Abstract
Bending with high curvature is one of the major mechanical properties of double‐stranded DNA (dsDNA) that is essential for its biological functions. The emergence of a kink arising from local melting in the middle of dsDNA has been suggested as a mechanism of releasing the energy cost of bending. Herein, we report that strong bending induces two types of short dsDNA deformations, induced by two types of local melting, namely, a kink in the middle and forks at the ends, which we demonstrate using D‐shaped DNA nanostructures. The two types of deformed dsDNA structures dynamically interconvert on a millisecond timescale. The transition from a fork to a kink is dominated by entropic contribution (anti‐Arrhenius behavior), while the transition from a kink to a fork is dominated by enthalpic contributions. The presence of mismatches in dsDNA accelerates kink formation, and the transition from a kink to a fork is removed when the mismatch size is three base pairs.
Keywords: bending, DNA structures, FRET, kinks, mismatched sequences
Understanding the mechanical properties of double‐stranded DNA (dsDNA), which is a semiflexible biopolymer, is crucial for discerning the biological processes that occur with dsDNA, and the application of DNA as a nanomaterial.1 The ability of dsDNA to bend with high curvature is important for many biological processes. For instance, approximately 147 base pairs (bp) of genomic DNA wraps 1.7 times around a nucleosome of 8.5 nm in diameter;1d, 2 viral DNA that is 16 μm long is packed into a tiny volume in viruses (ca. 5×104 nm3);1d and DNA looping with high curvature (a bending curvature of ca. 10 nm−1) occurs during transcriptional regulation.3 The bending properties of dsDNA have been described using the wormlike chain model, which is characterized by the persistence length L p (ca. 150 bp).4
Since the report of hyperflexibility during cyclization of short dsDNA (ca. 100 bp),5 dsDNA bending has been a central debate in the field.4b, 6 Recently, a single‐molecule study showed extreme dsDNA bendability at less than 100 bp by analyzing real‐time cyclization kinetics.7 Despite many investigations into short dsDNA bending, the mechanism that dsDNA uses to overcome high bending energy under strong bending conditions has been rarely investigated, leaving the origin of this phenomenon unexplained. DNA kinking with curvature‐induced local melting of dsDNA has been proposed as a mechanism for releasing high bending stress and increasing the flexibility of dsDNA, but the associated dynamic properties and other types of deformed structures remain unknown.8
We have investigated the mechanism by which short dsDNA (30 bp length) dynamically releases bending stress induced by a high compressive force. We investigated this mechanism through directly observing dsDNA conformational states and dynamics by using single‐molecule fluorescence resonance energy transfer (smFRET). A D‐shaped DNA nanostructure was constructed by hybridizing circular ssDNA with complementary ssDNA, which induces bending in the short dsDNA portion (Figure 1 in the Supporting Information).9 A compressive bending force was generated by the entropic spring of the ssDNA portion. Surprisingly, two D‐shaped dsDNA conformations were observed under a strong bending force; one was a kink state arising from local melting at the center of dsDNA portion, and the other conformation was a “fork” state arising from local melting at both ends of the dsDNA portion. Real‐time, single‐molecule FRET measurements showed that the two deformed structures dynamically interconverted on a 1–2 ms timescale. Our results suggest that short dsDNA releases bending stress through not only kink formation but also local end melting.
