Abstract
The honey bee (Apis mellifera L.) is the most important managed pollinator species worldwide and plays a critical role in the pollination of a diverse range of economically important crops. This species is important to agriculture and historically has been used as a surrogate species for pollinators to evaluate the potential adverse effects for conventional, biological, and microbial pesticides, as well as for genetically engineered plants that produce pesticidal products. As part of the ecological risk assessment of MON 87411 maize, which expresses a double‐stranded RNA targeting the Snf7 ortholog (DvSnf7) in western corn rootworm (Diabrotica virgifera virgifera), dietary feeding studies with honey bee larvae and adults were conducted. Based on the mode of action of the DvSnf7 RNA in western corn rootworm, the present studies were designed to be of sufficient duration to evaluate the potential for adverse effects on larval survival and development through emergence and adult survival to a significant portion of the adult stage. Testing was conducted at concentrations of DvSnf7 RNA that greatly exceeded environmentally relevant exposure levels based on expression levels in maize pollen. No adverse effects were observed in either larval or adult honey bees at these high exposure levels, providing a large margin of safety between environmental exposure levels and no‐observed–adverse‐effect levels. Environ Toxicol Chem 2016;35:287–294. © 2015 The Authors. Environmental Toxicology and Chemistry Published by Wiley Periodicals, Inc. on behalf of SETAC.
Keywords: Honey bee (Apis mellifera), DvSnf7 double‐stranded RNA, Ecological risk assessment
INTRODUCTION
The application of RNA interference as a plant incorporated protectant is a new tool for insect pest control in agriculture 1, 2, 3, 4, 5. Two important studies, 1 conducted using the western corn rootworm (Diabrotica virgifera virgifera) 6 and 1 with the cotton bollworm (Helicoverpa armigera) 7, demonstrated the practical application of RNA interference in insect pest control using sequence‐specific gene silencing to suppress genes critical for insect survival. The sensitivity of corn rootworm larvae to oral RNA interference 6, 8, 9 and the economic importance of this pest complex for maize production 10, 11 provide an opportunity to develop an insecticidal double‐stranded RNA as a plant incorporated product for corn rootworm control.
Monsanto Company has developed MON 87411, which confers protection against corn rootworm (Diabrotica spp.) by producing a double‐stranded RNA that targets the Snf7 gene ortholog in corn rootworm (DvSnf7). The gene Snf7 is part of the endosomal sorting complex required for transport (ESCRT)‐III, which is involved in the sorting of transmembrane proteins en route to lysosomal degradation through the endosomal–autophagic pathway in a number of organisms 12, 13, 14. Therefore, Snf7 is a key regulator of biological processes important for eukaryotic cell growth and survival. On consumption, the plant‐produced double‐stranded RNA in MON 87411 is recognized by the corn rootworm's RNA interference machinery, resulting in down‐regulation of the targeted DvSnf7 gene and leading to cellular death and corn rootworm mortality 8, 15, 16. Our previous research has demonstrated that a 100% complementary sequence length of a ≥21‐nucleotide contiguous sequence embedded in a ≥60‐nucleotide double‐stranded RNA is required for biological activity against corn rootworm 8, 9. The DvSnf7 double‐stranded RNA in ≥21‐nucleotide sequences acts as the functional unit incorporated into the RNA‐induced silencing complex, where the specific nucleotide sequences of the single‐strand RNA retained in the RNA‐induced silencing complex are required to guide the RNA‐induced silencing complex to locate and bind to target messenger RNA (mRNA) containing sequences complementary to the guide strand and finally block translation of the target mRNA through sequence‐specific mRNA cleavage 17.
The spectrum of activity for DvSnf7 RNA (hereafter DvSnf7) has been shown to be narrow 9. The activity of DvSnf7 was shown to be evident only in a subset of beetles within the Galerucinae subfamily of Chrysomelidae within the order Coleoptera (>90% identity with western corn rootworm Snf7 240 nucleotide), and no activity was found in those species that lack ≥21‐nucleotide sequence matches to the DvSnf7 ortholog 9. The high specificity of DvSnf7 greatly reduces the likelihood of adverse effects on nontarget organisms, including those beneficial to agriculture. The high specificity that can be achieved with double‐stranded RNAs has also been demonstrated with double‐stranded RNAs targeting vacuolar adenosinetriphosphatase transcripts 6, 18. The gene specificity inherent to the RNA interference mechanism allows for the design of double‐stranded RNA that can be used for the control of a single pest or a group of related species with the low likelihood of adversely affecting beneficial insects 18.
The honey bee (Apis mellifera L.) is the most important managed pollinator species worldwide and plays a critical role in providing pollination for many crops 19, 20. The economic value of honey bee pollination for agriculture has been estimated to be greater than $14 million in the United States 19 and $215 billion worldwide 20. Honey bee larvae and adults can be directly exposed to plant incorporated products expressed in genetically modified maize via pollen consumption. Therefore, the honey bee is routinely included as a surrogate species for insect pollinators in ecological risk assessment 21, 22, 23. Over a flowering period of approximately 2 wk, maize pollen is abundant and easily accessible to field honey bee workers 24. On average, 3.4 mg to 4.3 mg maize pollen could be consumed by a honey bee worker per day 25 and 1.52 mg to 2.04 mg maize pollen could be consumed during the larval stage 26. In most cases, however, the proportion of maize pollen as a total of all pollen collected and fed to larvae during summer will be low. It is therefore unlikely that maize pollen would regularly comprise more than 50% of the honey bee diet 27.
