid, glycerol and choline degradation pathways, confirms the importance of this nutrient source in vivo . PLoS ONE 9: e103778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan, B.K. , Tupper, B. , Sczyrba, A. , Lauro, F.M. , Martinez‐Garcia, M. , González J.M. et al (2013) Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA 110: 11463–11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. , and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thole, S. , Kalhoefer, D. , Voget, S. , Berger, M. , Engelhardt, T. , Liesegang, H. , et al (2012) Phaeobacter gallaeciensis genomes from globally opposite locations reveal high similarity of adaptation to surface life. ISME J 6: 2229–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueland, P.M. (2011) Choline and betaine in health and disease. J Inherit Metab Dis 34: 3–15. [DOI] [PubMed] [Google Scholar]
- Wu, J. , Sunda, W. , Boyle, E.A. , and Karl, D.M. (2000) Phosphate depletion in the western North Atlantic Ocean. Science 289: 759–762. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Phylogenetic analysis of the substrate binding proteins (SBPs) affiliated with the putative choline ABC‐type transporter found in marine bacteria. Reference sequences from characterized SBPs were added to the alignment. Characterized SBPs, related to osmolyte SBPs, based on the structural analysis conducted by Berntsson and colleagues (2010) were used as an outgroup. The tree was aligned in mega 5.2 using the neighbour‐joining method using 500 replications for bootstrapping. The scale bar represents the number of substitutions per amino acid. ChoX, SBP specific for choline; TmoX, SBP specific for trimethylamine N‐oxide; BetX, SBP specific for glycine betaine; CaiX, SBP specific for carnitine.
Fig. S2. Detailed phylogeny of ChoX from Fig. S1 showing strain names and their corresponding accession numbers (Gene ID in IMG/JGI).
Fig. S3. Phylogenetic analysis of choline dehydrogenase (BetA). The evolutionary history was inferred using the neighbour‐joining method. For the major nodes, the percentage (> 75%) of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p‐distance method and are in the units of the number of amino acid differences per site. The analysis involved 101 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 685 positions in the final dataset. Evolutionary analyses were conducted in mega6.
Fig. S4. Gene neighbourhoods of fhs1 and fhs2. The scale bar represents the number of bases. folD, 5,10‐methylene‐H4F dehydrogenase/methenyl‐H4F cyclohydrolase; tmm, trimethylamine monooxygenase; tdm, trimethylamine N‐oxide demethylase; fhs, formyl‐H4F synthetase; tmoR, putative regulator of tmm; amt, unspecified ammonium transporter; ftsH, ATP‐dependent metalloprotease; fhdA, formate dehydrogenase alpha subunit; fhdB, formate dehydrogenase beta subunit; PBP, uncharacterized HAAT family amino acid periplasmic binding protein.
Fig. S5. Growth of the R. pomeroyi fhs null mutant on GBT (red squares), homocysteine (purple crosses) or GBT and homocysteine (green triangles) as the carbon source respectively. A positive control consisted of glucose as a carbon source and the negative control had no added carbon. Cultures were grown in triplicate. Error bars denote SD. Hcy, homocysteine.
Fig. S6. Growth of R. pomeroyi wild‐type and the fhs null mutant on glucose and GBT as the carbon and energy source and ammonium as the nitrogen source. Tetrahydrofolate (1 mM) was added to wild‐type and mutant cultures at T = 0 h and T = 21 h, and GBT consumption was recorded. Cultures were grown in triplicate. Error bars denote SD.
Table S1. List of oligonucleotides used in this study.