To investigate short dsDNA bending, we prepared ssDNA rings of various lengths by ligating linear ssDNAs molecules (R34, R38, R40, R42, R44, R46, and R60); we then hybridized the ssDNA ring with a complementary 30 nucleotide (nt) ssDNA (S30) to produce D‐shaped DNA (D‐sample; Figure 1 a–c and Figure 1 in the Supporting Information). The D‐samples consisted of a 30 bp dsDNA portion and ssDNA string of different lengths (4–30 nt).9a,b For example, R38S30 denotes a D‐sample formed by hybridizing R38 and S30, which includes a 30 bp dsDNA portion and an 8 nt ssDNA string (Figure 1 c). ssDNA rings and S30 were labeled with Atto‐550 and Atto‐647N as donor and acceptor, respectively, which produced a 23 bp donor–acceptor distance for all D‐samples. We used the alternating‐laser excitation method (ALEX) to detect freely diffusing individual molecules (Figure 2 in the Supporting Information); this method measures FRET efficiency (E) and the stoichiometry parameter (S) simultaneously (Figure 1 d–f).10 E gives the distance between the donor and acceptor, while S describes the molecular species. For example, S≈1 indicates a donor‐only species, S≈0 indicates an acceptor‐only species, and S≈0.5 indicates a donor–acceptor species. As a result, the D‐samples, the hybridized form of the ssDNA ring, and S30 (orange box in Figure 1 d–f), can be distinguished from a nonhybridized ssDNA ring (donor‐only species, green ellipse in Figure 1 d) and S30 (acceptor‐only species, red ellipse in Figure 1 d) using the E–S graph.
Figure 1.
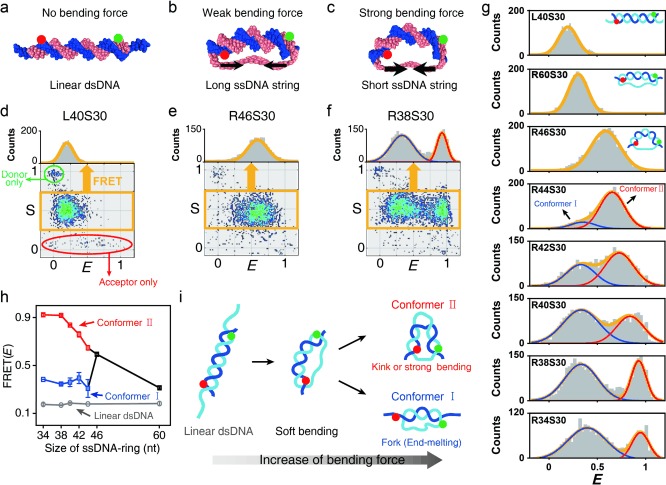
Observation of two types of dsDNA structures that are deformed by tension. a–c) Illustrations of linear dsDNA with an overhang (L40S30), D‐shaped DNA nanostructures with a 16 nt ssDNA string (R46S30) and an 8 nt ssDNA string (R38S30), respectively. d–f) Two‐dimensional E–S graphs obtained using ALEX single‐molecule measurements: d) L40S30, e) R46S30, and f) R38S30. The green and red ellipses denote donor‐only and acceptor‐only species, respectively. The yellow boxes represent doubly labeled D‐samples. The selected doubly labeled species (yellow boxes) were used to obtain one‐dimensional (1D) FRET histograms. g) 1D FRET histograms for various ssDNA string lengths. Histograms were fitted to a single or double Gaussian distribution. h) The FRET efficiencies of each conformer depending on the ssDNA ring size, which were obtained from Figure 1 g. Error bars were obtained from the three independent measurements. i) Model of the two conformers generated by strong ssDNA tension. Conformer I includes a fork structure with local end melting of the dsDNA portion (low E value), whereas conformer II includes a kink with a high E value.
We measured the E value of linear dsDNA with a 5 nt overhang at both ends (Figure 1 a) as a control; the efficiency was ca. 0.18 (Figure 1 d). However, the E value of R46S30 with a 16 nt string was ca. 0.59, which is significantly larger than that of linear dsDNA (Figure 1 b, e). This result implies that bending of dsDNA portion occurs because of the compressive force exerted by the entropic spring of the 16 nt ssDNA string.