A tiered approach for risk assessment for genetically modified crops has been successfully used to evaluate the potential for adverse effects of plant incorporated products to nontarget organisms for 2 decades 21, 28, 29. A key concept of the tiered approach is the use of worst‐case exposure scenarios to assess the potential for adverse effects to nontarget organisms in laboratory studies. If no adverse effects are observed in these representative species at high exposure levels, confidence in the conclusion of negligible ecological risk is greatly increased 21, 28, 29. The current ecological risk‐assessment approach used by the US Environmental Protection Agency for plant incorporated products provides a framework for the environmental risk assessment of RNA interference‐based insecticidal traits in selecting the required assessment and measurement endpoints to meet specific protection goals (e.g., pollination services, pollinator biodiversity) 30, 31. Protection of pollination services and pollinator biodiversity are typically assessed at the Tier 1 level for plant incorporated products by evaluating brood survival and development and adult survival following dietary exposure through a significant portion of a worker honey bee's life span. A colony‐level semi–field study would only need to be conducted if an unacceptable assessment could not be achieved with a laboratory study.
The present study was performed to assess the potential for adverse effects of DvSnf7 on the honey bee at both larval and adult stages under worst‐case exposure scenarios. Based on the time to effect of DvSnf7 in western corn rootworm and southern corn rootworm 8, 32, larval and adult studies were designed to be of sufficient duration to evaluate the potential for adverse effects on development and survival. The combination of larval and adult studies covered the portion of the life cycle from shortly after hatching through a major portion of the adult life span. To achieve a worst‐case exposure scenario for maize pollen consumption, honey bee larvae or adults were exposed to DvSnf7 at concentrations >10 times a worst‐case dietary exposure level. Historically for plant incorporated products, demonstrating less than 50% mortality in Tier 1 assays with a >10‐fold margin of safety has been used to indicate that no unacceptable adverse effects will occur to fauna in the environment 21. In addition, a bioinformatics analysis was conducted to evaluate shared sequence identity between DvSnf7 and the published A. mellifera genome 33 for possible 21‐nucleotide matches.
MATERIALS AND METHODS
Synthesis of DvSnf7 RNA
The test substance was in vitro T7 RNA polymerase‐transcribed 968‐nucleotide DvSnf7 RNA and suspended in ultrapure distilled water (Life Technologies). The DvSnf7 concentration was determined to be 2.21 mg/mL using NanoDrop 8000 (Thermo Scientific). The DvSnf7 sequence contains 2 DvSnf7 sequences of 240 nucleotides in an inverted orientation separated by an intervening sequence of 150 nucleotides (Figure 1A). When the DvSnf7 RNA is transcribed, the RNA expressed forms a hairpin loop, thereby allowing the formation of 240‐bp DvSnf7 double‐stranded RNA (Figure 1B), which is the “active” region within the larger 968‐nucleotide RNA molecule. Aliquots of the DvSnf7 sample were stored in a –80 °C freezer when not in use.
Figure 1.
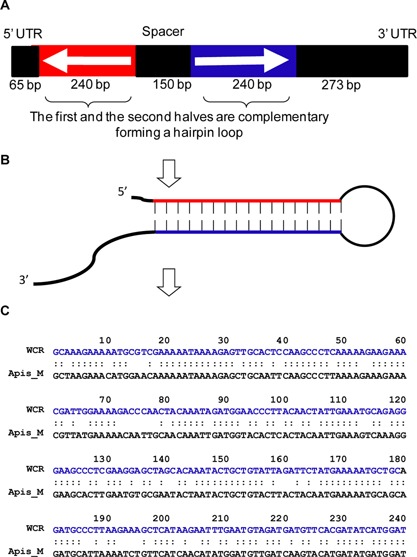
(A) A schematic diagram showing DvSnf7 RNA in 968‐nucleotide produced in MON87411 maize hybrid contains 2 DvSnf7 sequences of 240 nucleotide in an inverted orientation (1 in red and 1 in blue). (B) The 240‐bp DvSnf7 double‐stranded RNA in a hairpin loop is the “active” region that suppresses the targeted western corn rootworm Snf7 gene, resulting in cellular death and mortality on consumption by the corn rootworm. (C) The Snf7 orthologs of western corn rootworm and Apis mellifera share 72.5% identity, and no 21‐nucleotide matches exist. Apis M = Apis mellifera; UTR = untranslated region; WCR = western corn rootworm.
The DvSnf7 expression levels were determined in maize pollen from MON 87411 using a QuantiGene Plex 2.0 assay following the manufacturer's instructions (Affymetrix). A custom QuantiGene probe set was designed by the manufacturer to hybridize to the sequence within DvSnf7. The sequence‐specific recognition by the QuantiGene probe set has previously been described in detail 34. Mean measured DvSnf7 expression was 0.103 ng/g fresh weight pollen, with a range from 0.056 ng/g to 0.224 ng/g fresh weight pollen across 5 samples collected from 5 field sites representative of commercial maize‐producing regions during 2011 and 2012. Mean measured DvSnf7 expression was highest in over‐season whole plants at stages V3 and V4 at 10.5 ng/g fresh weight (range 6.78–23.1 ng/g fresh wt) and lowest in grain at 0.091 ng/g fresh weight (range 0.049–0.153 ng/g fresh wt).
An inorganic compound, potassium arsenate (KH2AsO4; Sigma‐Aldrich), was selected based on its mode of action as a positive control to confirm the effectiveness of diet feeding tests in the present study to detect the potential adverse effect of DvSnf7 through dietary exposure. Potassium arsenate is a stomach poison to many organisms, including insects, and has been frequently used as a positive control in bioassays with nontarget arthropods to assess plant incorporated products. It has been shown that DvSnf7 rapidly down‐regulates DvSnf7 mRNA expression in midgut tissue following dietary exposure 8.
Honey Bees
All hives used in the present study were created in April 2014 from adult honey bee queens purchased from Honeybee Genetics. Hives were maintained at field sites of Kerman, California, USA, and acclimated for a minimum of 30 d in the bee yard of California Agricultural Research. Larval and adult honey bees were obtained from hives that demonstrated good health and colony strength for inclusion in the present study.
Larval feeding test
The larval feeding test included an assay control treatment (negative control), a DvSnf7 treatment, and a positive control treatment, each with 4 replicates. All treatment diet solutions were prepared on the day of administration, including a 30% sucrose/purified water (w/v) solution as the assay control, the single DvSnf7 treatment diet solution at 1000 ng/g diet, and a positive control diet solution at 2000 µg potassium arsenate/mL diet.
On the day of test initiation, sets of 2 larval frames from 1 hive were transported to the laboratory inside an insulated container (i.e., ice chest) and covered with a damp towel. Two frames from the same hive were used in 1 replicate to minimize the variation among the larvae within the same replicate. Within each replicate, the DvSnf7 and assay control treatments were randomly assigned to 2 sides of 1 frame. The positive control was randomly assigned to 1 side of a separate frame. The 2 frames from 1 hive used for the 3 treatments in each replicate represent 1 block. The 3 treatments were overall arranged in a randomized complete block design. Twenty cells with larvae ≤2 d old were treated per replicate, and the location of each treated larvae was mapped on an acetate sheet.
Each treatment diet solution was administered in a 10‐μL volume with a pipette onto the bottom of each larval cell for oral consumption. During dosing, the laboratory was maintained at approximately 24 °C and 66% relative humidity. Treated frames were placed back in their original humidified container for at least 30 min to allow for undisturbed feeding, before the frames were returned to their original hive for care by the colony. On day 7 after treatment, all larvae in the locations assigned with the acetate paper were individually evaluated for capping or the presence or absence of larvae. Larvae were considered viable if cells were capped. After the final capping evaluation at day 10 after treatment, all treated frames were housed in an environmentally controlled growth chamber to monitor adult emergence until emergence was completed on day 17. Treated cells were covered with an adult emergence cage, and all cells not included in the treatment were destroyed within the caged area. The number of emerged adult bees was recorded daily. Temperature in the growth chamber ranged from 27 °C to 30 °C, with a relative humidity from 47.3% to 72.6% and a 0:24‐h light:dark photoperiod except during the time emergence assessments were conducted.
Adult feeding test
Two days prior to the test, 4 cleaned frames with mature broods were obtained from 4 separate hives. Each frame from 1 hive was moved into a screened hive box along with a food frame from the same hive and incubated in a growth chamber. On the day of the test, adult bees (≤2 d old) from the frame were placed in 3 cylinder‐shaped cages of approximately 60 × 155 mm, and each cage had 20 healthy adult bees impartially assigned to 1 of the 3 treatments before providing the appropriate treatment diet. Four replicates (4 cages) for each treatment were arranged as a randomized complete block design. Two 12‐mL glass vials (18 × 68 mm with 1‐mm holes in the caps) filled with the appropriate treatment diet solution were placed on the top of the cages and replaced every 2 d.
Treatment diets included an assay control diet of 50% sucrose (negative control), DvSnf7 at 1000 ng/g in 50% sucrose, and potassium arsenate at 20 µg/mL of 50% sucrose solution (positive control). All treatment diets were prepared as a single batch prior to test initiation and aliquoted for individual feedings. All aliquots were stored at –80 °C when not in use. All test bees were incubated in a growth chamber with a temperature range of 26 °C to 32 °C, 41% to 80% relative humidity, and a 0:24‐h light:dark photoperiod. All test adults were observed daily for mortality through the present 14‐d study.
Bioinformatic analysis
Orthologous Snf7 sequence alignments were prepared using the FASTA program 35 to demonstrate the sequence identity of Snf7 orthologs (240 nucleotide) of honey bee and western corn rootworm (Figure 1C). Alignments were analyzed for the percentage of sequence identity, the number of 21‐nucleotide matches, and the longest contiguous sequence match present. In addition, a BLAST search was performed for all possible 21‐bp matches from the DvSnf7 double‐stranded RNA (240 nucleotide) against the published genome for A. mellifera 33.
Dose confirmation of test substance
Two DvSnf7 diet samples collected at day 0 and day 14, respectively, for the adult test and 1 diet sample collected at the end of larval dosing were analyzed for biological activity. In addition, 1 sample was collected from the assay control diet in both larval and adult feeding tests. All diet samples were stored in –80 °C or on dry ice until analysis.
Biological activity of DvSnf7 in diet samples was evaluated in 12‐d diet incorporation bioassays using southern corn rootworm (Diabrotica undecimpunctata howardi) obtained from Crop Characteristics. The southern corn rootworm is a coleopteran species that is highly sensitive to DvSnf7 8, 9 and suitable for toxicity testing because of its relative ease of laboratory rearing and handling 36. Separate bioassays were conducted to analyze samples from the larval and adult stages using different southern corn rootworm batches. The biological activity of DvSnf7 in diet samples was compared with the DvSnf7 reference standard across 7 concentrations with a negative control to fully characterize the concentration responses and to estimate median lethal concentrations (LC50) along with associated 95% confidence intervals (CIs). Treatments were prepared by mixing a 2‐mL dosing solution, containing an aliquot of the DvSnf7 reference standard or DvSnf7 from diet samples, with an agar‐based southern corn rootworm diet (Bio‐Serv) to a final volume of 10 mL. Samples were vortex‐mixed until homogeneous, and 0.25‐mL aliquots were added to 48‐well plates (Falcon). Each concentration level tested 24 individually housed southern corn rootworm larvae (≤30 h posterior to the first observation of hatch). All assay plates were incubated at a target temperature of 27 °C, with 70% relative humidity, and in the dark. The number of insects infested and the number of survivors at each concentration level were recorded at the end of the 12‐d bioassay.
Data analyses
The PROBIT analysis was performed to estimate LC50 values and associated 95% CIs for dose confirmation assays using PROC PROBIT in SAS (Ver 9.4; SAS Institute). The OPTC option was used to correct for control mortality. Overlapping 95% CIs from estimated LC50 values was the criterion used to evaluate for potential difference in biological activity between the DvSnf7 in diet samples and the reference standard DvSnf7 for dose confirmations. If 95% CIs overlapped, no difference between treatments was concluded.