Next, we modulated the force that bends the dsDNA portion in the D‐sample by varying the ssDNA string length (string length=4, 8, 10, 12, 14, 16, and 30 nt) but maintaining the same length for the dsDNA portion (Figure 1 g). As expected, reducing the ssDNA string length from 30 nt to 16 nt (R60S30 to R46S30) induced an increase in E from 0.31 to 0.59 (the second and third panels in Figure 1 g), which implies that the distance between donor and acceptor decreased through bending the dsDNA portion. Surprisingly, however, two subpopulations in the FRET distributions appeared when the ssDNA string length was lower than 14 nt (R44S30, the fourth panel in Figure 1 g); conformer I (blue curve) had E<0.5 , and conformer II (red curve) had higher than E>0.6. The conformer I population significantly increased as the ssDNA string length decreased. In addition, the E value of conformer II increased as the ssDNA string length decreased, but the E value of conformer I was nearly independent of the ssDNA string length (Figure 1 h).
Although dsDNA hybridization is stable at room temperature, local melting in the dsDNA may occur through thermal fluctuations. In addition, local melting of dsDNA can be induced by external forces, such as a stretching force11 and torque.9c, 12 Thus, two dsDNA conformations we observed can be explained by two types of local dsDNA melting, which may reduce the bending force applied by the ssDNA string (Figure 1 i). One conformation is a kink or a sharply bent state. The kink has been previously suggested;8a,c,e,f in this conformation the distance between two ends of the dsDNA portion becomes close and thus, it may exhibit a high E value (Figure 1 i). In contrast, the second type of local melting reduces the E value, which implies that the distance between two ends of the dsDNA portion becomes longer compared with the simple bending state. The second type of local melting can be explained by the formation of a “fork”, that is, local melting at the ends of dsDNA portion (conformer I). The applied force can induce local melting in the dsDNA portion not only in the middle but also at both ends.13 Melting at both ends of the dsDNA portion can reduce the tension applied to the dsDNA portion, and thus, the dsDNA portion may exhibit a linear or weakly bent form (Figure 1 i). Therefore, the distance between the two probes will be greater than in the bent form, which may result in a low E value.
To verify this explanation, we introduced mismatched sequences in the middle of the dsDNA portion (Figure 2 a), which enhances kink formation by acting as a flexible hinge.8a,e We introduced a triple bp mismatch (S30m3) in the middle of the dsDNA portion as permanent local melt, which could induce a DNA kink (see Figure 3 in the Supporting Information for the effects of single and double bp mismatches).8a, 14 In the presence of the triple mismatch, all D‐samples, which otherwise gave the two conformers in Figure 1 g, showed only a high‐FRET conformer (Figure 2 a). This result indicates that conformer II, which exhibits the high E value shown in Figure 1 g, is induced by local melting in the middle of the dsDNA portion, which may produce a kink.
Figure 2.
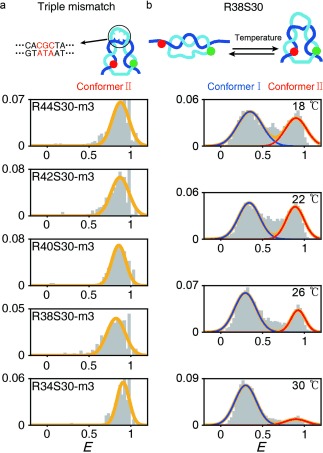
The effect of mismatches and temperature on the two conformer populations. a) FRET efficiency histograms of the D‐samples with a triple mismatch at the center of the dsDNA portion. b) FRET efficiency histograms of R38S30 as a function of temperature. The subpopulation of the low FRET species (conformer I) increased as the temperature increased.