All endpoints were reported as means along with standard errors. For adult survival in the adult feeding test, a pairwise comparison between the DvSnf7 treatment and assay control was defined within a generalized linear mixed model under a randomized complete block design, and the difference on the logit link scale was analyzed with a t test using SAS PROC GLIMMIX. The positive control was excluded from the analysis because survival was 0%. For survival in the larval feeding test, a statistical analysis was not performed because there was 100% survival in both DvSnf7 treatment and assay control. However, a pairwise comparison of time to 50% adult emergence between the DvSnf7 treatment and assay control was defined within a linear mixed model under a randomized complete block design and analyzed with a t test using SAS PROC MIXED. The level of statistical significance for all tests was set at α = 0.05.
RESULTS
Larval feeding test
The larval feeding test included an assay control, a 10‐μL volume of DvSnf7 at 1000 ng/g diet solution (nominally 11.3 ng DvSnf7/larva), and a positive control with potassium arsenate. All cells in the assay control and DvSnf7 treatments were observed as capped on day 7, indicating that 100% of the test larvae survived from dosing to capping (Figure 2A). In contrast, no treated cells were capped in the positive control treatment, indicating 100% mortality and confirming consumption of treatment solutions by test larvae (Figure 2A).
Figure 2.
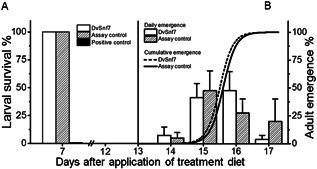
Survival of test larvae (A) and adult emergence from surviving honey bee larvae in the DvSnf7 and assay control treatments (B). Each data point represents the average of 4 replicates ± standard error. Bars in (B) represent the average daily adult emergence and standard errors from 4 replicate DvSnf7 treatments at a single dose of 11.3 ng/larva (gray bars) and for the larvae in the assay control treatment (open bars) from day 14 to day 17 after application of the treatment diet. Cumulative emergence distributions are shown as a solid Boltzmann fitting curve for the assay control and as a dotted Boltzmann fitting curve for DvSnf7 treatment.
In both the DvSnf7 and the assay control treatments adult emergence was initiated on day 14, reached approximately 50% emergence by day 15, and reached 100% emergence by day 17 (Figure 2B). Fifty percent of adult emergence was achieved across the 4 replicates at 15.6 ± 0.4 d in the assay control and at 15.5 ± 0.3 d in the DvSnf7 treatment, showing no significant difference (p = 0.668). Based on no adverse effects on survival and emergence, the no‐observed‐effect concentration (NOEC) was ≥11.3 ng DvSnf7/larva.
Adult feeding test
After 14 d of continuous feeding, there were no significant differences in adult worker bee survival between the DvSnf7 treatment and the assay control treatment (p = 0.821). Mean survival across the 4 replicates was nearly identical between the assay control and DvSnf7 treatments, with 92.50% (standard error of ±1.44) survival for DvSnf7 treatment and 91.25% (standard error of ±2.39) survival for assay control treatment (Figure 3). In contrast, there was no survival in the positive control by day 7, confirming the effectiveness of the dietary feeding exposure (Figure 3). Based on no adverse effects to survival of adult bees, the NOEC was ≥1000 ng/g diet.
Figure 3.
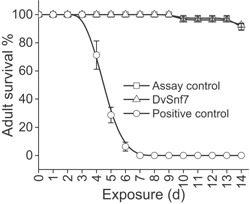
Survival of honey bee adults in 14‐d dietary feeding tests. Each data point represents the average of 4 replicates ± standard error.
Bioinformatics analysis
The alignment comparison of the Snf7 240‐nucleotide ortholog sequences showed only a 72.5% identity between western corn rootworm and A. mellifera (Figure 1C). In addition, there were no 21‐nucleotide contiguous matches between the 2 Snf7 orthologs with the longest contiguous match being only 13 nucleotides. A BLAST search for all possible 21‐bp matches from the DvSnf7 double‐stranded RNA against the genome published for A. mellifera indicated there is no single 21‐nucleotide contiguous match across the entire A. mellifera genome sequence.
Dose confirmation
Concentration–response curves and estimated LC50 values for DvSnf7 in diet samples collected from adult and larval feeding tests were comparable with the DvSnf7 reference standard with overlapping 95% CIs for the estimated LC50 values (Table 1 and Figure 4). The results confirm that DvSnf7 was biologically active in the test diets, was present in the treatment diets at the nominal concentration of 1000 ng/g diet solution, and was stable in the diet solution during storage. The biological activity measured in 2 sets of bioassays with diet samples from the adult feeding test and the larval feeding test showed some difference (Table 1), but this was not an unexpected result for the 2 sets of bioassays conducted on different days with different batches southern corn rootworm.
Table 1.
Median lethal concentration values (LC50) and associated 95% confidence intervals for reference standard DvSnf7 and the DvSnf7 in the diet samples collected from the honey bee adult feeding test and the larval feeding test
| Feeding test | DvSnf7 treatment | LC50 value (ng DvSnf7/mL diet) | 95% confidence interval |
|---|---|---|---|
| Adult feeding test | Reference standard | 4.22 | 2.84–5.99 |
| Sample 1 | 6.57 | 4.29–9.92 | |
| Sample 2 | 6.04 | 4.23–8.36 | |
| Larval feeding test | Reference standard | 13.75 | 9.10–20.96 |
| Diet collected at the end of dosing | 16.57 | 10.32–27.95 |
Figure 4.
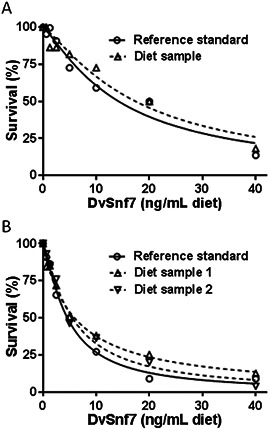
Concentration responses of reference standard DvSnf7 (solid circle) and the diet sample collected from the larval feeding test (A) and the adult feeding test (B) in 12‐d southern corn rootworm diet incorporation bioassays. Concentration–response curves were generated by plotting the percentage of survival against the concentration levels using GraphPad Prism 6. Median lethal concentration values and their associated 95% confidence intervals are presented in Table 1.
DISCUSSION
The use of a tiered approach has been recommended in nontarget organism risk assessment for plant incorporated products 21, 28, 29. In Tier I testing, risk is determined first from estimates of hazard under “worst‐case” exposure conditions. A lack of adverse effects under these conditions provides confidence that there is negligible risk, and no further testing is required 21. Critical to Tier I assessments is the incorporation of well‐designed laboratory assays that provide a high level of confidence in the results 21, 29. The hypothesis tested in the present study was that DvSnf7 expressed in MON 87411 maize has no adverse effects on larval survival and development and adult survival at a concentration that greatly exceeds field exposure conditions. This hypothesis was tested in experiments in line with recommendations for nontarget arthropod testing of plant incorporated products 29. These assays considered the exposure pathway, the known mode of action of DvSnf7, using a well‐characterized test substance delivered via artificial diet at concentrations thousands of times higher than expected in the environment, measuring larval and adult survival and larval development; using test durations appropriate for the DvSnf7 mode of action; and using appropriate positive and negative controls as described in Materials and Methods. Because dietary consumption of maize pollen is the main DvSnf7 exposure pathway for honey bee larvae in the hive or adults in the field, the hypothesis was tested in early‐tier laboratory dietary feeding assays with honey bee adults and in hive feeding assays with honey bee larvae under a worst‐case exposure level. To achieve the worst‐case exposure level, DvSnf7 synthesized in vitro was used; this was previously shown to be equivalent to plant‐produced DvSnf7 in sequence and biological activity (W. Urquhart, Monsanto Company, St. Louis, MO, USA, unpublished data). In addition, the nominal concentration of DvSnf7 and its biological activity and storage stability were confirmed in dose‐confirmation bioassays with southern corn rootworm, which is sensitive to DvSnf7 8, 9. The measurement endpoints included not only lethality but also sublethal effects on development of test larvae through emergence to adulthood. Adult bees in each replicate (cage) were allowed to feed ad libitum on DvSnf7 diet solution for 14 d, which covers a significant portion of the adult life span. In the larval feeding test, observations were made at day 7 and day 10 for larval survival and on day 13 through day 17 for adult emergence. Consumption of the treatment diet solution by larvae was visually confirmed using blue‐dyed diet solution in previous method development assays (K. Richards, California Agricultural Research, Kerman, CA, unpublished data). Both adult and larval feeding assays had a sufficient level of replication and individual specimens within a replicate based on current guidelines 37. These experimental design elements support the robustness of both adult and larval tests.
Adult honey bees may be directly exposed to DvSnf7 by pollen consumption, or larvae can be exposed by receiving royal jelly and pollen from nurse bees. Therefore, the maximum expected environmental concentration of DvSnf7 that honey bee adults or larvae would be exposed to under field conditions is characterized by the maximum expression level of DvSnf7 in maize pollen (0.224 ng/g fresh pollen) and the amount of pollen that could be consumed by honey bee adults or larvae. It has been reported that on average 3.4 mg to 4.3 mg of pollen was consumed per day per adult 25 and approximately 2 mg pollen could be consumed in the larval stage 26. Based on these expression and consumption levels, the dose exposed to honey bee in the larval stage was estimated to be 0.448 pg DvSnf7/larva, resulting in a 25 233 margin of exposure (Table 2). In the adult feeding test, a margin of exposure of 4464 was calculated simply based on the dietary test concentration of 1000 µg DvSnf7/g diet solution and the maximum expression level of DvSnf7 in pollen. Alternatively, a refined margin of exposure of 42 150 (Table 2) could be estimated, taking into consideration average daily adult pollen consumption and the daily dose of DvSnf7 ingested from DvSnf7‐treated diet solution 25, 38, 39. Margin of exposure values >10 demonstrate a large margin of safety between field exposure levels from pollen and the NOECs from larval honey bee in hive tests and adult honey bee in laboratory studies.
Table 2.
Maximum environmental exposure concentrations, no‐observed‐effect concentrations, and estimated margins of exposure of DvSnf7 RNA in the honey bee adult and larval feeding tests
| Feeding test | Maximum environmental exposure concentration | No‐observed‐effect concentration | Margin of exposure a |
|---|---|---|---|
| Adult | 0.224 ng/g fresh weight pollen | 1000 ng/g | 4464 b |
| 0.963 pg/adult/d | 40.59 ng | 42 150 c | |
| Larval | 0.448 pg/larva | 11.3 ng | 25 223 d |
Margin of exposure is the ratio of no‐observed‐effect concentration to maximum environmental exposure concentration.
The 4464 margin of exposure in the adult feeding test was estimated using the maximum expression level of DvSnf7 in fresh maize pollen (0.224 ng/g) as the maximum environmental exposure concentration and the DvSnf7 concentration (1000 ng/g diet solution) in the adult feeding test as the no‐observed‐effect concentration.
The 42 150 margin of exposure in the adult feeding test was calculated using the maximum environmental exposure concentration of 0.963 pg/d, which was estimated based on an average daily consumption of fresh pollen by a honey bee worker at 4.3 mg/adult 25 and a maximum expression level of DvSnf7 at 0.224 ng/g fresh weight pollen (0.224 ng DvSnf7/g fresh pollen × 4.3 mg pollen/worker bee/d = 0.963 pg) and the no‐observed‐effect concentration of 40.59 ng/adult/d, which was based on the daily consumption of diet solution (33 µL) by a single adult 38, 39 and the concentration of DvSnf7 in the treatment diet solution (33 µL × 1230 ng DvSnf7/mL diet solution = 40.59 ng).