Next, we tested the temperature and Mg2+‐concentration dependence of the FRET distribution of the D‐samples (Figure 2 b and Figure 4 in the Supporting Information). At high temperatures, DNA melting is facilitated by increasing the entropy of DNA. However, Mg2+ stabilizes dsDNA hybridization by reducing electrostatic repulsion between negatively charged DNA backbones15 and enhances the probability of kink formation.8e Thus, we expected that an increase in temperature would induce dsDNA melting, which may increase the fork population, whereas a high Mg2+ concentration would stabilize the dsDNA form and, thus, reduce the fork population. As expected, the conformer I population (R38S30) increased considerably compared with conformer II as the temperature increased (Figure 2 b). R40S30 and R42S30 also showed similar temperature‐dependent trends (Figure 5 in the Supporting Information). This result implies that conformer I is favored through destabilizing the hybridized dsDNA form. On the other hand, when we increased the Mg2+ concentration, the conformer II population increased in all D‐samples (Figure 4 in the Supporting Information). These results imply that conformer I is more favorable for dsDNA melting compared with conformer II. In addition, previous studies reported that Mg2+ facilitates a DNA kink8e and increases DNA flexibility,8f, 9c, 16 which is consistent with our results, wherein the conformer II population increases because of the presence of Mg2+ ions. Thus, all our results support that conformer I corresponds to a fork state, while conformer II corresponds to a kink state. The S1 endonuclease measurements support that conformer II is in a kink state with dsDNA melting (Figures 6 and 7 in the Supporting Information), which is consistent with previous studies that reported kink formation when dsDNA is highly bent.6b, 8e Our interpretation of the two deformed structures is further supported by transition‐barrier measurements below.
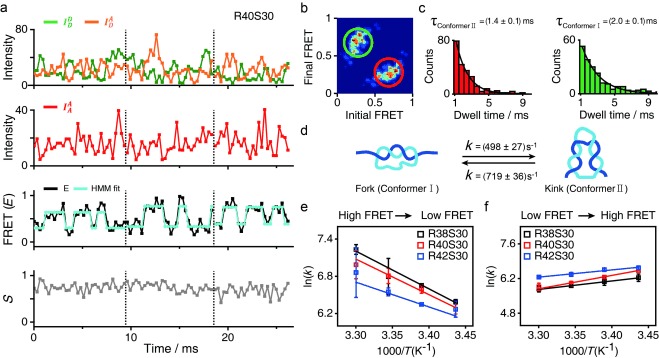
Even though we used a FRET distribution analysis to demonstrate the formation of two types of the deformed dsDNA portion under equilibrium conditions (Figure 1 g), it remains questionable whether these two conformations are permanently stable or dynamically interconvert with each other. To investigate their dynamic behavior, we immobilized the D‐samples on the surface of a quartz slide and obtained real‐time FRET traces for individual samples using total‐internal reflection microscopy (TIRF; Figure 8 in the Supporting Information): no transition in the FRET time trace of the D‐samples was observed. The E value observed in the time trace is similar to the average E value of conformers I and II, however, we suspected that the transition time between the two conformers was faster than 100 ms (the temporal resolution of TIRF). Thus, we performed diffusion‐based single‐molecule experiments using a high‐power laser (250 kW cm−2) with photoprotection buffer (see the Supporting Information).17 By using this approach, we could achieve a fluorescence intensity of approximately 100 photons ms−1, which is sufficient for 0.3 ms resolution on the FRET time trace (Figure 3). We selected time traces longer than 8 ms and then applied a hidden Markov model (HMM) analysis to the time traces.18 Figure 3 a shows typical time traces for R40S30 with a 0.3 ms temporal resolution. Although the fluorescent intensities of the donor emission 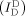 and FRET signal
and FRET signal 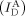 fluctuated considerably, the anticorrelation behavior between the two types of intensities was clearly observed (upper panel in Figure 3 a). All time traces (607 traces) showed dynamic fluctuation. The third panel in Figure 3 a gives FRET time traces with an HMM fitting curve, which shows the two major states at E=0.25 and 0.70. These two E values are consistent with the E values of two conformers from the FRET distributions (Figure 1 g). Despite the broad transitions, two main transitions appear in the transition density plot (Figure 3 b). The transition rates were generated from the dwell time distributions of each transition, k
I→II=(498±27) s−1 (i.e., the inverse of the average dwell time of conformer I≈2.0 ms) and k
II→I=(719±36) s−1 (i.e., the inverse of the average dwell time of conformer II≈1.4 ms), at room temperature (Figure 3 c). The transition rates of deformed dsDNA (Figure 3 d) are in a similar range as other DNA dynamics, such as DNA hairpin conformational dynamics.19 As for DNA hairpins, the dynamics on the microsecond scale were also observed,19 and cannot be resolved using the FRET measurement used in this work.