The 25 223 margin of exposure in the larval feeding test was calculated using the maximum environmental exposure concentration of 0.448 pg/larva, which was based on the maximum expression level of DvSnf7 in fresh maize pollen (0.224 ng DvSnf7/g) and the average consumption of pollen by honey bee larvae at 2.0 mg/larva 26 (0.224 ng/g fresh pollen × 2.0 mg pollen/larval stage = 0.448 pg) and the no‐observed‐effect concentration of 11.3 ng estimated by the volume of diet solution applied to each larva and the concentration of DvSnf7 in the diet solution (10 µL × 1130 ng DvSnf7/mL diet solution = 11.3 ng).
The margin of exposure estimated from the maximum expression level of DvSnf7 in maize pollen and pollen consumption by honey bee adults or larvae exceeds the worst‐case exposure scenario because there are many additional factors in the environment to confine the exposure of honey bee to DvSnf7. First, the flowering period for maize is approximately 2 wk, accessible for foraging by field honey bee workers 24. Second, the honey bee adults or larvae do not exclusively consume maize pollen, and the DvSnf7 RNA carried in maize pollen may be diluted by consumption of the pollen foraged from other plants. In most cases, the proportion of maize pollen as a total of all pollen collected and fed to larvae during summer will be low. It is therefore unlikely that maize pollen would regularly comprise more than 50% of the honey bee diet 27. In addition, insects have been shown to lack RNA‐dependent RNA polymerases 6, 40, 41. The absence of an endogenous amplification mechanism in insects suggests that potential RNA interference action will depend on exogenous double‐stranded RNA uptake. Furthermore, the double‐stranded RNA can potentially be degraded prior to possible RNA interference action 18. Extracellular degradation of exogenous double‐stranded RNA was demonstrated in the saliva of the tarnished plant bug (Lygus lineolaris) 42 and in the gut and hemolymph of the pea aphid (Acyrthosiphon pisum) 43 and of other insect species 44, 45. The rapid degradation of double‐stranded RNA by nucleases could create a barrier to uptake, decrease the persistence of double‐stranded RNA in insect hemolymph, or impede the RNA interference effect in insect species 42, 43, 44, 45.
The results of the present study are in line with previous research that characterized the spectrum of insecticidal activity for DvSnf7 in 14 insect species representing 10 families and 4 orders: Hemiptera, Hymenoptera, Lepidoptera, and Coleoptera 9. The insecticidal activity of DvSnf7 is narrow and only evident in a subset of beetles within the Galerucinae subfamily of Chrysomelidae (>90 % identity with western corn rootworm Snf7 240 nucleotide), with no activity in those species that lack ≥21‐nucleotide sequence matches to the DvSnf7 ortholog 9. Results from indirect feeding assays, with 2 species susceptible to oral RNA interference, western corn rootworm and Colorado potato beetle, demonstrated that a ≥21‐nucleotide contiguous sequence is required to observe biological activity in sensitive insects 9. A study on the mode of action of DvSnf7 also demonstrated that a minimum of a 100% complementary sequence length of ≥21 nucleotide embedded in a ≥60‐bp double‐stranded RNA is required for biological activity in diet incorporation bioassays with southern corn rootworm 8. The DvSnf7 used in the present study contained a 240‐bp double‐stranded region with a 98% sequence match to the southern corn rootworm ortholog and demonstrated a high level of activity against southern corn rootworm 8 (Figure 4). Bioinformatics analyses show that the Snf7 orthologs of western corn rootworm and A. mellifera share only 72.5% identity and that no 21‐nucleotide matches exist between the orthologs, with the longest sequence match of 13 nucleotides. In addition, a BLAST search indicated there is no single 21‐bp match against the honey bee (A. mellifera) genome 33. Bioinformatics analysis can be used as a supplementary tool to help identify nontarget organisms that could potentially be affected by DvSnf7 when 100% complementary sequence lengths of ≥21 nucleotides occur. However, bioinformatics cannot be reliably used as a standalone to predict the presence of RNA interference activity in nontarget organism species because there are physical and chemical barriers to achieving an RNA interference‐mediated effect in many species and consequently not all species are susceptible to ingested double‐stranded RNA 4, 5. Conversely, when bioinformatics data are available and indicate that the minimum sequence requirements for RNA interference activity in insects are not met, toxicity testing in nontarget insects may not be necessary as the likelihood of adverse effects is low. The results from the present study support the use of bioinformatics when available as a supplementary tool in nontarget insect assessments. Based on the lack of ≥21‐nucleotide complementary sequences between the DvSnf7 sequence and the honey bee genome, the likelihood of adverse effects of DvSnf7 on honey bee was considered to be low, which was consistent with the results of the present study.
The results and conclusions in the present study were generated under conditions meeting all of the recommendations from a tripartite panel of experts 29 regarding the test substance, method of delivery, concentration, measurement endpoints, test duration, control substances, and statistical analysis. The results from larval and adult honey bee bioassays demonstrate that DvSnf7 does not induce adverse effects on development, emergence, and survival of honey bees at exposure levels that greatly exceed environmentally realistic concentrations.
Data availability
The data set is included in 2 study files, which were archived in the Regulatory Library of Monsanto Company. All studies described herein were in compliance with good laboratory practice requirements. All data can be accessed by the person who has been authorized by the Regulatory Library. Contact the authors for further information (jianguo.tan@monsanto.com and steven.l.levine@monsanto.com).
Acknowledgment
The authors thank K. Giddings and C. Lawrence for comments on earlier versions of the present study, S. Carleton for the description of synthesis of DvSnf7_968 RNA and the schematic of the DvSnf7_968 RNA molecule and hairpin loop, A. Silvanovich and Y. Cao for bioinformatics analysis of Snf7 orthologs, and the anonymous reviewers for helpful comments.
REFERENCES
- 1. Gatehouse JA, Price DRG. 2011. Protection of crops against insect pests using RNA interference. Insect Biotechnology 2:145–168. [Google Scholar]
- 2. Burand JP, Hunter WB. 2013. RNAi: Future in insect management. J Invest Pathol 112:S68–S74. [DOI] [PubMed] [Google Scholar]
- 3. Katoch R, Sethi A, Thakur N, Murdock LL. 2013. RNAi for insect control: Current perspective and future challenges. Appl Biochem Biotechnol 171:847–873. [DOI] [PubMed] [Google Scholar]
- 4. Scott JG, Michel K, Bartholomay L, Siegfried BD, Hunter WB, Smagghe G, Zhu KY, Douglas AE. 2013. Towards the elements of successful insect RNAi. J Insect Physiol 59:1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baum JA, Roberts JK. 2014. Progress towards RNAi‐mediated insect pest management. Adv Insect Physiol 47:249–295. [Google Scholar]
- 6. Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M, Vaughn T, Roberts J. 2007. Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25:1322–1326. [DOI] [PubMed] [Google Scholar]
- 7. Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY. 2007. Silencing a cotton bollworm P450 monooxygenase gene by plant‐mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25:1307–1313. [DOI] [PubMed] [Google Scholar]
- 8. Bolognesi R, Ramaseshadri P, Anderson J, Bachman P, Clinton W, Flannagan R, Ilagan O, Lawrence C, Levine S, Moar W, Mueller G, Tan J, Uffman J, Wiggins E, Heck G, Segers G. 2012. Characterizing the mechanism of action of double‐stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS One 7:e47534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bachman PM, Bolognesi RJ, Moar W, Mueller GM, Paradise MS, Ramaseshadri P, Tan J, Uffman JP, Warren J, Wiggins BE, Levine SL. 2013. Characterization of the spectrum of insecticidal activity of a double‐stranded RNA with targeted activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). Transgenic Res 22:1207–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gray ME, Sappington TW, Miller NJ, Moeser J, Bohn MO. 2009. Adaptation and invasiveness of western corn rootworm: Intensifying research on a worsening pest. Annu Rev Entomol 54:303–321. [DOI] [PubMed] [Google Scholar]
- 11. Wesseler J, Fall EH. 2010. Potential damage costs of Diabrotica virgifera virgifera infestation in Europe—The “no control.” J Appl Entomol 134:385–394. [Google Scholar]
- 12. Roxrud I, Stenmark H, Malerod L. 2010. ESCRT & Co. Biol Cell 102:293–318. [DOI] [PubMed] [Google Scholar]
- 13. Henne WM, Buchkovich NJ, Emr SD. 2011. The ESCRT pathway. Dev Cell 21:77–91. [DOI] [PubMed] [Google Scholar]
- 14. Wegner CS, Rodahl LMW, Stenmark H. 2011. ESCRT proteins and cell signalling. Traffic 12:1291–1297. [DOI] [PubMed] [Google Scholar]
- 15. Ramaseshadri P, Segers G, Flannagan R, Wiggins E, Clinton W, Ilagan O, McNulty B, Clark T, Bolognesi R. 2013. Physiological and cellular responses caused by RNAi‐mediated suppression of Snf7 orthologue in western corn rootworm (Diabrotica virgifera virgifera) larvae. PLoS One 8:e54270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koči J, Ramaseshadri P, Bolognesi R, Segers G, Flannagan R, Park Y. 2014. Ultrastructural changes caused by Snf7 RNAi in larval enterocytes of western corn rootworm Diabrotica virgifera virgifera Le Conte. PLoS One 9:e83985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hannon GJ. 2002. RNA interference. Nature 418:244–251. [DOI] [PubMed] [Google Scholar]
- 18. Whyard S, Singh AD, Wong S. 2009. Ingested double‐stranded RNAs can act as species‐specific insecticides. Insect Biochem Mol Biol 39:824–832. [DOI] [PubMed] [Google Scholar]
- 19. Morse RA, Calderone NW. 2001. The value of honey bees as pollinators of US crops in 2000. Bee Culture 128:1–15. [Google Scholar]
- 20. Gallaia N, Sallesc J‐M., Setteled J, Vaissièrea BE. 2009. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecological Economics 68:810–821. [Google Scholar]
- 21.US Environmental Protection Agency. 2007. White paper on tier‐based testing for the effects of proteinaceous insecticidal plant‐incorporated protectants on non‐target arthropods for regulatory risk assessments. [cited 2015 January 5]. Available from: http://www.epa.gov/oppbppd1/biopesticides/pips/non‐target‐arthropods.pdf
- 22.European Food Safety Authority. 2013. Guidance on the risk assessment of plant protection products on bees (Apis mellifera, Bombus spp. and solitary bees). EFSA Journal 11:3295. [cited 2015 January 5]. Available from: http://www.efsa.europa.eu/en/efsajournal/pub/3295.htm [DOI] [PMC free article] [PubMed]
- 23. Romeis J, Raybould A, Bigler F, Candolfi MP, Hellmich RL, Huesing JE, Shelton AM. 2013. Deriving criteria to select arthropod species for laboratory tests to assess the ecological risks from cultivating arthropod‐resistant genetically engineered crops. Chemosphere 90:901–909. [DOI] [PubMed] [Google Scholar]
- 24. Goss JA. 1968. Development, physiology, and biochemistry of corn and wheat pollen. Bot Rev 34:333–355. [Google Scholar]