fluctuated considerably, the anticorrelation behavior between the two types of intensities was clearly observed (upper panel in Figure 3 a). All time traces (607 traces) showed dynamic fluctuation. The third panel in Figure 3 a gives FRET time traces with an HMM fitting curve, which shows the two major states at E=0.25 and 0.70. These two E values are consistent with the E values of two conformers from the FRET distributions (Figure 1 g). Despite the broad transitions, two main transitions appear in the transition density plot (Figure 3 b). The transition rates were generated from the dwell time distributions of each transition, k
I→II=(498±27) s−1 (i.e., the inverse of the average dwell time of conformer I≈2.0 ms) and k
II→I=(719±36) s−1 (i.e., the inverse of the average dwell time of conformer II≈1.4 ms), at room temperature (Figure 3 c). The transition rates of deformed dsDNA (Figure 3 d) are in a similar range as other DNA dynamics, such as DNA hairpin conformational dynamics.19 As for DNA hairpins, the dynamics on the microsecond scale were also observed,19 and cannot be resolved using the FRET measurement used in this work.
Figure 3.
Real‐time measurement of the D‐sample conformational dynamics under a strong bending force. a) Time traces of single diffusing D‐samples (R40S30) obtained using a high‐power laser with photo‐protection buffer. Three individual traces are displayed. Top panel: fluorescence intensity of the donor through donor excitation (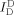 , green line) and fluorescence intensity of the acceptor through donor excitation (
, green line) and fluorescence intensity of the acceptor through donor excitation (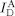 , orange line), which is a FRET signal. Second panel: fluorescence intensity of the acceptor through acceptor excitation (
, orange line), which is a FRET signal. Second panel: fluorescence intensity of the acceptor through acceptor excitation (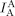 , red line), which indicates that the acceptor dye is present. Third panel: FRET efficiency (E) time traces with the HMM fitting curve, where two state transitions are shown. Fourth panel: time traces for the stoichiometry values (S) between the donor and acceptor. b) Transition density plot of the two main transitions between the E values of 0.25 and 0.70 obtained from the third panel of Figure 3 a (607 traces total). c) Dwell time (τ) distributions of conformer I and conformer II. The transition rate (1/τ) was obtained from the single exponential decay fit. d) The transition rates between the two d‐sample conformers. e–f) Arrhenius plot for the transition rate of d‐samples depending on the temperature: e) transition from conformer II to conformer I and f) transition from conformer I to conformer II. The activation energy barrier for the conformer II to conformer I transition was measured at (51±3), (48±5), and (33±7) kJ mol−1 for R38S30, R40S30, and R42S30, respectively.
, red line), which indicates that the acceptor dye is present. Third panel: FRET efficiency (E) time traces with the HMM fitting curve, where two state transitions are shown. Fourth panel: time traces for the stoichiometry values (S) between the donor and acceptor. b) Transition density plot of the two main transitions between the E values of 0.25 and 0.70 obtained from the third panel of Figure 3 a (607 traces total). c) Dwell time (τ) distributions of conformer I and conformer II. The transition rate (1/τ) was obtained from the single exponential decay fit. d) The transition rates between the two d‐sample conformers. e–f) Arrhenius plot for the transition rate of d‐samples depending on the temperature: e) transition from conformer II to conformer I and f) transition from conformer I to conformer II. The activation energy barrier for the conformer II to conformer I transition was measured at (51±3), (48±5), and (33±7) kJ mol−1 for R38S30, R40S30, and R42S30, respectively.
We next studied the temperature dependence of the transition rates to understand the nature of the free‐energy barrier between the two conformations (Figure 3 e, f). Based on an Arrhenius plot 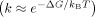 ,19a the slope of ln(k) with respect to 1/T for the transition from conformer II to conformer I is negative (Figure 3 e), and the free‐energy barrier (ΔG=ΔH−TΔS) was measured at 33–51 kJ mol−1 for R38S30, R40S30, and R42S30. This result implies that the transition from a kink state to a fork state is activated by crossing the free‐energy barrier, which is majorly contributed by enthalpy change (ΔH>0). In contrast, the Arrhenius plot slope for the transition from conformer I to conformer II is positive (Figure 3 f), which indicates that the free‐energy barrier from a fork state to a kink state, although reduced by the negative enthalpy change (ΔH<0), is majorly influenced by the entropic contribution. This type of anti‐Arrhenius behavior, namely when the slope in the Arrhenius plot is positive, was previously observed for a DNA hairpin; the transition from an open conformation to a closed conformation showed anti‐Arrhenius behavior.19a The anti‐Arrhenius behavior of DNA hairpin was postulated to occur because hybridization (or zipping) between two ends of a DNA hairpin is entropically unfavorable. Similarly, the transition from a fork state to a kink state also requires hybridization (or zipping) between two separated ssDNA portions. High temperatures increase the configuration entropy of the fork state, which lowers the zipping rate. In contrast, the transition from a kink state to a fork state requires dehybridization at both ends of the dsDNA portion; thus, increased thermal energy at higher temperatures aids in overcoming the free‐energy barrier. Although an increase in temperature also facilitates melting in the middle of the dsDNA portion and could be favorable for kink state, the thermal energy seems to contribute more favorably to melting the end parts of the dsDNA. Overall, the Arrhenius and anti‐Arrhenius behaviors of the transitions within the D‐samples further support our explanation that conformer I is in a fork state, and conformer II is in a kink state.
,19a the slope of ln(k) with respect to 1/T for the transition from conformer II to conformer I is negative (Figure 3 e), and the free‐energy barrier (ΔG=ΔH−TΔS) was measured at 33–51 kJ mol−1 for R38S30, R40S30, and R42S30. This result implies that the transition from a kink state to a fork state is activated by crossing the free‐energy barrier, which is majorly contributed by enthalpy change (ΔH>0). In contrast, the Arrhenius plot slope for the transition from conformer I to conformer II is positive (Figure 3 f), which indicates that the free‐energy barrier from a fork state to a kink state, although reduced by the negative enthalpy change (ΔH<0), is majorly influenced by the entropic contribution. This type of anti‐Arrhenius behavior, namely when the slope in the Arrhenius plot is positive, was previously observed for a DNA hairpin; the transition from an open conformation to a closed conformation showed anti‐Arrhenius behavior.19a The anti‐Arrhenius behavior of DNA hairpin was postulated to occur because hybridization (or zipping) between two ends of a DNA hairpin is entropically unfavorable. Similarly, the transition from a fork state to a kink state also requires hybridization (or zipping) between two separated ssDNA portions. High temperatures increase the configuration entropy of the fork state, which lowers the zipping rate. In contrast, the transition from a kink state to a fork state requires dehybridization at both ends of the dsDNA portion; thus, increased thermal energy at higher temperatures aids in overcoming the free‐energy barrier. Although an increase in temperature also facilitates melting in the middle of the dsDNA portion and could be favorable for kink state, the thermal energy seems to contribute more favorably to melting the end parts of the dsDNA. Overall, the Arrhenius and anti‐Arrhenius behaviors of the transitions within the D‐samples further support our explanation that conformer I is in a fork state, and conformer II is in a kink state.