- 25. Crailsheim K, Schneider LHW, Hrassnigg N, Bühlmann G, Brosch U, Gmeinbauer R, Schöffmann B. 1992. Pollen consumption and utilization in worker honeybees (Apis mellifera carnica): Dependence on individual age and function. J Insect Physiol 38:409–419. [Google Scholar]
- 26. Babendreier D, Kalberer N, Romeis J, Fluri P, Bigler F. 2004. Pollen consumption in honey bee larvae: A step forward in the risk assessment of transgenic plants. Apidologie 35:293–300. [Google Scholar]
- 27. Devos Y, De Schrijver A, De Clercq P, Kiss J, Romeis J. 2012. Bt‐maize event MON 88017 expressing Cry3Bb1 does not cause harm to non‐target organisms. Transgenic Res 21:1191–1214. [DOI] [PubMed] [Google Scholar]
- 28. Romeis J, Bartsch D, Bigler F, Candolfi MP, Gielkens MMC, Hartley SE, Hellmich RL, Huesing JE, Jepson PC, Layton R, Quemada H, Raybould A, Rose RI, Schiemann J, Sears MK, Shelton AM, Sweet J, Vaituzis Z, Wolt JD. 2008. Assessment of risk of insect‐resistant transgenic crops to nontarget arthropods. Nat Biotechnol 26:203–208. [DOI] [PubMed] [Google Scholar]
- 29. Romeis J, Hellmich RL, Candolfi MP, Carstens K, De Schrijver A, Gatehouse AMR, Herman RA, Huesing JE, McLean MA, Raybould A, Shelton AM, Waggoner A. 2011. Recommendations for the design of laboratory studies on non‐target arthropods for risk assessment of genetically engineered plants. Transgenic Res 20:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Auer C, Frederick R. 2009. Crop improvement using small RNAs: Applications and predictive ecological risk assessments. Trends Biotechnol 27:644–651. [DOI] [PubMed] [Google Scholar]
- 31.International Life Sciences Institute, Center for Environmental Risk Assessment. 2011. Problem formulation for the environmental risk assessment of RNAi plants. Proceedings, International Life Sciences Institute, Center for Environmental Risk Assessment, Washington, DC, USA, June 1–3, 2011. [cited 2015 January 5]. Available from: http://www.cera‐gmc.org/files/cera/uploads/pub_08_2011.pdf
- 32. Levine SL, Tan J, Mueller GM, Bachman PM, Jensen PD, Uffman JP. 2015. Independent action between DvSnf7 RNA and Cry3Bb1 protein in southern corn rootworm, Diabrotica undecimpunctata howardi and Colorado potato beetle, Leptinotarsa decemlineata . PLoS One 10:e0118622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honeybee Genome Sequencing Consortium. 2006. Insights into social insects from the genome of the honeybee Apis mellifera . Nature 443:931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Armstrong TA, Chen H, Ziegler TE, Iyadurai KR, Gao A‐G, Wang Y, Song Z, Tian Q, Zhang Q, Ward JM, Segers GC, Heck GR, Staub JM. 2013. Quantification of transgene‐derived double‐stranded RNA in plants using the QuantiGene nucleic acid detection platform. J Agric Food Chem 61:12557–12564. [DOI] [PubMed] [Google Scholar]
- 35. Pearson WR, Lipman DJ. 1988. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marrone PG, Ferri FD, Mosley TR, Meinke LJ. 1985. Improvements in laboratory rearing of the southern corn rootworm, Diabrotica undecimpunctata howardi Barber (Coleoptera: Chrysomelidae), on an artificial diet and corn. J Econ Entomol 78:290–293. [Google Scholar]
- 37.Organisation for Economic Co‐operation and Development. 2013. Test No. 237: Honey bee (Apis mellifera) larval toxicity test, single exposure. OECD Guidelines for the Testing of Chemicals, Paris, France.
- 38. Decourtye A, Lacassie E, Pham‐Delgue MH. 2003. Learning performances of honeybees (Apis mellifera L.) are differentially affected by imidacloprid according to the season. Pest Manag Sci 59:269–278. [DOI] [PubMed] [Google Scholar]
- 39. Babendreier D, Kalberer NM, Romeis J, Fluri P, Mulligan E, Bigler F. 2005. Influence of Bt‐transgenic pollen, Bt‐toxin and protease inhibitor (SBTI) ingestion on development of the hypopharyngeal glands in honeybees. Apidologie 36:585–594. [Google Scholar]
- 40. Tomoyasu Y, Miller SC, Tomita S, Schoppmeier M, Grossmann D, Bucher G. 2008. Exploring systemic RNA interference in insects: A genome‐wide survey for RNAi genes in Tribolium . Genome Biol 9:R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miller SC, Miyata K, Brown SJ, Tomoyasu Y. 2012. Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: Parameters affecting the efficiency of RNAi. PLoS One 7:e47431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Allen ML, Walker WB. 2012. Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J Insect Physiol 58:391–396. [DOI] [PubMed] [Google Scholar]
- 43. Christiaens O, Swevers L, Smagghe G. 2014. DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 53:307–314. [DOI] [PubMed] [Google Scholar]
- 44. Liu J, Swevers L, Iatrou K, Huvenne H, Smagghe G. 2012. Bombyx mori DNA/RNA non‐specific nuclease: Expression of isoforms in insect culture cells, subcellular localization and functional assays. J Insect Physiol 58:1166–1176. [DOI] [PubMed] [Google Scholar]
- 45. Garbutt JS, Bellés X, Richards EH, Reynolds SE. 2013. Persistence of double‐stranded RNA in insect hemolymph as a potential determiner of RNA interference success: Evidence from Manduca sexta and Blattella germanica . J Insect Physiol 59:171–178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data set is included in 2 study files, which were archived in the Regulatory Library of Monsanto Company. All studies described herein were in compliance with good laboratory practice requirements. All data can be accessed by the person who has been authorized by the Regulatory Library. Contact the authors for further information (jianguo.tan@monsanto.com and steven.l.levine@monsanto.com).