We have demonstrated that two types of deformed conformations of D‐shaped dsDNA co‐exist upon strong bending tension. One is a kink state with local melting in the central region of dsDNA, and the other is a fork state that includes local melting at both ends of the dsDNA portion. In this work, R40S30 is in a boundary structure that includes a DNA kink. Using a simple approximation (Figure 9 in the Supporting Information), the R40S30 curvature radius was estimated to be around 3 nm, which corresponds to approximately 56–65 bp dsDNA for cyclization.8f, 20
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
miscellaneous_information
This work was supported by grants from the National Research Foundation of Korea (NRF‐2013R1A1A2063302 and NRF‐2013R1A1A2008900) and Nano Material Technology Development Program (NRF‐2014M3A7B6034580). dsDNA=double‐stranded DNA.
References
- 1.
- 1a. J. P. Peters III , Maher L. J., Q. Rev. Biophys. 2010, 43, 23–63; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b. Rothemund P. W. K., Nature 2006, 440, 297–302; [DOI] [PubMed] [Google Scholar]
- 1c. Seeman N. C., Q. Rev. Biophys. 2005, 38, 363–371; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1d. Garcia H. G., Grayson P., Han L., Inamdar M., Kondev J., Nelson P. C., Phillips R., Widom J., Wiggins P. A., Biopolymers 2007, 85, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Richmond T. J., Davey C. A., Nature 2003, 423, 145–150. [DOI] [PubMed] [Google Scholar]
- 3. Wong O. K., Guthold M., Erie D. A., Gelles J., Plos Biol. 2008, 6, 2028–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.
- 4a. Smith S. B., Cui Y. J., Bustamante C., Science 1996, 271, 795–799; [DOI] [PubMed] [Google Scholar]
- 4b. Mastroianni A. J., Sivak D. A., Geissler P. L., Alivisatos A. P., Biophys. J. 2009, 97, 1408–1417; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4c. Vologodskaia M., Vologodskii A., J. Mol. Biol. 2002, 317, 205–213; [DOI] [PubMed] [Google Scholar]
- 4d. Bustamante C., Bryant Z., Smith S. B., Nature 2003, 421, 423–427; [DOI] [PubMed] [Google Scholar]
- 4e. Bustamante C., Smith S. B., Liphardt J., Smith D., Curr. Opin. Struct. Biol. 2000, 10, 279–285. [DOI] [PubMed] [Google Scholar]
- 5. Cloutier T. E., Widom J., Mol. Cell 2004, 14, 355–362. [DOI] [PubMed] [Google Scholar]
- 6.
- 6a. Wiggins P. A., Van der Heijden T., Moreno‐Herrero F., Spakowitz A., Phillips R., Widom J., Dekker C., Nelson P. C., Nat. Nanotechnol. 2006, 1, 137–141; [DOI] [PubMed] [Google Scholar]
- 6b. Vologodskii A., Frank‐Kamenetskii M. D., Nucleic Acids Res. 2013, 41, 6785–6792; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6c. Yuan C. L., Lou X. W., Rhoades E., Chen H. M., Archer L. A., Nucleic Acids Res. 2007, 35, 5294–5302; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6d. Du Q., Smith C., Shiffeldrim N., Vologodskaia M., Vologodskii A., Proc. Natl. Acad. Sci. USA 2005, 102, 5397–5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vafabakhsh R., Ha T., Science 2012, 337, 1097–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.
- 8a. Yan J., Marko J. F., Phys. Rev. Lett. 2004, 93, 108108; [DOI] [PubMed] [Google Scholar]
- 8b. Wiggins P. A., Phillips R., Nelson P. C., Phys. Rev. E 2005, 71, 021909; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8c. Lankaš F., Lavery R., Maddocks J. H., Structure 2006, 14, 1527–1534; [DOI] [PubMed] [Google Scholar]
- 8d. Curuksu J., Zacharias M., Lavery R., Zakrzewska K., Nucleic Acids Res. 2009, 37, 3766–3773; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8e. Fields A. P., Meyer E. A., Cohen A. E., Nucleic Acids Res. 2013, 41, 9881–9890; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8f. Du Q., Kotlyar A., Vologodskii A., Nucleic Acids Res. 2008, 36, 1120–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.
- 9a. Shroff H., Reinhard B. M., Siu M., Agarwal H., Spakowitz A., Liphardt J., Nano Lett. 2005, 5, 1509–1514; [DOI] [PubMed] [Google Scholar]
- 9b. Shroff H., Sivak D., Siegel J. J., McEvoy A. L., Siu M., Spakowitz A., Geissler P. L., Liphardt J., Biophys. J. 2008, 94, 2179–2186; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9c. Qu H., Wang Y., Tseng C. Y., Zocchi G., Phys. Rev. X 2011, 1, 021008. [Google Scholar]
- 10.
- 10a. Kapanidis A. N., Lee N. K., Laurence T. A., Doose S., Margeat E., Weiss S., Proc. Natl. Acad. Sci. USA 2004, 101, 8936–8941; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10b. Kim C., Kim J. Y., Kim S. H., Lee B. I., Lee N. K., Chem. Commun. 2012, 48, 1138–1140. [DOI] [PubMed] [Google Scholar]
- 11.
- 11a. Shokri L., McCauley M. J., Rouzina L., Williams M. C., Biophys. J. 2008, 95, 1248–1255; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11b. Santosh M., Maiti P. K., J. Phys. Condens. Matter 2009, 21, 037443; [DOI] [PubMed] [Google Scholar]
- 11c. Murade C. U., Subramaniam V., Otto C., Bennink M. L., Nucleic Acids Res. 2010, 38, 3423–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.
- 12a. Sarkar A., Leger J. F., Chatenay D., Marko J. F., Phys. Rev. E 2001, 63, 051903; [DOI] [PubMed] [Google Scholar]
- 12b. Jeon J. H., Sung W., Biophys. J. 2008, 95, 3600–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee O. C., Jeon J. H., Sung W., Phys. Rev. E 2010, 81, 021906. [DOI] [PubMed] [Google Scholar]
- 14. Zeng Y., Zocchi G., Biophys. J. 2006, 90, 4522–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.
- 15a. Owczarzy R., Moreira B. G., You Y., Behlke M. A., Walder J. A., Biochemistry 2008, 47, 5336–5353; [DOI] [PubMed] [Google Scholar]
- 15b. Williams L. D., Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 497–521. [DOI] [PubMed] [Google Scholar]
- 16. Fenley M. O., Manning G. S., Marky N. L., Olson W. K., Biophys. Chem. 1998, 74, 135–152. [DOI] [PubMed] [Google Scholar]
- 17.
- 17a. Campos L. A., Liu J. W., Wang X., Ramanathan R., English D. S., Munoz V., Nat. Methods 2011, 8, 143–146; [DOI] [PubMed] [Google Scholar]
- 17b. Kim J. Y., Kim C., Lee N. K., Nat. Commun. 2015, 6, 6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bronson J. E., Fei J., Hofman J. M., R. L. Gonzalez, Jr. , Wiggins C. H., Biophys. J. 2009, 97, 3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.
- 19a. Wallace M. I., Ying L. M., Balasubramanian S., Klenerman D., Proc. Natl. Acad. Sci. USA 2001, 98, 5584–5589; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19b. Nayak R. K., Peersen O. B., Hall K. B., Van Orden A., J. Am. Chem. Soc. 2012, 134, 2453–2456; [DOI] [PubMed] [Google Scholar]
- 19c. Jung J., Van Orden A., J. Am. Chem. Soc. 2006, 128, 1240–1249; [DOI] [PubMed] [Google Scholar]
- 19d. Kim J., Doose S., Neuweiler H., Sauer M., Nucleic Acids Res. 2006, 34, 2516–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Grayson P., Evilevitch A., Inamdar M. M., Purohit P. K., Gelbart W. M., Knobler C. M., Phillips R., Virology 2006, 348, 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
miscellaneous_information


